Abstract
Phage Mu transposase (A-protein) is primarily responsible for transposition of the Mu genome. The protein binds to six att sites, three at each end of Mu DNA. At most att sites interaction of a protein monomer with DNA is seen to occur over three minor and two consecutive major grooves and to result in bending up to about 90 degrees. To probe the directionality and locus of these A-protein-induced bends, we have used the antitumor antibiotic (+)-CC-1065 as a structural probe. As a consequence of binding within the minor groove, (+)-CC-1065 is able to alkylate N3 of adenine in a sequence selective manner. This selectivity is partially determined by conformational flexibility of the DNA sequence, and the covalent adduct has a bent DNA structure in which narrowing of the minor groove has occurred. Using this drug in experiments in which either gel retardation or DNA strand breakage are used to monitor the stability of the A-protein--DNA complex or the (+)-CC-1065 alkylation sites on DNA (att site L3), we have demonstrated that of the three minor grooves implicated in the interaction with A-protein, the peripheral two are 'open' or accessible to drug bonding following protein binding. These drug-bonding sites very likely represent binding at at least two A-protein-induced bending sites. Significantly, the locus of bending at these sites is spaced approximately two helical turns apart, and the bending is proposed to occur by narrowing of the minor groove of DNA. The intervening minor groove between these two peripheral sites is protected from (+)-CC-1065 alkylation. The results are discussed in reference to a proposed model for overall DNA bending in the A-protein att L3 site complex. This study illustrates the utility of (+)-CC-1065 as a probe for protein-induced bending of DNA, as well as for interactions of minor groove DNA bending proteins with DNA which may be masked in hydroxyl radical footprinting experiments.
Full text
PDF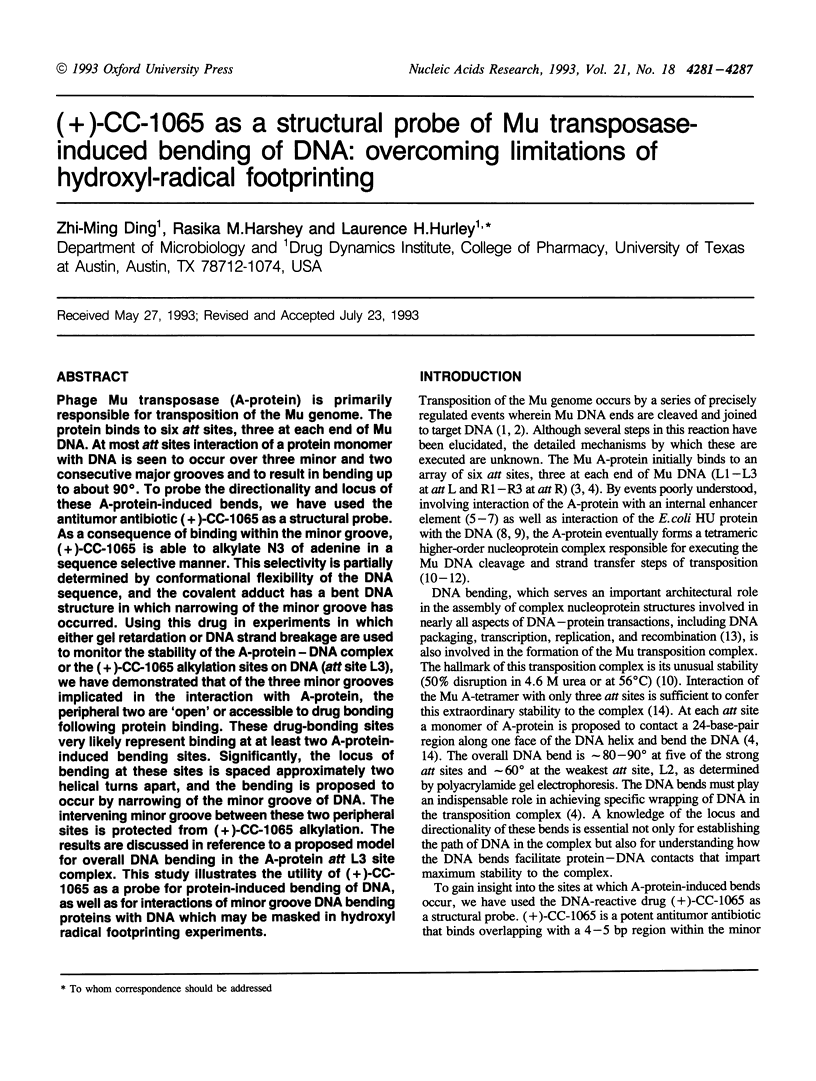




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison R. G., Chaconas G. Role of the A protein-binding sites in the in vitro transposition of mu DNA. A complex circuit of interactions involving the mu ends and the transpositional enhancer. J Biol Chem. 1992 Oct 5;267(28):19963–19970. [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hurley L. H., Lee C. S., McGovren J. P., Warpehoski M. A., Mitchell M. A., Kelly R. C., Aristoff P. A. Molecular basis for sequence-specific DNA alkylation by CC-1065. Biochemistry. 1988 May 17;27(10):3886–3892. doi: 10.1021/bi00410a054. [DOI] [PubMed] [Google Scholar]
- Hurley L. H., Reynolds V. L., Swenson D. H., Petzold G. L., Scahill T. A. Reaction of the antitumor antibiotic CC-1065 with DNA: structure of a DNA adduct with DNA sequence specificity. Science. 1984 Nov 16;226(4676):843–844. doi: 10.1126/science.6494915. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kuo C. F., Zou A. H., Jayaram M., Getzoff E., Harshey R. DNA-protein complexes during attachment-site synapsis in Mu DNA transposition. EMBO J. 1991 Jun;10(6):1585–1591. doi: 10.1002/j.1460-2075.1991.tb07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J Biol Chem. 1990 Jan 25;265(3):1623–1627. [PubMed] [Google Scholar]
- Lavoie B. D., Chan B. S., Allison R. G., Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991 Oct;10(10):3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Sun D., Kizu R., Hurley L. H. Determination of the structural features of (+)-CC-1065 that are responsible for bending and winding of DNA. Chem Res Toxicol. 1991 Mar-Apr;4(2):203–213. doi: 10.1021/tx00020a013. [DOI] [PubMed] [Google Scholar]
- Leung P. C., Teplow D. B., Harshey R. M. Interaction of distinct domains in Mu transposase with Mu DNA ends and an internal transpositional enhancer. Nature. 1989 Apr 20;338(6217):656–658. doi: 10.1038/338656a0. [DOI] [PubMed] [Google Scholar]
- Lin C. H., Sun D. Y., Hurley L. H. (+)-CC-1065 produces bending of DNA that appears to resemble adenine/thymine tracts. Chem Res Toxicol. 1991 Jan-Feb;4(1):21–26. doi: 10.1021/tx00019a003. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker T. A., Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992 Jul 24;70(2):303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell. 1989 Jul 28;58(2):399–408. doi: 10.1016/0092-8674(89)90854-4. [DOI] [PubMed] [Google Scholar]
- Reynolds V. L., Molineux I. J., Kaplan D. J., Swenson D. H., Hurley L. H. Reaction of the antitumor antibiotic CC-1065 with DNA. Location of the site of thermally induced strand breakage and analysis of DNA sequence specificity. Biochemistry. 1985 Oct 22;24(22):6228–6237. doi: 10.1021/bi00343a029. [DOI] [PubMed] [Google Scholar]
- Sun D., Lin C. H., Hurley L. H. A-tract and (+)-CC-1065-induced bending of DNA. Comparison of structural features using non-denaturing gel analysis, hydroxyl-radical footprinting, and high-field NMR. Biochemistry. 1993 May 4;32(17):4487–4495. doi: 10.1021/bi00068a003. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Warpehoski M. A., Hurley L. H. Sequence selectivity of DNA covalent modification. Chem Res Toxicol. 1988 Nov-Dec;1(6):315–333. doi: 10.1021/tx00006a001. [DOI] [PubMed] [Google Scholar]
- Zou A. H., Leung P. C., Harshey R. M. Transposase contacts with mu DNA ends. J Biol Chem. 1991 Oct 25;266(30):20476–20482. [PubMed] [Google Scholar]