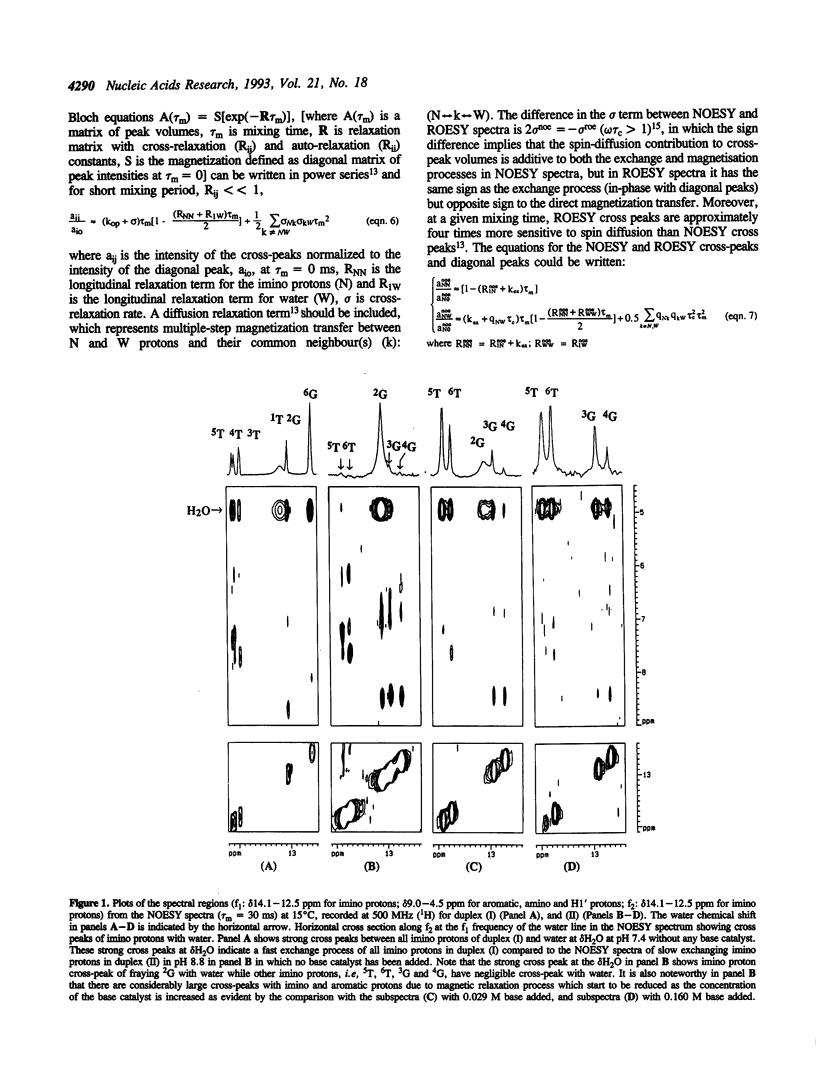
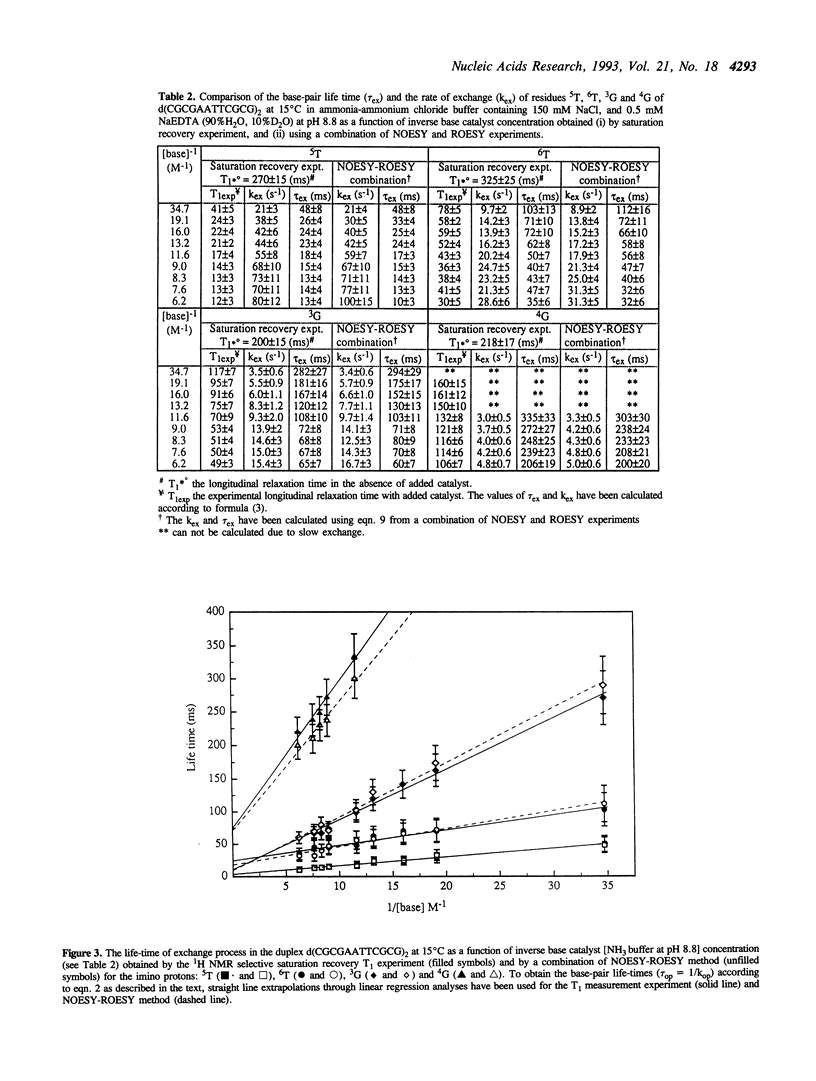
Abstract
A new method for the determination of the kinetics of exchange of the imino protons of DNA duplex is reported using a combination NOESY and ROESY experiments at short mixing times (< or = 20 ms). These results have been compared with the commonly used longitudinal relaxation approach through the T1 measurement. To calculate kex and pi ex by ROESY-NOESY experiment, the volume of the cross-peaks between imino protons and water in the NOESY and ROESY spectra have been measured separately from the magnetization term. This work shows that the present approach for the measurement of the kinetics of slow exchanging imino protons of DNA duplex is comparable to the saturation recovery experiment in which the exchange rate can be accelerated by the addition of a base catalyst. The present ROESY-NOESY approach has been found to be particularly useful and reasonably accurate for the measurement of exchange kinetics of both the fast- and slow-exchanging imino protons in DNA duplex both under non-physiological and physiological condition where the saturation recovery method can not be used.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Guéron M., Kochoyan M., Leroy J. L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987 Jul 2;328(6125):89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Study of transfer ribonucleic acid unfolding by dynamic nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3996–4006. doi: 10.1021/bi00517a008. [DOI] [PubMed] [Google Scholar]
- Leijon M., Gräslund A. Effects of sequence and length on imino proton exchange and base pair opening kinetics in DNA oligonucleotide duplexes. Nucleic Acids Res. 1992 Oct 25;20(20):5339–5343. doi: 10.1093/nar/20.20.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy J. L., Kochoyan M., Huynh-Dinh T., Guéron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J Mol Biol. 1988 Mar 20;200(2):223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- Liepinsh E., Otting G., Wüthrich K. NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Res. 1992 Dec 25;20(24):6549–6553. doi: 10.1093/nar/20.24.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepinsh E., Otting G., Wüthrich K. NMR spectroscopy of hydroxyl protons in aqueous solutions of peptides and proteins. J Biomol NMR. 1992 Sep;2(5):447–465. doi: 10.1007/BF02192808. [DOI] [PubMed] [Google Scholar]
- Lycksell P. O., Gräslund A., Claesens F., McLaughlin L. W., Larsson U., Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987 Nov 11;15(21):9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltseva T., Sandström A., Ivanova I. M., Sergeyev D. S., Zarytova V. F., Chattopadhyaya J. Structural studies of the 5'-phenazinium-tethered matched and G-A-mismatched DNA duplexes by NMR spectroscopy. J Biochem Biophys Methods. 1993 May;26(2-3):173–236. doi: 10.1016/0165-022x(93)90046-q. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Farmer B. T., 2nd, Wüthrich K. Protein hydration studied with homonuclear 3D 1H NMR experiments. J Biomol NMR. 1991 Jul;1(2):209–215. doi: 10.1007/BF01877232. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of the imino protons of the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix in complexes with the antibiotics netropsin and/or actinomycin. Biochemistry. 1983 Mar 1;22(5):1107–1113. doi: 10.1021/bi00274a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Ikuta S., Kozlowski S., Itakura K. Sequence dependence of hydrogen exchange kinetics in DNA duplexes at the individual base pair level in solution. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2184–2188. doi: 10.1073/pnas.80.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Pardi A., Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982 May 7;216(4546):581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Pardi A., Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982 May 7;216(4546):581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992 Nov;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]


