Abstract
MicroRNAs (miRNAs) play important roles during development and also in adult organisms by regulating the expression of multiple target genes. Here, we studied the function of miR-133b during zebrafish spinal cord regeneration and show upregulation of miR-133b expression in regenerating neurons of the brainstem after transection of the spinal cord. miR-133b has been shown to promote tissue regeneration in other tissue, but its ability to do so in the nervous system has yet to be tested. Inhibition of miR-133b expression by anti-sense morpholino (MO) application resulted in impaired locomotor recovery and reduced regeneration of axons from neurons in the NMLF (nucleus of the medial longitudinal fascicle), SRF (superior reticular formation), and IMRF (intermediate reticular formation). miR-133b targets the small GTPase RhoA, which is an inhibitor of axonal growth, as well as other neurite outgrowth-related molecules. Our results indicate that miR-133b is an important determinant in spinal cord regeneration of adult zebrafish through reduction of RhoA protein levels by direct interaction with its mRNA. While RhoA has been studied as a therapeutic target in spinal cord injury, this is the first demonstration of endogenous regulation of RhoA by a microRNA that is required for spinal cord regeneration in zebrafish. The ability of miR-133b to suppress molecules that inhibit axon regrowth may underlie the capacity for adult zebrafish to recover locomotor function after spinal cord injury (SCI).
Keywords: miRNA, axon regeneration, SCI, RhoA
Introduction
The zebrafish is capable of regenerating several types of tissue after injury including the central nervous system (CNS). Its unique capacity for regeneration allowed the identification of regulatory mechanisms involved in the regeneration process (Nakatani et al., 2007), including those implicated in regeneration of supraspinal axons after complete spinal cord transection (Becker et al., 1997; Becker et al., 2004). Unlike growth-associated genes whose contribution to axon regeneration has been well studied, very little is known about the role of miRNAs during this process. However, recent studies have indicated that miRNAs do indeed have a crucial role in tissue regeneration. For example, the microRNA, miR-133b, enhances muscle regeneration and reduces the formation of fibrotic tissue, demonstrating the feasibility of miRNAs to promote functional recovery (Nakasa et al., 2009).
miRNAs are short, non-coding RNAs, found in a wide variety of organisms ranging from roundworms to humans (Ambros, 2001; Bartel, 2004). They regulate gene expression primarily by imperfect binding to miRNA response elements (RE) in mRNA targets (Bartel, 2004), leading to attenuation of protein production by mechanisms such as mRNA cleavage (Meister et al., 2004), mRNA de-adenylation (Wu et al., 2006), inhibition of translation initiation (Humphreys et al., 2005), and/or mRNA sequestration to P-bodies (Liu et al., 2005). Their ability to simultaneously regulate the expression of several genes makes miRNAs ideal candidates for coordinating complex gene expression programs, such as those required for spinal cord regeneration.
miR-133 has been shown to play an important role in several regulatory processes. For example, in cardiomyocytes, miR-133 serves an anti-apoptotic role by inhibiting caspase-9 (Xu et al., 2007). Among its multiple targets, miR-133 down-regulates RhoA protein expression (Care et al., 2007; Chiba et al., 2009). RhoA is strongly upregulated following SCI (Conrad et al., 2005b; Erschbamer et al., 2005), and inhibition of RhoA enhances regrowth of the corticospinal tract and promotes neuroprotection by decreasing tissue damage and cavity formation that develop after SCI (Dergham et al., 2002; Fournier et al., 2003; Tanaka et al., 2004; Hoffmann et al., 2008; Holtje et al., 2009). Because miR-133 promotes regeneration in other tissues and zebrafish do not display this same pathophysiology after SCI, we hypothesized that the regulation of miR-133b might contribute to functional recovery, potentially through its interaction with RhoA, one of many predicted targets of miR-133b.
In this study, we show that miR-133b plays a critical role in regulating the regenerative capacity of zebrafish after SCI. Following complete spinal cord transection, miR-133b expression is increased in supraspinal neurons. Anti-sense morpholino mediated inhibition of endogenous miR-133b reduces functional recovery and regeneration of brainstem axons across the injury site. In addition, we show that RhoA mRNA is a target of miR-133b and that RhoA expression is increased at the lesion site after SCI when miR-133b expression is inhibited. Since multiple cellular and molecular pathways are regulated by miRNAs and since the targets of miR-133b are conserved from zebrafish to mammals, we believe that our results may guide the development of novel strategies for improving functional recovery after SCI in humans.
Materials and Methods
Animals
Adult zebrafish (Danio rerio, body length >2.5 cm, age >6 months) were purchased from Aquatica Tropicals Inc. (Plant City, FL). The fish were kept on a 14 hour light and 10 hour dark cycle at 28°C. All experiments were approved by the Rutgers University Animal Care and Facilities Committee and conformed to NIH guidelines.
Spinal cord injury
Fish were anesthetized by immersion in 0.4% aminobenzoic acid ethyl methyl ester (MS-222, Sigma, St. Louis, MO) dissolved in phosphate buffered saline (PBS), pH 7.3, until active respiration was stopped. The fish were then placed on ice under a dissecting microscope and received a longitudinal incision through the axial muscles to expose the vertebral column. A complete transection of the spinal cord was performed between two vertebrae with microscissors, 4 mm caudal to the brainstem-spinal cord junction (thoracic level). For sham-lesioned fish, the axial muscle surrounding the spinal cord was cut at the same level as spinal cord transected animals, but the spinal cord was left intact. The incision was sealed with Histoacryl (TissueSeal, Ann Arbor, MI) and fish were then placed into two-liter tanks containing filtered water at room temperature until the time of sacrifice.
Cloning and mutagenesis of the green fluorescent protein (GFP) reporter
Zebrafish RhoA sequences (NM213137) were amplified by RT-PCR with RhoA primers (Table 2) and subcloned (GFP-RhoA) downstream of the GFP open reading frame inserted into the EcoRI-BamHI restriction site of pCS2+ (a kind gift from Dr. James G. Patton, Vanderbilt University). Deletion of the miR-133b recognition elements (RE) in the RhoA 3’ UTR (GFP-mtRhoA) was created using QuickChange Site-Directed Mutagenesis kit (Stratagene, Cedar Creek, TX) with mtRhoA primers (Table 2). The Red Fluorescent Protein (RFP) mRNA expression vector, pCS2+RFP, was purchased from Addgene (Cambridge, MA).
Table 2.
Summary of DNA and RNA oligonucleotides
| PCR primers | |
| RhoA | 5’-GCGAATTCAAACGTGGCAAGAAGAATGC-3’ |
| 5’-GCCTCGAGCAAAGGGGCCAAAGAATACA-3’ | |
|
mtRhoA_RE1 deletion |
5’-GAAAATGAGAAGAACGAGGGAACACGGCCAAGGAAAATGAAAG-3’ |
| 5’-CTTTCATTTTCCTTGGCCGTGTTCCCTCGTTCTTCTCATTTTC-3’ | |
|
mtRhoA_RE2 deletion |
5’- GATTGACGTGGGTTCACAGTGATATGCCATGTATTCTTTGGCC-3’ |
| 5’- GGCCAAAGAATACATGGCATATCACTGTGAACCCACGTCAATC-3’ | |
| RNA Oligonucleotides and Probes | |
| miR-133b duplex |
5’-UAGCUGGUUGAAGGGGACCAAA-3’ |
| 5’-UUUGGUCCCCUUCAACCAGCUA-3’ | |
| miR-133 MO | 5'-CTGACTTGGTTCCATTTGACCAGCC-3' |
| Control MO | 5'-CCTCTTACCTCAGTTACAATTTATA-3' |
| In situ hybridization |
5’ TAGCTGGTTGAAGGGGACCAAA 3’ |
Microinjection
Zebrafish embryos were collected at the one-cell stage and injected with 100 ng/µl of capped GFP and RFP mRNAs that were synthesized in vitro using mMessage Machine SP6 Kit (Ambion, Austin, TX), with or without 10 µM miR-133b RNA duplex (IDT Inc., see Table 2).
Morpholino application
miR-133b anti-sense morpholino (MO) and standard control MO (Table 2, Gene Tools, Philomath, OR) were dissolved in Danieau solution (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca (NO3)2, 5 mM HEPES, pH 7.6). The solubilized MOs (4µl of a 250 µM solution) were then soaked in small pieces of Gelfoam (Upjohn, Kalamazoo, MI) that were further divided into 15 smaller pieces to yield 600 ng of MO per piece and were allowed to dry. Each of the dried Gelfoam pieces containing the MO (one per fish) was then applied to the lesion site immediately after the transection. The wound was then sealed with a drop of Histoacryl (Becker et al., 2004).
Locomotor analysis
Swimming capabilities of fish that received standard control or miR-133b MO were analyzed at six weeks after spinal cord transection. At this time point after injury, fish have regained most of their capacity in functional recovery (on average about 80%) when compared to non-lesioned fish (set to 100%). This post-injury time point has proven to reach asymptotic saturation in regaining locomotor function and was thus used in the present and all previous experiments (Becker et al., 2004). Each fish was placed in a brightly illuminated glass tank (50 × 30 cm and 30 cm high) filled with aquarium water and allowed to acclimate for 5–10 minutes before video recording. Each five-minute trial was recorded using a video camera placed directly above the tank. Total swimming distance (total distance moved) was tracked and analyzed using ANY-maze software (Stoelting Co., Wood Dale, IL), with the analyst blinded to the treatment of the individual fish.
Retrograde tracing
Retrograde axonal tracing was performed by application of the axonal tracer biocytin (Sigma, St. Louis, MO), 4 mm caudal to the spinal lesion site, 6 weeks after complete transection (Becker et al., 1997). Gelfoam pieces soaked in a saturated solution of biocytin were left at the lesion site to be taken up by cut axons and retrogradely transported to their cell bodies of origin. Biocytin labeling in sectioned tissue was then detected by Alexa Fluor 488 Streptavidin (Molecular Probes, Eugene, OR). Labeled cells were counted in brainstem sections with an Axiophot microscope (Zeiss, Thornwood, NY), with the experimenter blinded to the experimental treatment.
Immunofluorescence staining
Animals were euthanized with an overdose of MS-222. Spinal cords were then removed with microscissors, immersion fixed in 4% paraformaldehyde, and subsequently cryoprotected in 30% sucrose. Spinal cord tissue was next embedded in optimal cutting temperature medium (OCT) and transversely sectioned onto Superfrost Plus slides (Fisher Scientific, Waltham, MA) at 30 µm. Antigen retrieval (for RhoA) was performed by incubating slides with trypsin (TrypLE ™) at 37°C for two minutes, then washing in PBS for fifteen minutes, changing the solution every five minutes. All slides were blocked in 10% normal goat serum for two hours at room temperature, then incubated with polyclonal anti-RhoA primary antibody (1:200, SC-179, Santa Cruz Biotechnology, Santa Cruz, CA) or monoclonal anti-NeuN (1:400, A-60, Millipore, Billerica, MA) diluted in 5% fetal calf serum (FCS) overnight at 4°C. The slides were then washed in three times in PBS (five minutes per wash) and detected by incubating for two hours at room temperature with Alexafluor 546 or 647 secondary antibodies (1:1000) in 5% FCS (Invitrogen, Carlsbad, CA). A ten-minute incubation in Hoechest (1:1000) was performed to stain nuclei, followed by mounting with Fluoromount G (Southern Biotech, Birmingham, AL). All fluorescent images were taken with the Zeiss Axiovert 200M fluorescence microscope equipped with deconvolution software. RhoA expression was quantified by converting color images to gray-scale in Photoshop, and measuring the suprathreshold stained area of spinal cord tissue with NIH Image J software. Three sections per animal were averaged for each animal, using three animals per condition.
Western blot analysis
Embryos used in GFP reporter experiments were taken at two days post-fertilization (dpf), dechorionated, deyolked, and homogenized in Passive Lysis Buffer (Promega, Madison, WI). Nine days after spinal cord injury, brain slices of approximately 2 mm thickness (including the NMLF area) and 2 mm from spinal cords, centered on the transection site, were quickly dissected from anesthetized fish. Two mm brain slices from three separate animals or spinal cords from four separate animals (encompassing the transection site) were pooled and homogenized in RIPA Lysis Buffer supplemented with protease inhibitors (Roche Diagnostics, Indianapolis, IN). Homogenates were immunoblotted following a standard Western blot protocol. Antibodies were obtained from the following sources: rabbit anti-GFP, 1:1000 (TP401, Torrey Pines, Houston, TX); chicken anti-RFP, 1:1000 (AB3528, Millipore, Bedford, MA); mouse anti-RhoA, 1:100 (ARH03, Cytoskeleton, Denver, CO); and mouse anti-α-tubulin, 1:1000 (T9026, Sigma). A secondary antibody (1:1,000, Milipore) from the appropriate host, conjugated with horseradish peroxidase was used along with Pierce ECL chemiluminescent substrate (Thermo, Rockford, IL) for signal development on Kodak Biomax XAR film (Sigma). Quantitative analysis was performed using Kodak molecular imaging software version 4.0 (Carestream Molecular Imaging, New Haven, CT).
Antibody characterization
The antibodies used for immunofluorescence staining of tissue were obtained from their proprietary sources listed above. Each antibody used was tested on tissue with a primary antibody omission control that confirmed the absence of the observed signal and pattern of staining. The anti-RhoA antibody produces a single band on a Western blot and has been used in previous studies (Erschbamer et al., 2005). Anti-NeuN antibody has been used successfully in previous studies to specifically detect the nuclei of mouse hippocampal neurons (Goodman et al., 2010). Additionally, all of the antibodies used for Western blot analysis were obtained from proprietary sources that provided confirmation of a single band on a Western blot at the appropriate molecular weight, indication of affinity purification where applicable, and/or their use in published studies (Jefford et al., 2004; Eguchi et al., 2008; Ford-Speelman et al., 2009).
In situ hybridization
Locked nucleic acid (LNA)-based in situ hybridization was performed as described (Obernosterer et al., 2007) miR-133b was detected by in situ hybridization using digoxigenin-labeled LNA probes (Exiqon, Vedbaek, Denmark). The probes (Table 2) were visualized by using an alkaline phosphatase conjugated anti-digoxygenin antibody Fab fragment (1:500, Roche) enzymatically reacted with NBT/BCIP substrate (Roche) until the signal was detectable above background. For quantification, miR-133b positive cells were counted with the experimenter blinded to the treatments.
Cell counting
Counting of the total number of miR-133b positive cells in histological serial coronal sections (30 µm) of the whole nucleus of the medial longitudinal fascicle and oculomotor nucleus per animal brain was performed using a Zeiss Axiophot microscope. To avoid counting a single neuron more than once in adjacent sections, only neurons containing nuclei were counted. These quantifications were performed by optical dissection using stereological analysis (Reimer et al., 2008).
Statistical analysis
All data are presented as the mean ± SEM. Depending on the number of the groups and independent factors, data were analyzed with Student’s t-test or one-way ANOVA followed by Tukey post-hoc test when appropriate. All tests were two-tailed and the level of significance was set at p < 0.05.
RESULTS
Expression of miR-133b in the central nervous system of adult zebrafish
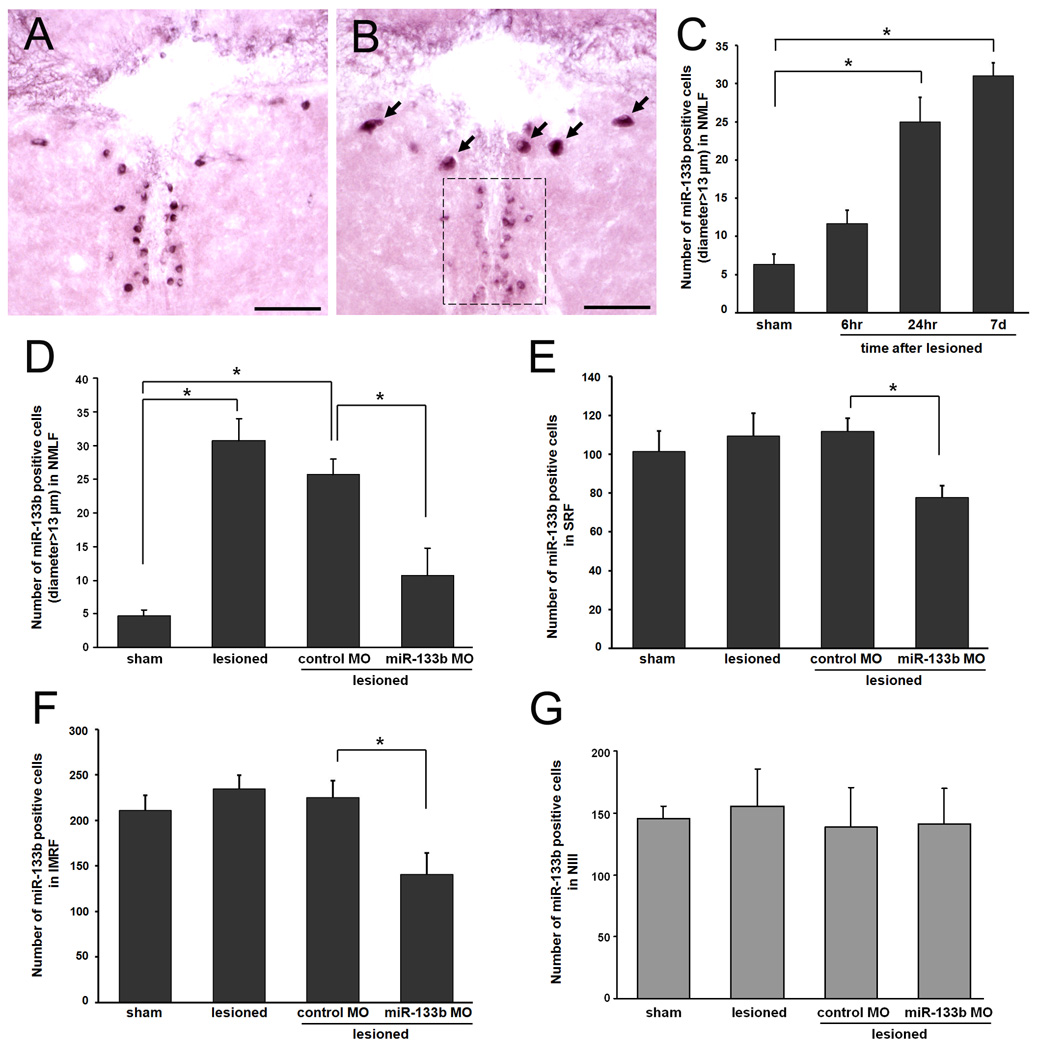
Since previous studies identified miR-133b to play an important role in muscle regeneration (Nakasa et al., 2009), we wanted to determine if miR-133b was involved in spinal cord regeneration as well. The expression pattern of miR-133b in the central nervous system of adult zebrafish was examined by in situ hybridization with an LNA miR-133b probe. Figure 1 shows coronal sections of the brain and spinal cord (n=6) with miR-133b positive cells indicated by dark purple staining. miR-133b was detected only in neuron cell bodies in the following nuclei: NMLF (Fig. 1A); ventrolateral nucleus of torus semicircularis (TSvl); oculomotor nucleus (NIII, Fig. 1A) of the diencephalon; medial division of valvula cerebelli (Vam); SRF (Fig.1B); IMRF (Fig.1C); magnocellular octaval nucleus (MaON, Fig. 1D) of the hindbrain; and in the gray matter of the spinal cord (Fig. 1E). No labeling was detected in sections incubated with a control (scrambled-sequence) LNA probe (data not shown). These cells displayed the typical spatial distribution and morphology that we have observed in previous studies that characterized the regenerative capacity of CNS neurons (Becker et al., 1997). For instance, miR-133b–positive neurons of the NMLF had large cell bodies while neurons in the NIII were considerably smaller and symmetrically arranged along the border of the ventricle. Cells in the spinal cord stained more intensely for miR-133b than those in brain regions (Fig. 1E). Whereas in mammals the gray matter of the spinal cord has an H-shaped appearance, in fish it has an inverted Y shape and contains motor or sensory neurons. Stronger expression of miR-133b was observed in motor neurons, which were identified by their typical location and size (Westerfield et al., 1986). We also performed immunofluorescence labeling of spinal motor neurons, which showed an identical staining pattern to the miR-133b–positive cells (supplemental Fig. 1). Unfortunately, we were unable to combine in situ hybridization with immunofluorescence in a single image because the enzymatic treatment of tissue necessary to detect miR-133b negatively affected our immunofluorescence staining. Interestingly, many of the mirR-133b expressing neurons are injured by spinal cord lesion and are capable of regenerating their axons after injury in adult zebrafish (Becker et al., 1997).
Figure 1. miR-133b is expressed in brainstem nuclei and the spinal cord of adult zebrafish.
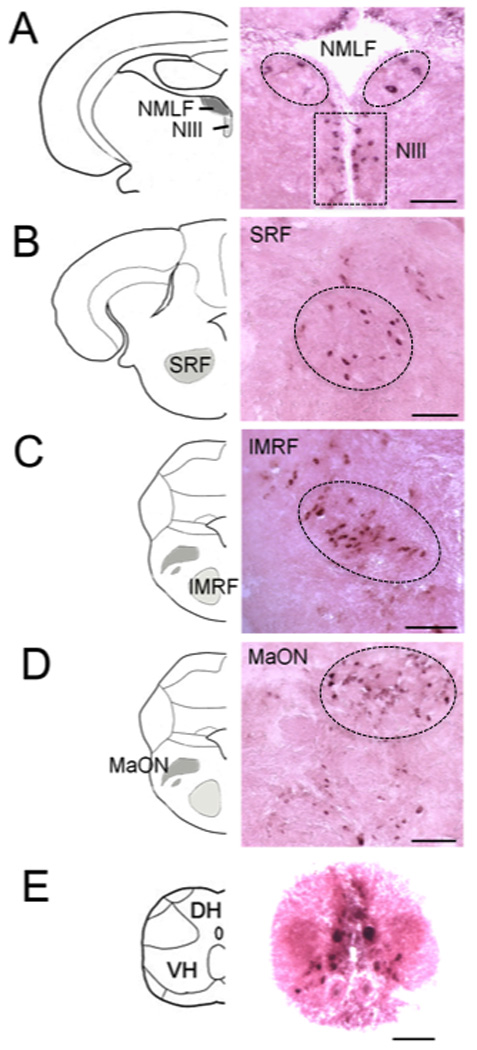
In situ hybridization with an LNA probe for miR-133b was performed on coronal sections of the brainstem and localized to neurons in the NMLF (A, dashed box contains neurons of NIII), SRF (B), IMRF (C), MaON (D), and of the spinal cord (E) of adult uninjured zebrafish (n=6). Brain and spinal cord were fixed with 4% formaldehyde in PBS overnight and cryoprotected in 30% sucrose. Serial coronal sections from brain and spinal cords were cryosectioned at 30 µm thickness. Scale bars, 50 µm. NMLF, nucleus of the medial longitudinal fascicle; SRF, superior reticular formation; IMRF, intermediate reticular formation; MaON, magnocellular octaval nucleus; NIII, the oculomotor nucleus; DH, dorsal horn of the spinal cord; VH, ventral horn of the spinal cord.
Expression of miR-133b after spinal cord injury
To determine whether miR-133b expression is regulated following spinal cord injury (SCI), we performed a time-course study on the most robustly expressing miR-133b nucleus in the brainstem, the NMLF. Zebrafish brains were assayed at six hours, twenty-four hours, and seven days after SCI using in situ hybridization (Fig. 2A–B). In sham-lesioned fish, the expression of miR-133b was similar to that which was observed in non-lesioned fish (data not shown). Conversely, miR-133b expression was increased in the NMLF as soon as six hours (11.7 ±1.76 cells in the total nucleus, n=3) after spinal cord transection, with significant increases occurring at twenty-four hours (25.0±3.21 cells in the total nucleus, n=3), and seven days (Fig. 2B, C, 30.7±3.33 cells in the total nucleus, n=3, p=0.001), with sham-lesioned fish serving as a control group (Fig. 2A, 4.7±0.88 cells in the total nucleus, n=3). Upregulation of miR-133b expression was limited to larger-diameter cells (arrows, diameter >13µm) in the NMLF, located bilaterally and symmetrically in the ventral midbrain. These results agree with those of our other studies in which the expression of spinal cord regeneration-conducive molecules L1 and tenascin-C were increased in the NMLF after SCI (Becker et al., 2004; Yu et al., in preparation).
Figure 2. miR-133b expression is upregulated in NMLF neurons 7 days after SCI in zebrafish and reduced after application of miR-133b morpholino (MO).
In situ hybridization with an LNA probe for miR-133b was performed on coronal sections of brainstem at the level of the NMLF in sham-lesioned (sham) (A) and lesioned (B) fish, and quantified at six hours, twenty-four hours, and seven days after injury (C). Scale bars, 50 µm. Arrows indicate miR-133b expressing cells in the NMLF, boxed area indicates neurons of the NIII. Quantitation of miR-133b positive cells in the NMLF (C, D), SRF (E), IMRF (F), and NIII (G) was performed on serial sections; numbers refer to all cells in each nucleus in sham-lesioned (sham) and lesioned fish and in lesioned fish treated with control MO or miR-133b MO (n=3 per group; *p<0.05, one-way ANOVA with Tukey’s post-hoc test).
We then examined all other nuclei at seven days after SCI, when miR-133b expression was significantly increased. We observed no upregulation of miR-133b in the NIII, TSvl, and Vam, SRF, and IMRF (Fig. 2D–G, data not shown for TSvl and Vam). Unlike axons originating from the NMLF and reticular nuclei, NIII, TSvl and Vam axons were not transected in our injury model, as they do not project to the spinal cord (Becker et al., 1997), and therefore do not directly contribute to the regeneration of transected spinal cord axons. Since a significant change in miR-133b expression was observed in the NMLF, it was the nucleus to which we directed our focus.
To identify the miR-133b–expressing neurons that project their axons to the transection site, we knocked down miR-133b expression by anti-sense miR-133b MO application to the spinal cord lesion site immediately after SCI, which is then taken up by the cut axons and retrogradely transported to the neurons of origin. Compared with standard control MO application, miR-133b MO application reduced the number of miR-133b–expressing cells detected by in situ hybridization in the post-injury NMLF, SRF, and IMRF (Fig. 2D–F respectively, n=3). However, we did not observe a reduced expression in the NIII (Fig. 2G). Taken together, the sequence specificity of the LNA probe and reduced miR-133b expression with MO application indicate that neurons from the NMLF, SRF, and IMRF extend axons to the spinal cord that are transected in our injury model, but axotomy alone is not sufficient to increase expression of miR-133b in every neuron in which miR-133b is present. Therefore, the increased miR-133b expression in the NMLF appears to be specific to that nucleus and so we focused on these neurons to find evidence of miR-133b–regulated regeneration.
Anti-sense miR-133b MO treatment decreases both locomotor recovery and the number of brainstem neurons with regenerated axons
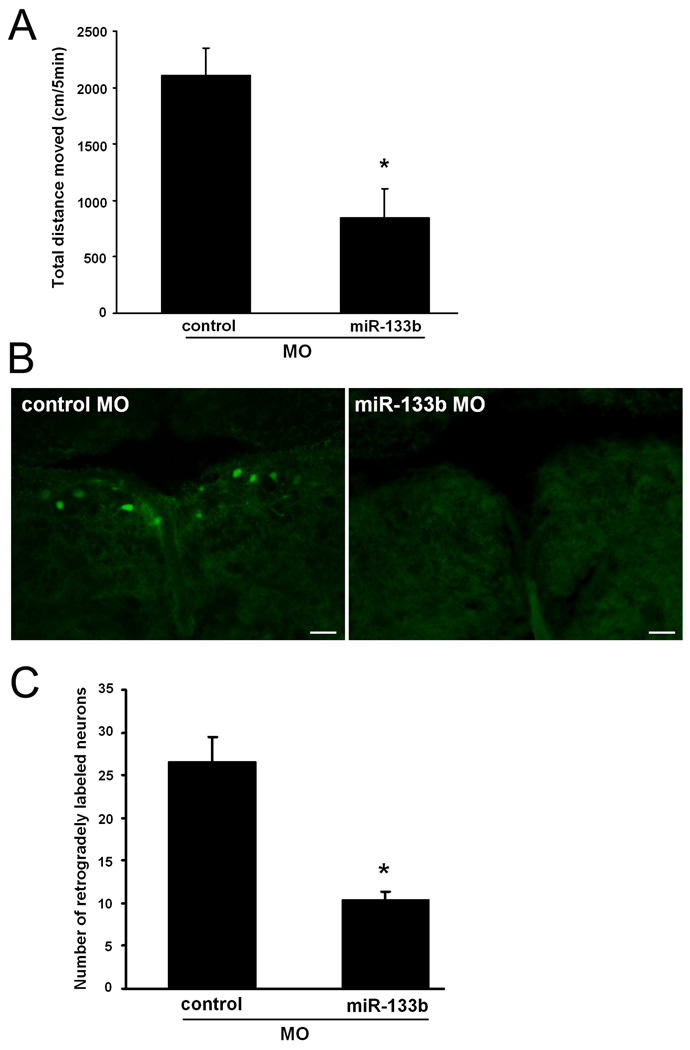
To study the contribution of miR-133b to locomotor recovery from SCI, we examined swimming performance as the total distance that fish moved using video-tracking at six weeks after SCI and application of anti-sense miR-133b MO or a control MO (Fig. 3A). For aquatic vertebrates, there is no formal grading system to assess locomotor recovery after injury. Total distance moved is an accepted parameter that measures the extent of functional recovery after SCI and has been used extensively by our lab in previous studies (Becker et al., 2004; Guo et al., 2010). Fish are paralyzed immediately after spinal cord transection and spontaneously recovery swimming ability by six weeks after SCI, a time at which recovered values are nearly identical to sham-lesioned fish in which the axial muscles required for swimming are cut, but the spinal cords were not transected (Becker et al., 2004). Thus, at six weeks after spinal cord injury, total distance moved was compared between control MO treated fish and miR-133b MO treated fish. As observed in previous studies, no difference was detected between control MO treated fish and untreated, or sham-lesioned fish (Becker et al., 2004). However, the total distance moved by miR-133b MO treated fish (847.3±253.37 cm/5min., n=5, p=0.0057) was reduced by 60% relative to the control MO treated fish (2111.7±240.09 cm/5min., n=6). These data indicate that miR-133b is required for complete recovery of locomotor function after SCI.
Figure 3. Functional and anatomical recovery after SCI.
(A) Fish were anesthetized and the spinal cord transected at 4 mm caudal to the brain stem-spinal cord junction. A control or anti-sense miR-133b MO soaked in Gelfoam was applied immediately after SCI at the lesion site. Locomotor activity was measured as total swimming distance moved of undisturbed fish during five minute trial periods by automatic video-tracking at six weeks after spinal cord transection.(n=6; *p<0.01, t-test). (B) Retrograde tracing were performed with biocytin at six weeks after SCI, with control or miR-133b MO treatment, by application of biocytin 4 mm caudal to the site of transection. Retrogradely labeled neurons in the total NMLF were detected with 488-Streptavidin (green). Scale bars, 50 µm. (C) Numbers of retrogradely labeled neurons were determined in the total NMLF of fish treated with control MO or miR-133b MO (*p<0.01, t-test).
NMLF and reticular formation neurons are known to mediate swimming activity (Uematsu & Todo, 1997) and to project regenerated axons into the spinal cord after injury (Becker et al., 1997). To investigate whether axonal regeneration from the NMLF and reticular formation depends on miR-133b, we performed retrograde tracing of brainstem neurons by applying biocytin at a second spinal lesion site that was 4 mm caudal from the original transection site, six weeks after SCI. Brainstem sections were probed twenty-four hours later with fluorescent streptavidin to detect biocytin, indicating neurons whose axons regenerated across the injury site after transection. Fish treated with miR-133b MO showed reduced numbers of streptavidin-labeled neurons compared to control MO treated fish, indicating impaired regeneration of the axons from NMLF (Fig. 3B), SRF, and IMRF (data not shown) neurons. The most dramatic reduction of retrogradely labeled neurons was observed in the NMLF where miR-133b MO treated fish (10.5 ± 0.87 cells, n=4, p=0.0018) showed 60% fewer labeled neurons relative to the number in control MO treated fish (26.6 ± 2.82 cells, n=5) (Fig. 3C). Thus, miR-133b expression in NMLF, SRF, and IMRF neurons is required for both locomotor recovery and axon regrowth.
miR-133b targets RhoA after SCI
As a first approach to identify potential mRNA targets of miR-133b, we used computational target prediction algorithms such as Targetscan and Miranda (John et al., 2004; Friedman et al., 2009). Noting that these programs do not include target predictions specific for zebrafish transcripts, we decided to focus on validated mammalian targets and searched for conserved recognition element (RE) sites specific for miR-133b in homologous zebrafish genes. One of these predicted targets, RhoA, which contains a conserved miR-133b RE, has been identified in mammals as a target of miR-133b (Care et al., 2007; Chiba et al., 2009). Inhibition of RhoA has been found to enhance locomotor recovery and axonal regrowth after SCI in mammals (Dergham et al., 2002; Fournier et al., 2003; Tanaka et al., 2004). Thus, targeting of RhoA by miR-133b may contribute to functional recovery after SCI in zebrafish.
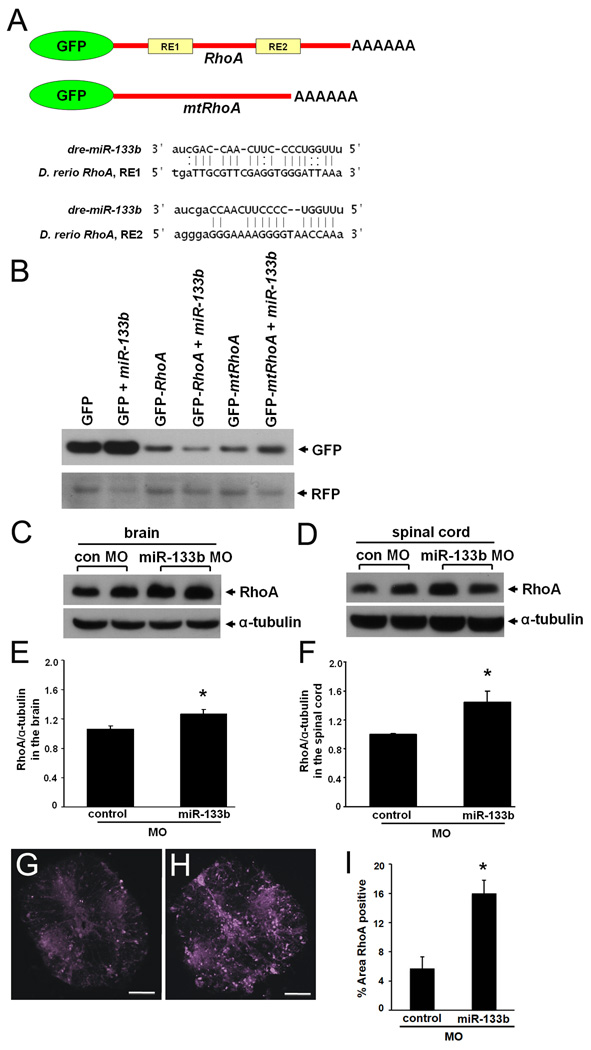
To test whether RhoA mRNA is targeted by miR-133b in zebrafish, we constructed a GFP reporter plasmid containing a region of RhoA 3’UTR, including two predicted RhoA miRNA-133b RE (GFP-RhoA) and a control plasmid lacking these RE sequences (GFP-mtRhoA; Fig. 4A). In vitro transcribed GFP fusion transcripts (GFP, GFP-RhoA, GFP-mtRhoA) were injected into single cell embryos in the presence or absence of exogenous miR-133b and GFP levels were examined in the embryos 2 days post-fertilization (dpf) (Fig. 4B). As a control, we co-injected RFP fusion transcript lacking RhoA sequences. Western blot analyses of lysates from twelve pooled embryos were performed using antibodies against GFP and RFP. The expression level of GFP in GFP-RhoA mRNA injected embryos was decreased when co-injected with miR-133b compared to those without added miR-133b (Fig. 4B). However, GFP expression levels from either control GFP mRNA or from GFP-mtRhoA mRNA injected embryos were not suppressed by exogenous miR-133b. Inhibition of the GFP signal only in the presence of the RhoA RE demonstrates the sequence-specific targeting of RhoA mRNA by miR-133b.
Figure 4. miR-133b targets RhoA and reduces RhoA expression after SCI.
(A) Diagram indicating GFP reporters. (B) One-cell-stage zebrafish embryos were injected with RNA encoding GFP or the GFP-RhoA fusion construct (see A) in the presence or absence of exogenous miR133b. At two days post-fertilization, Western blot analyses were performed using antibodies against GFP or RFP as an internal control. (C, D) Western blot analyses RhoA and α-tubulin expression in the NMLF of brain (C) and spinal cord (D) of adult zebrafish at 9 days after SCI. Quantitation of RhoA/α-tubulin ratios from Western blot analyses (E, F) shown in (C) and (D). Immunofluorescence images stained for RhoA in spinal cord coronal sections after treatment with control MO (G) or miR-133b MO (H), and quantified (I) as the percent area above the set threshold. All values were normalized to control MO (*p<0.05, t-test).
To determine whether miR-133b regulates RhoA during spinal cord regeneration in adult zebrafish, we examined the RhoA expression levels in the brain and lesioned spinal cord after treatment with anti-sense miR-133b or control MO at 9 days after SCI by Western blot analysis and immunofluorescence. In slices of brain including the NMLF, RhoA relative expression levels increased in miR-133b MO treated fish (1.3±0.06, p=0.008) compared to control MO treated fish (1.0±0.04) (Fig. 4C, E). In the spinal cord, RhoA levels also increased in miR-133b MO treated fish (1.4±0.15, p=0.004) when compared to control MO treated fish (1.0±0.01) (Fig. 4D, F). Sections of spinal cord tissue taken from the point of morpholino application also demonstrated an increased level of RhoA (Fig. 4G–I). In agreement with Western blot results, the miR-133 MO treated sections (16.0±1.8, n=3) had a significantly increased percent of RhoA positive area than control MO treated (5.7±1.6, p=0.001, n=3) fish. Taken together, these results are consistent with the hypothesis that RhoA is targeted by endogenous miR-133b in adult zebrafish CNS after SCI.
DISCUSSION
Our results demonstrate that miR-133b is necessary for spontaneous regeneration of supraspinal axons from the NMLF, SRF, and IMRF neurons following SCI in adult zebrafish. miR-133b is expressed in a number of neuronal populations in uninjured zebrafish brain and spinal cord (Fig. 1), including motor neurons in the NMLF, SRF, IMRF, and MaON, all of which are brainstem nuclei that are important for controlling swimming behavior (Sankrithi & O'Malley, 2010) and are also known to reliably regenerate their axons after SCI (Becker et al., 1997). In addition, we detected miR-133b in the NIII, TSvl, and Vam. Since these nuclei do not project axons to the spinal cord, their regenerative capacity was not tested in our spinal cord transection model. In agreement with this view, the inhibition of miR-133b expression after SCI reduces both locomotor recovery and the number of NMLF, SRF, and IMRF axons that cross the injury site.
Lower vertebrates, such as fish and amphibians, are able to regenerate several injured organs since these animals have reduced levels of inhibitory molecules as compared to mammals (Becker, 2007; Becker & Becker, 2007). An alternative hypothesis is that the loss of regenerative capacity could be due to the differential expression of regulatory agents such as miR-133b. For example, a recent report shows that miR-133b expression is increased four hours after SCI in the rat, but then is rapidly diminished by one day and remains repressed below baseline levels at seven days after SCI (Liu et al., 2009). This expression pattern is the opposite of what we observed in zebrafish, where miR-133b increases after SCI and remains high for at least seven days after SCI (Fig. 2). This observation also suggests that the mammalian CNS attempts to mount a response to overcome inhibitory signals after SCI, but for reasons yet to be uncovered, fails to do so. Additionally, it is clear that altering the expression of a single gene is insufficient to overcome inhibitory signals in the mammalian CNS after injury (Yiu & He, 2006). Since a single miRNA may inhibit the expression of dozens to thousands of genes (Miranda et al., 2006), miR-133b may represent one mechanism that globally suppresses numerous inhibitory molecules to promote spinal cord regeneration. This difference in the regulation of miR-133b expression between zebrafish and mammals could at least in part explain the lack of regenerative capacity in mammals after SCI.
As an initial demonstration of the ability of miR-133b to reduce the expression of one of its targets that inhibit axon regrowth after injury, we showed that RhoA mRNA is directly targeted by miR-133b in a zebrafish embryo reporter system (Fig. 4). It is clear from the literature that RhoA is a convergence point of several deleterious signaling pathways after SCI, and that its increased expression leads to a reduced functional outcome after SCI. RhoA GTPase signaling through Rho kinase (ROCK) promotes growth cone collapse, inhibits regeneration (Luo et al., 1997), and has previously been shown to inhibit functional recovery after SCI in mammals (Dergham et al., 2002; Ellezam et al., 2002; Dubreuil et al., 2003; Fournier et al., 2003; Monnier et al., 2003; Sung et al., 2003). RhoA also plays a role in infiltration of neutrophils and macrophages into the nervous system (Ridley, 2001) by regulating endothelial permeability via the integrity of tight junctions (Essler et al., 1998; Carbajal & Schaeffer, 1999; Hordijk et al., 1999; Schreibelt et al., 2007). Following SCI, inhibitory molecules in CNS myelin and glial scar-associated chondroitin sulfate proteoglycans induce RhoA signaling (McGee & Strittmatter, 2003; Kubo et al., 2007), resulting in reduced dendrite and axon outgrowth (McKerracher, 2001; Dergham et al., 2002; McKerracher & Winton, 2002; Qiu et al., 2002; Fournier et al., 2003; Domeniconi & Filbin, 2005; McKerracher & Higuchi, 2006). After spinal cord injury in mammals, the application of C3 transferase, a bacterial exoenzyme that inhibits RhoA function, enhances regrowth of the corticospinal tract beyond the lesion site and promotes neuroprotection by decreasing tissue damage and cavity formation. Intrathecal administration of RhoA siRNA improved functional recovery including white matter sparing, serotonergic fiber growth, and reduced allodynia (Otsuka, 2011). We found that levels of RhoA protein are increased in injured spinal cords when miR-133b is knocked-down, and that inhibition of miR-133b reduced both locomotor recovery and axonal regrowth from regenerating brainstem nuclei. Inducing the expression of a single molecular regulator capable of simultaneously affecting several cellular processes, such as miR-133b, would be an effective way to target many pathways that negatively affect regeneration. Therefore, induction of miR-133b expression promotes regeneration, at least through repression of RhoA translation, and likely through several additional targets as well.
Additional biochemically validated targets of miR-133b include several genes that have inhibitory roles in SCI (Table 1). For example, calcineurin, a protein phosphatase, promotes oligodendrocyte apoptosis after SCI through the activation of caspase-3 (Nottingham et al., 2002). Inhibitors of calcineurin, such as cyclosporine A, FK506, and tacrolimus, serve beneficial roles after SCI and traumatic brain injury (Madsen et al., 1998b; Diaz-Ruiz et al., 2005). Another predicted target is the pro-apoptotic gene caspase-9 (Xu et al., 2007). Rats treated with the caspase-9 inhibitor z-LEHD-fmk after SCI show reduced apoptosis and improved functional outcome (Colak et al., 2005). miR-133 has also been shown to regulate connective tissue growth factor (CTGF) in mammals (Duisters et al., 2009). CTGF has been linked to glial scar formation, a major inhibitor of axonal growth at injury sites in the CNS (Hertel et al., 2000; Conrad et al., 2005a; White & Jakeman, 2008). TGF-beta is targeted by miR-133b in a nicotine-induced atrial fibrillation model in canines (Shan et al., 2009). Intrathecal administration of a neutralizing antibody to TGF-beta1 in rats with thoracic spinal cord contusion enhanced growth and/or preservation of axons after injury (Kohta et al., 2009), resulting in reduced glial scar formation and significant functional recovery. These are only a few examples of the inhibitory molecules that miR-133b is likely to suppress in the regenerating zebrafish system, according to studies validating their direct targeting by miR-133. In addition, computational prediction of miR-133b targets yields a large list containing the above-mentioned genes, and many others that have yet to be validated, such as the Nogo-66 receptor homologue (NgR3); these have been excluded from Table 1. It is also worth mentioning that the majority of the validated targets shown in Table 1 serve an inhibitory role in spinal cord regeneration after SCI, indicating that miR-133b is likely to serve as a negative regulator of several genes that are inhibitory after SCI.
Table 1. Validated miR-133b mammalian targets.
We performed a literature search for miR-133b targets in mammalian systems and show only those with proposed roles in acute and chronic central nervous system trauma. Only two additional validated mammalian targets exist that have no identified role in the CNS or SCI.
| Mammalian miR-133 Targets |
Role in SCI and/or CNS | ||
|---|---|---|---|
| RhoA | (Care et al., 2007) | Rho GTPase, inhibits neurite outgrowth in vitro and axonal regrowth after injury in vivo |
(Luo et al., 1997; Dergham et al., 2002; Ellezam et al., 2002; Dubreuil et al., 2003; Fournier et al., 2003; Monnier et al., 2003; Sung et al., 2003) |
| Calcineurin | (Dong et al., 2010) | Phosphatase, pro-apoptotic | (Madsen et al., 1998a; Nottingham et al., 2002; Diaz-Ruiz et al., 2005) |
| Caspase9 | (Xu et al., 2007) | Pro-apoptotic | (Colak et al., 2005) |
| TGF-beta1 | (Shan et al., 2009) | Multiple pro-inflammatory activities |
(Shan et al., 2009) |
| CTGF | (Duisters et al., 2009) | Participate glial scar formation | (Hertel et al., 2000; Conrad et al., 2005a; White & Jakeman, 2008) |
| HCN2 | (Luo et al., 2008) | Role in DRG in neuropathic pain, mechanical allodynia |
(Jiang et al., 2008a; Jiang et al., 2008b) |
| KLF15 | (Horie et al., 2009) | Induces glial cell differentiation |
(Fu et al., 2009) |
| nPTB | (Boutz et al., 2007) | Inhibits the splicing of neuron specific alterative exons |
(Makeyev et al., 2007) |
| NFATc4 | (Li et al., 2010) | Exhibits pro- and anti- apoptotic activities |
(Luoma & Zirpel, 2008; Vashishta et al., 2009) |
|
NELF-A/ WHSC2 |
(Care et al., 2007) | N/A | |
| KCNQ1 | (Luo et al., 2007) | N/A | |
| Runx2 | (Li et al., 2008) | Differentiation of GABA phenotype |
(Benes et al., 2007) |
| SRF | (Chen et al., 2006) | Cooperatively regulates expression of cytoskeletal genes and neurite outgrowth |
(Mokalled, 2010) |
Surprisingly, the logic of miR-133b regulation in SCI is directly opposite to that in fin regeneration. Downregulation of miR-133b expression after fin lesion was suggested to enhance efficient fin regeneration, while increasing the levels of miR-133b reduced regeneration of injured fin in zebrafish (Yin et al., 2008). By contrast, miR-133b expression increases upon SCI, at least in the NMLF, and is required for regeneration of axons from brainstem neurons. It is thus noteworthy that this same pattern of increased miR-133 expression is observed in regenerating muscle (Nakasa et al., 2009). We attribute these opposing roles in regenerating tissues to differences in cellular context or regeneration mechanisms. These different lesion paradigms have unique regeneration inhibitors and enhancers that may not necessarily be the targets of miR-133b in fin versus CNS and muscle and therefore, we do not believe that the opposing pattern of miR-133 expression in fin regeneration conflicts with our interpretation of its role in SCI.
The identification of miR-133b as an endogenous, injury-regulated inhibitor targeting RhoA helps to explain, at least in part, how zebrafish retain the ability to recover locomotor functions after SCI while mammals do not. The mechanism causing long-term induction of miR-133b after CNS injury has clearly been lost in mammals, leading to derepressed inhibitors that serve to reduce effective regeneration of injured axons and reformation of functional neuronal networks. The regulatory molecule miR-133b has the potential for targeting a broad range of these inhibitory genes that negatively affect spinal cord regeneration in mammals. This makes miR-133b an attractive therapeutic candidate for treating spinal cord injured humans since small RNAs can be delivered intravenously and intrathecally, without genetically altering the treated tissue (Dorn et al., 2004). Instead of reducing the expression of only one inhibitory molecule at a time, as would be the case for traditional siRNA, a single miRNA has the ability to reduce the expression of several genes that inhibit spinal cord regeneration. We predict that exogenous application of miR-133b will be an effective treatment for SCI in rodent models and are currently testing this hypothesis in our laboratory.
Supplementary Material
Acknowledgements
We are grateful to Dr. James G. Patton for his gift of the pCS2+GFP vector, and to Samantha Thomas for her assistance with image analysis. This work was supported by grants from the New Jersey Commission on Spinal Cord Research [08-3079-SCR-E-0 and 05-3048-SCR-E-0 to MS]. JD and RPH were supported by NIH R21 MH085088. MS is a consultant at the Center for Neuroscience of Shantou University Medical College. KMG is a fellow of the New Jersey Commission on Spinal Cord Research.
Abbreviations
- CNS
central nervous system
- GFP
green fluorescent protein
- IMRF
intermediate reticular formation
- LNA
locked nucleic acid probe
- MaON
magnocellular octaval nucleus
- miRNA
microRNA
- MO
morpholino
- MS-222
aminobenzoic acid ethyl methyl ester
- NIII
oculomotor nucleus
- NMLF
nucleus of the medial longitudinal fascicle
- PBS
phosphate buffered saline
- RFP
red fluorescent protein
- SCI
spinal cord injury
- SRF
superior reticular formation
- TSvl
ventrolateral nucleus of the torus semicircularis
- UTR
untranslated region
- Vam
medial division of the valvula cerebelli
REFERENCES
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Becker CG, Becker T. Growth and pathfinding of regenerating axons in the optic projection of adult fish. J Neurosci Res. 2007;85:2793–2799. doi: 10.1002/jnr.21121. [DOI] [PubMed] [Google Scholar]
- Becker CG, Becker T. Model Organisms in Spinal Cord Regeneration. Weinheim: Wiley; 2007. [Google Scholar]
- Becker CG, Lieberoth BC, Morellini F, Feldner J, Becker T, Schachner M. L1.1 is involved in spinal cord regeneration in adult zebrafish. J Neurosci. 2004;24:7837. doi: 10.1523/JNEUROSCI.2420-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377:577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes & development. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal JM, Schaeffer RC., Jr RhoA inactivation enhances endothelial barrier function. The American journal of physiology. 1999;277:C955–C964. doi: 10.1152/ajpcell.1999.277.5.C955. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, 2nd, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- Colak A, Karaoglan A, Barut S, Kokturk S, Akyildiz AI, Tasyurekli M. Neuroprotection and functional recovery after application of the caspase-9 inhibitor z-LEHD-fmk in a rat model of traumatic spinal cord injury. J Neurosurg Spine. 2005;2:327–334. doi: 10.3171/spi.2005.2.3.0327. [DOI] [PubMed] [Google Scholar]
- Conrad S, Schluesener HJ, Adibzahdeh M, Schwab JM. Spinal cord injury induction of lesional expression of profibrotic and angiogenic connective tissue growth factor confined to reactive astrocytes, invading fibroblasts and endothelial cells. J Neurosurg Spine. 2005a;2:319–326. doi: 10.3171/spi.2005.2.3.0319. [DOI] [PubMed] [Google Scholar]
- Conrad S, Schluesener HJ, Trautmann K, Joannin N, Meyermann R, Schwab JM. Prolonged lesional expression of RhoA and RhoB following spinal cord injury. J Comp Neurol. 2005b;487:166–175. doi: 10.1002/cne.20561. [DOI] [PubMed] [Google Scholar]
- Dergham P, Ellezam B, Essagian C, Avedissian H, Lubell WD, McKerracher L. Rho signaling pathway targeted to promote spinal cord repair. J Neurosci. 2002;22:6570–6577. doi: 10.1523/JNEUROSCI.22-15-06570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz A, Vergara P, Perez-Severiano F, Segovia J, Guizar-Sahagun G, Ibarra A, Rios C. Cyclosporin-A inhibits constitutive nitric oxide synthase activity and neuronal and endothelial nitric oxide synthase expressions after spinal cord injury in rats. Neurochem Res. 2005;30:245–251. doi: 10.1007/s11064-005-2447-0. [DOI] [PubMed] [Google Scholar]
- Domeniconi M, Filbin MT. Overcoming inhibitors in myelin to promote axonal regeneration. J Neurol Sci. 2005;233:43–47. doi: 10.1016/j.jns.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Dong DL, Chen C, Huo R, Wang N, Li Z, Tu YJ, Hu JT, Chu X, Huang W, Yang BF. Reciprocal Repression Between MicroRNA-133 and Calcineurin Regulates Cardiac Hypertrophy. A Novel Mechanism for Progressive Cardiac Hypertrophy. Hypertension. 2010 doi: 10.1161/HYPERTENSIONAHA.109.139519. [DOI] [PubMed] [Google Scholar]
- Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, Martin P, Bevan S, Fox A, Ganju P, Wishart W, Hall J. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil CI, Winton MJ, McKerracher L. Rho activation patterns after spinal cord injury and the role of activated Rho in apoptosis in the central nervous system. The Journal of cell biology. 2003;162:233–243. doi: 10.1083/jcb.200301080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M. Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol Cell Biol. 2008;28:2391–2413. doi: 10.1128/MCB.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellezam B, Dubreuil C, Winton M, Loy L, Dergham P, Selles-Navarro I, McKerracher L. Inactivation of intracellular Rho to stimulate axon growth and regeneration. Progress in brain research. 2002;137:371–380. doi: 10.1016/s0079-6123(02)37028-6. [DOI] [PubMed] [Google Scholar]
- Erschbamer MK, Hofstetter CP, Olson L. RhoA, RhoB, RhoC, Rac1, Cdc42, and Tc10 mRNA levels in spinal cord, sensory ganglia, and corticospinal tract neurons and long-lasting specific changes following spinal cord injury. J Comp Neurol. 2005;484:224–233. doi: 10.1002/cne.20471. [DOI] [PubMed] [Google Scholar]
- Essler M, Amano M, Kruse HJ, Kaibuchi K, Weber PC, Aepfelbacher M. Thrombin inactivates myosin light chain phosphatase via Rho and its target Rho kinase in human endothelial cells. The Journal of biological chemistry. 1998;273:21867–21874. doi: 10.1074/jbc.273.34.21867. [DOI] [PubMed] [Google Scholar]
- Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009;20:3905–3917. doi: 10.1091/mbc.E08-10-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM. Rho kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci. 2003;23:1416–1423. doi: 10.1523/JNEUROSCI.23-04-01416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, Stiles CD. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–11408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman T, Trouche S, Massou I, Verret L, Zerwas M, Roullet P, Rampon C. Young hippocampal neurons are critical for recent and remote spatial memory in adult mice. Neuroscience. 2010;171:769–778. doi: 10.1016/j.neuroscience.2010.09.047. [DOI] [PubMed] [Google Scholar]
- Guo Y, Ma L, Cristofanilli M, Hart RP, Hao A, Schachner M. Transcription factor Sox11b is involved in spinal cord regeneration in adult zebrafish. Neuroscience. 2010;172:329–341. doi: 10.1016/j.neuroscience.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel M, Tretter Y, Alzheimer C, Werner S. Connective tissue growth factor: a novel player in tissue reorganization after brain injury? Eur J Neurosci. 2000;12:376–380. doi: 10.1046/j.1460-9568.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Hofmann F, Just I, Lehnardt S, Hanisch UK, Bruck W, Kettenmann H, Ahnert-Hilger G, Holtje M. Inhibition of Rho-dependent pathways by Clostridium botulinum C3 protein induces a proinflammatory profile in microglia. Glia. 2008;56:1162–1175. doi: 10.1002/glia.20687. [DOI] [PubMed] [Google Scholar]
- Holtje M, Djalali S, Hofmann F, Munster-Wandowski A, Hendrix S, Boato F, Dreger SC, Grosse G, Henneberger C, Grantyn R, Just I, Ahnert-Hilger G. A 29-amino acid fragment of Clostridium botulinum C3 protein enhances neuronal outgrowth, connectivity, and reinnervation. FASEB J. 2009;23:1115–1126. doi: 10.1096/fj.08-116855. [DOI] [PubMed] [Google Scholar]
- Hordijk PL, Anthony E, Mul FP, Rientsma R, Oomen LC, Roos D. Vascular-endothelial-cadherin modulates endothelial monolayer permeability. J Cell Sci. 1999;112(Pt 12):1915–1923. doi: 10.1242/jcs.112.12.1915. [DOI] [PubMed] [Google Scholar]
- Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, Kuwabara Y, Takanabe R, Hasegawa K, Kita T, Kimura T. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389:315–320. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefford CE, Feki A, Harb J, Krause KH, Irminger-Finger I. Nuclear-cytoplasmic translocation of BARD1 is linked to its apoptotic activity. Oncogene. 2004;23:3509–3520. doi: 10.1038/sj.onc.1207427. [DOI] [PubMed] [Google Scholar]
- Jiang YQ, Sun Q, Tu HY, Wan Y. Characteristics of HCN channels and their participation in neuropathic pain. Neurochem Res. 2008a;33:1979–1989. doi: 10.1007/s11064-008-9717-6. [DOI] [PubMed] [Google Scholar]
- Jiang YQ, Xing GG, Wang SL, Tu HY, Chi YN, Li J, Liu FY, Han JS, Wan Y. Axonal accumulation of hyperpolarization-activated cyclic nucleotide-gated cation channels contributes to mechanical allodynia after peripheral nerve injury in rat. Pain. 2008b;137:495–506. doi: 10.1016/j.pain.2007.10.011. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohta M, Kohmura E, Yamashita T. Inhibition of TGF-beta1 promotes functional recovery after spinal cord injury. Neurosci Res. 2009;65:393–401. doi: 10.1016/j.neures.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Kubo T, Hata K, Yamaguchi A, Yamashita T. Rho-ROCK inhibitors as emerging strategies to promote nerve regeneration. Current pharmaceutical design. 2007;13:2493–2499. doi: 10.2174/138161207781368657. [DOI] [PubMed] [Google Scholar]
- Li Q, Lin X, Yang X, Chang J. NFATc4 is negatively regulated in miR-133a–mediated cardiomyocyte hypertrophic repression. Am J Physiol Heart Circ Physiol. 2010 doi: 10.1152/ajpheart.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Jan LY, Jan YN. Rho family GTP-binding proteins in growth cone signalling. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. The Journal of biological chemistry. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- Luo X, Xiao J, Lin H, Li B, Lu Y, Yang B, Wang Z. Transcriptional activation by stimulating protein 1 and post-transcriptional repression by muscle-specific microRNAs of IKs-encoding genes and potential implications in regional heterogeneity of their expressions. J Cell Physiol. 2007;212:358–367. doi: 10.1002/jcp.21030. [DOI] [PubMed] [Google Scholar]
- Luoma JI, Zirpel L. Deafferentation-induced activation of NFAT (nuclear factor of activated T-cells) in cochlear nucleus neurons during a developmental critical period: a role for NFATc4-dependent apoptosis in the CNS. J Neurosci. 2008;28:3159–3169. doi: 10.1523/JNEUROSCI.5227-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JR, MacDonald P, Irwin N, Goldberg DE, Yao GL, Meiri KF, Rimm IJ, Stieg PE, Benowitz LI. Tacrolimus (FK506) increases neuronal expression of GAP-43 and improves functional recovery after spinal cord injury in rats. Exp.Neurol. 1998a;154:673. doi: 10.1006/exnr.1998.6974. [DOI] [PubMed] [Google Scholar]
- Madsen JR, MacDonald P, Irwin N, Goldberg DE, Yao GL, Meiri KF, Rimm IJ, Stieg PE, Benowitz LI. Tacrolimus (FK506) increases neuronal expression of GAP-43 and improves functional recovery after spinal cord injury in rats. Exp Neurol. 1998b;154:673–683. doi: 10.1006/exnr.1998.6974. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends in neurosciences. 2003;26:193–198. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- McKerracher L. Spinal cord repair: strategies to promote axon regeneration. Neurobiol Dis. 2001;8:11–18. doi: 10.1006/nbdi.2000.0359. [DOI] [PubMed] [Google Scholar]
- McKerracher L, Higuchi H. Targeting Rho to stimulate repair after spinal cord injury. J Neurotrauma. 2006;23:309–317. doi: 10.1089/neu.2006.23.309. [DOI] [PubMed] [Google Scholar]
- McKerracher L, Winton MJ. Nogo on the go. Neuron. 2002;36:345–348. doi: 10.1016/s0896-6273(02)01018-8. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mokalled MHea. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development. 2010;14:2365–2374. doi: 10.1242/dev.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Schwab JM, Henke-Fahle S, Mueller BK. The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Molecular and cellular neurosciences. 2003;22:319–330. doi: 10.1016/s1044-7431(02)00035-0. [DOI] [PubMed] [Google Scholar]
- Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Kawakami A, Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev Growth Differ. 2007;49:145–154. doi: 10.1111/j.1440-169X.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- Nottingham S, Knapp P, Springer J. FK506 treatment inhibits caspase-3 activation and promotes oligodendroglial survival following traumatic spinal cord injury. Exp Neurol. 2002;177:242–251. doi: 10.1006/exnr.2002.7975. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Otsuka S, Adamson C, Sankar V, Gibbs KM, Kane-Goldsmith N, Ayer JJ, Babiarz J, Kalinski H, Ashush H, Alpert E, Lahav R, Feinstein E, Grumet M. Delayed intrathecal delivery of RhoA siRNA to the contused spinal cord inhibits allodynia, preserves white matter and increases serotonergic fiber growth. J Neurotrauma. 2011 Mar 29; doi: 10.1089/neu.2010.1568. [Epub ahead of print] PMID: 21443453. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Reimer MM, Sorensen I, Kuscha V, Frank RE, Liu C, Becker CG, Becker T. Motor neuron regeneration in adult zebrafish. J Neurosci. 2008;28:8510–8516. doi: 10.1523/JNEUROSCI.1189-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho proteins, PI 3-kinases, and monocyte/macrophage motility. FEBS letters. 2001;498:168–171. doi: 10.1016/s0014-5793(01)02481-4. [DOI] [PubMed] [Google Scholar]
- Sankrithi NS, O'Malley DM. Activation of a multisensory, multifunctional nucleus in the zebrafish midbrain during diverse locomotor behaviors. Neuroscience. 2010;166:970–993. doi: 10.1016/j.neuroscience.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Schreibelt G, Kooij G, Reijerkerk A, van Doorn R, Gringhuis SI, van der Pol S, Weksler BB, Romero IA, Couraud PO, Piontek J, Blasig IE, Dijkstra CD, Ronken E, de Vries HE. Reactive oxygen species alter brain endothelial tight junction dynamics via RhoA, PI3 kinase, and PKB signaling. Faseb J. 2007;21:3666–3676. doi: 10.1096/fj.07-8329com. [DOI] [PubMed] [Google Scholar]
- Shan H, Zhang Y, Lu Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y, Yang B. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- Sung JK, Miao L, Calvert JW, Huang L, Louis Harkey H, Zhang JH. A possible role of RhoA/Rho-kinase in experimental spinal cord injury in rat. Brain Res. 2003;959:29–38. doi: 10.1016/s0006-8993(02)03717-4. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yamashita T, Yachi K, Fujiwara T, Yoshikawa H, Tohyama M. Cytoplasmic p21(Cip1/WAF1) enhances axonal regeneration and functional recovery after spinal cord injury in rats. Neuroscience. 2004;127:155–164. doi: 10.1016/j.neuroscience.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Uematsu K, Todo T. Identification of the midbrain locomotor nuclei and their descending pathways in the teleost carp, Cyprinus carpio. Brain Res. 1997;773:1–7. doi: 10.1016/s0006-8993(97)00619-7. [DOI] [PubMed] [Google Scholar]
- Vashishta A, Habas A, Pruunsild P, Zheng JJ, Timmusk T, Hetman M. Nuclear factor of activated T-cells isoform c4 (NFATc4/NFAT3) as a mediator of antiapoptotic transcription in NMDA receptor-stimulated cortical neurons. J Neurosci. 2009;29:15331–15340. doi: 10.1523/JNEUROSCI.4873-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M, McMurray JV, Eisen JS. Identified motoneurons and their innervation of axial muscles in the zebrafish. J Neurosci. 1986;6:2267–2277. doi: 10.1523/JNEUROSCI.06-08-02267.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RE, Jakeman LB. Don't fence me in: harnessing the beneficial roles of astrocytes for spinal cord repair. Restor Neurol Neurosci. 2008;26:197–214. [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.