Abstract
A predominant pathway of xenobiotic-induced toxicity is initiated by bioactivation. Characterizing reactive intermediates will provide information on the structure of reactive species, thereby defining a potential bioactivation mechanism. Because most reactive metabolites are not stable, it is difficult to detect them directly. Reactive metabolites can form adducts with trapping reagents, such as glutathione, which makes the reactive metabolites detectable. However, it is challenging to “fish” these adducts out from a complex biological matrix, especially for adducts generated via uncommon metabolic pathways. In this regard, we developed a novel approach based upon metabolomic technologies to screen trapped reactive metabolites. The bioactivation of pulegone, acetaminophen, and clozapine were reexamined by using this metabolomic approach. In all these cases, a large number of trapped reactive metabolites were readily identified. These data indicate that this metabolomic approach is an efficient tool to profile xenobiotic bioactivation.
INTRODUCTION
Most xenobiotics are transformed into more polar and stable metabolites by metabolizing enzymes and then are excreted from the body. However, some xenobiotics undergo metabolic activation to generate the reactive electrophiles capable of covalent binding to protein, DNA, and other biomolecules (1–3). Although the mechanistic relationship between reactive metabolites and toxicity is still unclear, ample evidence suggests their association (2, 4). The critical proteins modified by reactive metabolites may alter the biological process and further result in the direct toxicities demonstrated by tissue necrosis and/or apoptosis (5). The binding of reactive metabolites to DNA has the potential to produce genotoxicity (6, 7). Thus, identifying reactive metabolites is crucial in assessing the toxicity of chemicals that are exposed to humans (8–10).
In general, reactive metabolites are classified into soft and hard electrophiles. Classic soft electrophiles include epoxides, α,β-unsaturated carbonyls, quinones, quinone imines, quinone methides, imine methide, isocyanate, isothiocynates, aziridinium, and episulfonium (11, 12). Aldehydes and iminium ions belong to the group of hard electrophiles. Most reactive metabolites are unstable and cannot be detected directly. GSH and its analogues are commonly used to trap soft electrophiles (5, 13–15). Hard electrophiles can be trapped by semicarbazide, methoxylamine, or cyanide ions (16–20). The reaction of trapping reagents and reactive metabolites will form adducts that are stable. However, it is challenging to separate these adducts from a complex biological matrix. Multiple mass spectrometry (MS) methodologies, including neutral loss (21), precursor ion (22), multiple reaction monitoring (MRM) (23), and mass defect filtering (MDF) (24), have been developed to screen the reagent-trapped reactive metabolites. All these methods are effective. However, they are all biased methods and pre-set parameters are needed to conduct data analysis. For example, transition lists are required in the analysis using MRM, which will detect the predicted reactive metabolites, but miss the unexpected metabolites (23). An unbiased approach is needed for the screening of trapped reactive metabolites.
Metabolomics refers to the systemic investigation of metabolites in living organisms (25). Recently, metabolomic approach has been successfully adopted in drug metabolism and proved to be a powerful tool for rationally “fishing” the stable metabolites from biological matrices (26–30). However, this approach has not been used for studying unstable reactive metabolites. In the current study, we introduced this unbiased approach for screening of trapped reactive metabolites for the first time. This approach was validated by analyzing the bioactivation of pulegone, acetaminophen, and clozapine.
MATERIALS AND METHODS
Overall strategy
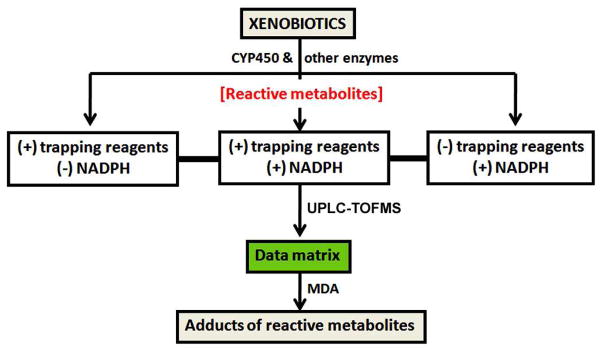
Our experimental design is illustrated in Figure 1. The xenobiotics were incubated with human liver microsomes (HLM) that contained Cytochromes P450 (CYP450) and other enzymes. The incubation groups without NADPH or trapping reagent served as controls, which were used to identify NADPH and trapping reagent-dependent biomarkers of xenobiotic bioactivation. All incubated samples were analyzed by ultra performance liquid chromatography (UPLC) and time of flight mass spectrometry (TOFMS). The acquired data were processed by MarkerLynx software (Waters Corp., Milford, MA) to produce a data matrix. Multivariate data analysis (MDA) was then conducted to screen trapped reactive metabolites of xenobiotics. To be noted, TOFMS has some limitations. One of the main limitations is the quantitation due to matrix ion suppression in electrospray ionization (31). In the current study, we did not quantify trapped reactive metabolites.
Figure 1. Schematic illustration of the metabolomic strategy for the screening of trapped reactive metabolites.
Xenobiotics were incubated in a reaction system containing enzymes, with or without NADPH and trapping reagents. The incubated samples were analyzed by UPLC-TOFMS. A data matrix was generated by MarkerLynx. Multivariate data analysis (MDA) was then conducted to screen trapped reactive metabolites.
Materials
Acetaminophen (APAP), pulegone (PLG), clozapine (CLP), glutathione (GSH), semicarbazide, potassium cyanide (KCN), and NADPH were obtained from Sigma-Aldrich (St. Louis, MO). HLM was purchased from XenoTech (Lenexa, KS). All the solvents for liquid chromatography and mass spectrometry were of the highest grade commercially available.
Microsomal incubations
Examined compounds (30 μM) were incubated separately in phosphate buffered saline (100 mM, pH 7.4) containing HLM (1.0 mg/ml), NADPH (1.0 mM), and different trapping reagents, such as GSH (2.5 mM), semicarbazide (2.5 mM), or potassium cyanide (1.5 mM). The total incubation volume was 400 μl. The incubations in the absence of trapping reagent or NADPH were used as controls. The incubations were conducted for 45 min. The reactions were initiated by the addition of NADPH and were stopped by the addition of 100 μl of formic acid (10%). After centrifugation at 18,000 rcf for 10 min, supernatants were loaded onto solid-phase extraction cartridges (Oasis HLB cartridges, Waters Corp., Milford, MA). The cartridges were washed with 1.0 ml of H2O and then eluted with 1.0 ml of MeOH. The methanol fractions were dried and reconstituted with 100 μl of CH3CN/H2O (1:1). Aliquots (5 μl) of the reconstituted solutions were injected into a system combining UPLC and TOFMS for metabolite analysis. All incubations were conducted in quadruplicates.
Animals and treatments
Wild-type mice (male, 2–4 months old) were maintained under a standard 12-hr dark and 12-hr light cycle with water and chow provided ad libitum. Handling was in accordance with study protocols approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee. Mice were treated with vehicle or 300 mg/kg APAP (ip, n=4). Thirty min after treatment, livers were collected. Livers were homogenized in water (100 mg liver in 400 μl of H2O), and then 200 μl of acetonitrile was added to 200 μl of liver homogenates. After vortex and centrifugation at 18,000 rcf for 10 min, the supernatant was transferred to a clean Eppendoff vial for a second centrifugation (18,000 rcf for 10 min). Each supernatant was transferred to an auto sampler vial, and 5.0 μl was injected to a UPLC-TOFMS (Waters, Milford, MA) for metabolite analysis.
UPLC-TOFMS analysis
Metabolites were separated using a 100 mm × 2.1 mm (Acquity 1.7 μm) UPLC BEH C-18 column (Waters, Milford, MA). The flow rate of the mobile phase was 0.3 ml/min with a gradient ranging from 2% to 98% aqueous acetonitrile containing 0.1% formic acid in a 10-min run. TOFMS was operated in positive mode with electrospray ionization. The source temperature and desolvation temperature were set at 120 °C and 350 °C, respectively. Nitrogen was applied as cone gas (10 L/hour) and desolvation gas (700 L/hour). Argon was applied as collision gas. TOFMS was calibrated with sodium formate and monitored by the intermittent injection of lock mass leucine enkeophalin in real time. The capillary voltage and the cone voltage were set at 3.5 kV and 35 V in positive ion mode. The structures of the targeted metabolites were elucidated by mass fragmentation with a collision energy ramp ranging from 10 to 40 eV.
Data analysis
Mass chromatograms and mass spectra were acquired by MassLynx software in centroid format from m/z 50 to m/z 1000. Centroid and integrated mass chromatographic data were processed by MarkerLynx software to generate a multivariate data matrix. The mass window for processing of MarkerLynx was set as molecular weight (MW) of tested xenobiotic minus 100 Da to MW plus 400 Da. The corresponding data matrices were then exported into SIMCA-P+12 (Umetrics, Kinnelon, NJ) for MDA analysis. Principal component analysis (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA) were conducted on Pareto-scaled data. The applications of PCA and OPLS-DA in metabolite identification have been described in previous reports (26, 32). In the current study, all the control groups were combined to compare with the trapped group, and all detected ions were ranked based upon their relative abundance and the correlation to the model. Screening of the trapped reactive metabolites was performed based on ion ranking in the S-plot generated from OPLS-DA analysis.
RESULTS AND DISCUSSION
PLG bioactivation
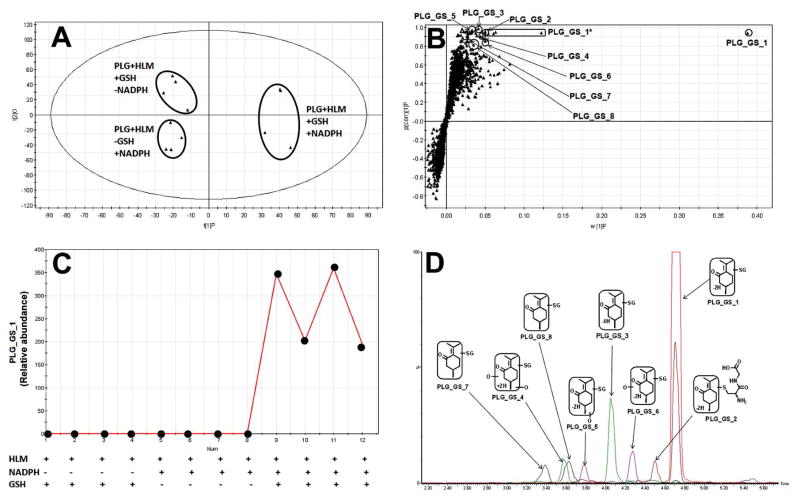
PLG is a naturally occurring organic compound present in a variety of plants such as nepeta cataria and mentha piperita (33). PLG-induced hepatotoxicity has been reported, and bioactivation plays a key role in this toxic event (34, 35). GSH was used to trap reactive metabolites of PLG. Three groups of incubation were conducted in the absence or presence of NADPH and GSH, and the groups without NADPH or GSH served as the controls. Supervised OPLS-DA analysis clearly separated the three groups (Figure 2A). The S-plot generated from OPLS-DA analysis displayed the ion contribution to the group separation. The top ranking ions were identified as GSH conjugated PLG metabolites (Figure 2B). Structures of these adducts were elucidated by MS/MS analyses (Supplemental Figure 1). A representative line score plot of PLG_GS_1 is presented in Figure 2C. In the control groups, PLG_GS_1 was not observed, indicating that its formation is NADPH and GSH dependent. The chromatograms of GSH conjugated PLG metabolites are displayed in Figure 2D. PLG_GS_1 is the most abundant adduct, and the abundances of other adducts are relatively low (Figure 2D). In a previous report, a similar number of GSH conjugated PLG metabolites were identified by using isotope-labeled trapping agents and UPLC-TOFMS coupled with an MDF technique (2). By comparison with that method, the major advantage of the metabolomic approach lies in its avoidance of the isotope-labeled trapping reagents.
Figure 2. GSH-trapped reactive metabolites of PLG.
The method of PLG incubation with HLM is detailed in Materials and Methods. All samples were analyzed by UPLC-TOFMS. (A) Separation of each incubation group in OPLS-DA score plot. The t[1]P and t[2]O values represent the score of each sample in principal component 1 and 2, respectively. (B) Loading S-plot generated by OPLS-DA analysis. The X-axis is a measure of the relative abundance of ions and the Y-axis is a measure of the correlation of each ion to the model. *, in-source fragment. (C) Line score plot of PLG_GS_1 generated by OPLS-DA analysis. (D) The chromatograms and structures of GSH conjugated PLG metabolites.
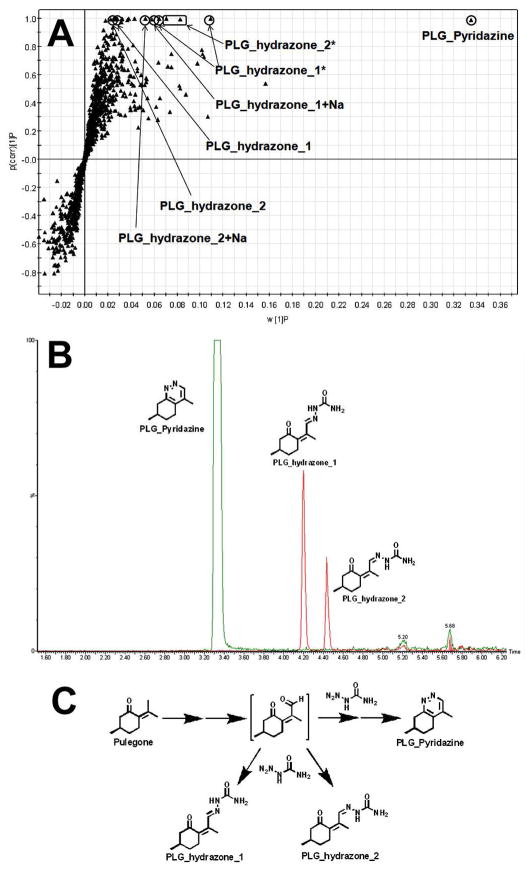
In the metabolomic analysis of the incubation samples with semicarbazide, the top ranking ions were identified as PLG-pyridazine and PLG_hydrazones (Figure 3A). Structures of these adducts were elucidated by MS/MS analyses (Supplemental Figure 2), and their chromatograms are shown in Figure 3B. The mechanism of these PLG adduct formations is proposed in Figure 3C. Semicarbazide is a commonly used trapping reagent to confirm aldehyde formation (16, 17). The identification of the adducts of PLG with semicarbazide indicates that aldehyde was formed in PLG metabolism. Aldehydes are commonly encountered reactive metabolites that are generally undetectable by MS. Semicarbazide and methoxylamine can be used to trap aldehydes (16, 17). However, screening for the trapped aldehydes is difficult, especially when the structures of the trapped aldehydes are significantly changed from the parent chemical, for example, PLG_pyridazine (Figure 3C). In the current study, we readily identified PLG-pyridazine by using a metabolomic approach. Our findings suggest that a metabolomic approach is a rational way to uncover unpredictable metabolites. A similar approach has been used in studying atazanavir bioactivation, and two aldehydes were identified (36).
Figure 3. Semicarbazide-trapped aldehyde of PLG.
The method of PLG incubation with HLM is detailed in Materials and Methods. All samples were analyzed by UPLC-TOFMS. (A) Loading S-plot generated by OPLS-DA analysis. The X-axis is a measure of the relative abundance of ions and the Y-axis is a measure of the correlation of each ion to the model. *, in-source fragment; +Na, sodium adduct. (B) The chromatograms and structures of PLG_Pyridazine and PLG_hydrazones. (C) The proposed mechanism of PLG_Pyridazine and PLG_hydrazone formation.
APAP bioactivation
APAP is an over-the-counter analgesic and antipyretic. Overdose of APAP causes serious liver injury. The role of CYP450-mediated bioactivation in APAP hepatotoxicity has been established (37). By using a metabolomic approach, we identified four GSH adducts of APAP in the incubation with HLM (Figure 4). Structures of these adducts were elucidated by MS/MS analyses (Figure 5). The chromatograms of these adducts are displayed in Figure 4B. APAP_GS_1 was the most abundant adduct, which is consistent with a previous report (23). APAP is a widely used chemical for studying bioactivation. Multiple trapped reactive metabolites of APAP have been reported (2, 23, 38). In the current study, a novel GSH-trapped reactive metabolite was characterized, which was APAP_GS_3, a deacetylated APAP+GS (Figure 4B). In addition, we analyzed same APAP incubation samples using the current method and MRM method (38). Four APAP adducts were uncovered in the metabolomic analysis, while MRM analysis only revealed the most abundant APAP adduct (Supplemental Figure 3). Previous studies have revealed that overdose of APAP depletes GSH in the liver (39–41). In the current study, mice were treated with APAP and liver extracts were analyzed using a metabolomic approach. APAP_GS_1 and GSH stood out and ranked the 1st among the increased and decreased ions, respectively, in the APAP-treated group (Figure 4C). The chromatograms of the APAP adducts detected in liver extracts are displayed in Figure 4D. This finding indicates that a metabolomic approach can be applied to both in vitro and in vivo analysis of xenobiotic bioactivation.
Figure 4. GSH-trapped reactive metabolites of APAP.
All samples were analyzed by UPLC-TOFMS. (A) Loading S-plot generated by OPLS-DA analysis of the incubation samples with HLM. The method of APAP incubation with HLM is detailed in Materials and Methods. The X-axis is a measure of the relative abundance of ions and the Y-axis is a measure of the correlation of each ion to the model. *, in-source fragment. (B) The chromatograms and structures of GSH conjugated APAP. (C) Loading S-plot generated by OPLS-DA analysis of the liver extracts. Mice were treated with vehicle or 300 mg/kg APAP (ip, n=4). Thirty min after treatment, livers were collected for metabolomic analysis. The inlaid figure presents the relative abundances of GSH and APAP_GS_1 in control and APAP-treated group, respectively. N.D., not detected. (D) The chromatograms of the APAP adducts detected in liver extracts of mice treated with APAP.
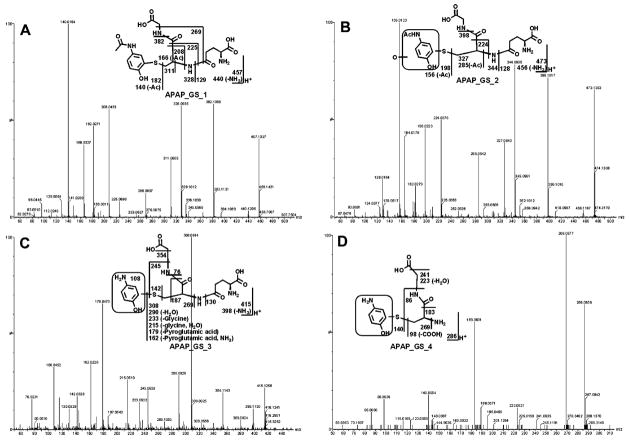
Figure 5. The MS/MS spectra of APAP_GS_1 to APAP_GS_4 and their structural elucidations.
The structures of metabolites were elucidated by mass fragmentation with a collision energy ramp ranging from 10 to 40 eV. (A) APAP_GS_1. APAP_GS_1 was eluted at 2.82 min, having a mass of [M+H]+ = 457 m/z. MS/MS analysis of APAP_GS_1 showed the fragment ions at m/z 440 (loss of NH3), 382 (loss of glycine unit), 328 (loss of pyroglutamic acid), 182, and 140. (B) APAP_GS_2. APAP_GS_2 was eluted at 2.72 min, having a mass of [M+H]+ = 473 m/z, 16 Dalton higher than that of APAP_GS_1. MS/MS analysis of APAP_GS_2 showed the ions at m/z 456 (loss of NH3), 398 (loss of glycine unit), 344 (loss of pyroglutamic acid), 198, and 156. Compared with the fragment ion at m/z 440, 382, 328, 182, and 140 generated from APAP_GS_1, the fragment ions of APAP_GS_2 suggested that monooxidation took place on this framed unit. (C) APAP_GS_3. APAP_GS_3 was eluted at 0.98 min and had a mass of [M+H]+ = 415 m/z, 42 mass units less than that of APAP_GS_1. MS/MS of APAP_GS_3 produced the fragment ions at 398 (loss of NH3), 308 (GSH), 142, and 108. The fragment ion at m/z 108 suggested that the acetyl group was lost in the framed unit. (D) APAP_GS_4. APAP_GS_4 (3.22 min) corresponded to a protonated molecule at m/z 286. MS/MS of APAP_GS_4 produced the fragment ions at m/z 269 (loss of NH3), 183, and 140. Compared with the fragment ion at m/z 182 generated in APAP_GS_1, the ion at m/z 140 suggested that deacetylation occurred in encircled unit.
CLP bioactivation
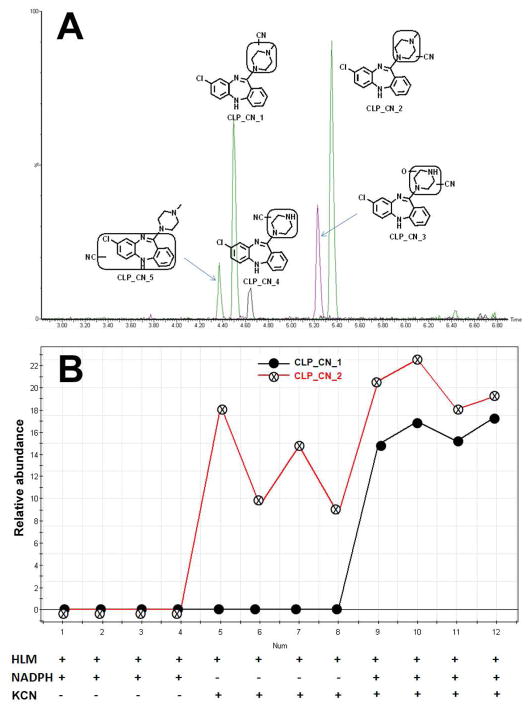
CLP is an antipsychotic used for the treatment of schizophrenia. CLP-induced liver injury is associated with its bioactivation (42, 43). Iminium ions are generated in CLP metabolism. Iminium ions are hard electrophiles that can be trapped by cyanide ions (20). Metabolomic analysis of the incubation samples of CLP with cyanide ions revealed five CN adducts, including two novel and three known adducts (2). The structures of these adducts were characterized by tandem MS/MS (Supplemental Figure 4). Their chromatograms are shown in Figure 6A. CLP_CN_1, 3, 4 and 5 are NADPH-dependent. CLP_CN_2 is NADPH-independent (Figure 6B), suggesting that this metabolic pathway is not mediated by CYP450. Except iminium ions, nitrenium ions are generated in CLP metabolism. Nitrenium ions can be trapped by GSH (2, 38). Metabolomic analysis of the incubation samples of CLP with GSH revealed five adducts. Their structures and chromatograms were present in Supplemental Figures 5 and 6.
Figure 6. Cyanide ion-trapped reactive metabolites of CLP.
The method of CLP incubation with HLM is detailed in Materials and Methods. All samples were analyzed by UPLC-TOFMS. (A) The chromatograms and structures of CN conjugated CLP metabolites. (B) The line score plots of CLP_CN_1 and CLP_CN_2.
CONCLUSIONS AND PERSPECTIVES
In the current study, we developed an unbiased approach for screening trapped reactive metabolites based upon metabolomic technologies. This method was validated using PLG, APAP, and CLP as model compounds. In all these cases, a large number of trapped reactive metabolites were readily identified (Supplementary Table 1), which suggests that the metabolomic approach is an effective tool for screening trapped reactive metabolites. This approach has several advantages: (1) it avoids the laborious process of predicting possible reactive metabolites and provides information on unexpected reactive metabolites; (2) it avoids the use of isotope-labeled compounds; and (3) it is not restricted to the GSH conjugated classes, but is also useful for other type of electrophiles, such as aldehydes and iminium ions. In the future studies on the role of reactive metabolites in toxicity, we propose (1) treatment with reactive metabolite(s) in mice or other animal models, (2) structural modification of a drug to selectively block the metabolic pathway(s) that generate reactive metabolite(s), and (3) genetic knockout or knockdown of the enzyme(s) that involve in the generation of reactive metabolite(s).
Supplementary Material
Acknowledgments
Funding support. This work was supported by the National Institutes of Health National Center for Research Resources [COBRE 5P20-RR021940].
We thank Dr. Martha Montello for editing the manuscript.
ABBREVIATIONS
- APAP
acetaminophen
- CLP
clozapine
- GSH
glutathione
- KCN
potassium cyanide
- CYP450
Cytochromes P450
- HLM
human liver microsomes
- PLG
pulegone
- MDA
multivariate data analysis
- MDF
mass defect filtering
- MRM
multiple reaction monitoring
- OPLS-DA
orthogonal projection to latent structures-discriminant analysis
- PCA
principal component analysis
- TOFMS
time-of-flight mass spectrometry
- UPLC
ultraperformance liquid chromatography
Footnotes
Supporting information available: Supplemental Table 1 and Figures 1–6.
References
- 1.Kumar GN, Surapaneni S. Role of drug metabolism in drug discovery and development. Med Res Rev. 2001;21:397–411. doi: 10.1002/med.1016. [DOI] [PubMed] [Google Scholar]
- 2.Rousu T, Pelkonen O, Tolonen A. Rapid detection and characterization of reactive drug metabolites in vitro using several isotope-labeled trapping agents and ultra-performance liquid chromatography/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:843–855. doi: 10.1002/rcm.3953. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien PJ, Siraki AG, Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit Rev Toxicol. 2005;35:609–662. doi: 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- 4.Wen B, Fitch WL. Analytical strategies for the screening and evaluation of chemically reactive drug metabolites. Expert Opin Drug Metab Toxicol. 2009;5:39–55. doi: 10.1517/17425250802665706. [DOI] [PubMed] [Google Scholar]
- 5.Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 6.Farmer PB, Brown K, Tompkins E, Emms VL, Jones DJ, Singh R, Phillips DH. DNA adducts: mass spectrometry methods and future prospects. Toxicol Appl Pharmacol. 2005;207:293–301. doi: 10.1016/j.taap.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 7.Koc H, Swenberg JA. Applications of mass spectrometry for quantitation of DNA adducts. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 8.Baillie TA. Future of toxicology-metabolic activation and drug design: challenges and opportunities in chemical toxicology. Chem Res Toxicol. 2006;19:889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- 9.Guengerich FP, MacDonald JS. Applying mechanisms of chemical toxicity to predict drug safety. Chem Res Toxicol. 2007;20:344–369. doi: 10.1021/tx600260a. [DOI] [PubMed] [Google Scholar]
- 10.Holt MP, Ju C. Mechanisms of drug-induced liver injury. AAPS J. 2006;8:E48–54. doi: 10.1208/aapsj080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang W, Lu AY. Metabolic bioactivation and drug-related adverse effects: current status and future directions from a pharmaceutical research perspective. Drug Metab Rev. 2010;42:225–249. doi: 10.3109/03602530903401658. [DOI] [PubMed] [Google Scholar]
- 12.Ma S, Subramanian R. Detecting and characterizing reactive metabolites by liquid chromatography/tandem mass spectrometry. J Mass Spectrom. 2006;41:1121–1139. doi: 10.1002/jms.1098. [DOI] [PubMed] [Google Scholar]
- 13.Leblanc A, Shiao TC, Roy R, Sleno L. Improved detection of reactive metabolites with a bromine-containing glutathione analog using mass defect and isotope pattern matching. Rapid Commun Mass Spectrom. 2010;24:1241–1250. doi: 10.1002/rcm.4507. [DOI] [PubMed] [Google Scholar]
- 14.Soglia JR, Contillo LG, Kalgutkar AS, Zhao S, Hop CE, Boyd JG, Cole MJ. A semiquantitative method for the determination of reactive metabolite conjugate levels in vitro utilizing liquid chromatography-tandem mass spectrometry and novel quaternary ammonium glutathione analogues. Chem Res Toxicol. 2006;19:480–490. doi: 10.1021/tx050303c. [DOI] [PubMed] [Google Scholar]
- 15.Gan J, Harper TW, Hsueh MM, Qu Q, Humphreys WG. Dansyl glutathione as a trapping agent for the quantitative estimation and identification of reactive metabolites. Chem Res Toxicol. 2005;18:896–903. doi: 10.1021/tx0496791. [DOI] [PubMed] [Google Scholar]
- 16.Zhang KE, Naue JA, Arison B, Vyas KP. Microsomal metabolism of the 5-lipoxygenase inhibitor L-739,010: evidence for furan bioactivation. Chem Res Toxicol. 1996;9:547–554. doi: 10.1021/tx950183g. [DOI] [PubMed] [Google Scholar]
- 17.Chauret N, Nicoll-Griffith D, Friesen R, Li C, Trimble L, Dube D, Fortin R, Girard Y, Yergey J. Microsomal metabolism of the 5-lipoxygenase inhibitors L-746,530 and L-739,010 to reactive intermediates that covalently bind to protein: the role of the 6,8-dioxabicyclo[3.2.1]octanyl moiety. Drug Metab Dispos. 1995;23:1325–1334. [PubMed] [Google Scholar]
- 18.Gorrod JW, Whittlesea CM, Lam SP. Trapping of reactive intermediates by incorporation of 14C-sodium cyanide during microsomal oxidation. Adv Exp Med Biol. 1991;283:657–664. doi: 10.1007/978-1-4684-5877-0_82. [DOI] [PubMed] [Google Scholar]
- 19.Kalgutkar AS, Dalvie DK, O’Donnell JP, Taylor TJ, Sahakian DC. On the diversity of oxidative bioactivation reactions on nitrogen-containing xenobiotics. Curr Drug Metab. 2002;3:379–424. doi: 10.2174/1389200023337360. [DOI] [PubMed] [Google Scholar]
- 20.Argoti D, Liang L, Conteh A, Chen L, Bershas D, Yu CP, Vouros P, Yang E. Cyanide trapping of iminium ion reactive intermediates followed by detection and structure identification using liquid chromatography-tandem mass spectrometry (LC-MS/MS) Chem Res Toxicol. 2005;18:1537–1544. doi: 10.1021/tx0501637. [DOI] [PubMed] [Google Scholar]
- 21.Wen B, Fitch WL. Screening and characterization of reactive metabolites using glutathione ethyl ester in combination with Q-trap mass spectrometry. J Mass Spectrom. 2009;44:90–100. doi: 10.1002/jms.1475. [DOI] [PubMed] [Google Scholar]
- 22.Dieckhaus CM, Fernandez-Metzler CL, King R, Krolikowski PH, Baillie TA. Negative ion tandem mass spectrometry for the detection of glutathione conjugates. Chem Res Toxicol. 2005;18:630–638. doi: 10.1021/tx049741u. [DOI] [PubMed] [Google Scholar]
- 23.Zheng J, Ma L, Xin B, Olah T, Humphreys WG, Zhu M. Screening and identification of GSH-trapped reactive metabolites using hybrid triple quadruple linear ion trap mass spectrometry. Chem Res Toxicol. 2007;20:757–766. doi: 10.1021/tx600277y. [DOI] [PubMed] [Google Scholar]
- 24.Zhu M, Ma L, Zhang H, Humphreys WG. Detection and structural characterization of glutathione-trapped reactive metabolites using liquid chromatography-high-resolution mass spectrometry and mass defect filtering. Anal Chem. 2007;79:8333–8341. doi: 10.1021/ac071119u. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindon JC, Holmes E, Nicholson JK. Metabonomics in pharmaceutical R&D. FEBS J. 2007;274:1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- 28.Schneider RP, Zhang H, Mu L, Kalgutkar AS, Bonner R. Utility of multivariate analysis in support of in vitro metabolite identification studies: retrospective analysis using the antidepressant drug nefazodone. Xenobiotica. 2010;40:262–274. doi: 10.3109/00498251003592691. [DOI] [PubMed] [Google Scholar]
- 29.Ma X, Chen C, Krausz KW, Idle JR, Gonzalez FJ. A metabolomic perspective of melatonin metabolism in the mouse. Endocrinology. 2008;149:1869–1879. doi: 10.1210/en.2007-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Wang L, Guo GL, Ma X. Metabolism-mediated drug interactions associated with ritonavir-boosted tipranavir in mice. Drug Metab Dispos. 38:871–878. doi: 10.1124/dmd.109.030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrer I, Thurman EM, Fernandez-Alba AR. Quantitation and accurate mass analysis of pesticides in vegetables by LC/TOF-MS. Anal Chem. 2005;77:2818–2825. doi: 10.1021/ac048458x. [DOI] [PubMed] [Google Scholar]
- 32.Patterson AD, Gonzalez FJ, Idle JR. Xenobiotic metabolism: a view through the metabolometer. Chem Res Toxicol. 2010;23:851–860. doi: 10.1021/tx100020p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grognet J. Catnip: Its uses and effects, past and present. Can Vet J. 1990;31:455–456. [PMC free article] [PubMed] [Google Scholar]
- 34.Madyastha KM, Moorthy B. Pulegone mediated hepatotoxicity: evidence for covalent binding of R(+)-[14C]pulegone to microsomal proteins in vitro. Chem Biol Interact. 1989;72:325–333. doi: 10.1016/0009-2797(89)90007-0. [DOI] [PubMed] [Google Scholar]
- 35.Thomassen D, Slattery JT, Nelson SD. Menthofuran-dependent and independent aspects of pulegone hepatotoxicity: roles of glutathione. J Pharmacol Exp Ther. 1990;253:567–572. [PubMed] [Google Scholar]
- 36.Li F, Lu J, Wang L, Ma X. CYP3A-Mediated Generation of Aldehyde and Hydrazine in Atazanavir Metabolism. Drug Metab Dispos. 2011;39:394–401. doi: 10.1124/dmd.110.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bessems JG, Vermeulen NP. Paracetamol (acetaminophen)-induced toxicity: molecular and biochemical mechanisms, analogues and protective approaches. Crit Rev Toxicol. 2001;31:55–138. doi: 10.1080/20014091111677. [DOI] [PubMed] [Google Scholar]
- 38.Wen B, Ma L, Nelson SD, Zhu M. High-throughput screening and characterization of reactive metabolites using polarity switching of hybrid triple quadrupole linear ion trap mass spectrometry. Anal Chem. 2008;80:1788–1799. doi: 10.1021/ac702232r. [DOI] [PubMed] [Google Scholar]
- 39.Han D, Hanawa N, Saberi B, Kaplowitz N. Mechanisms of liver injury. III. Role of glutathione redox status in liver injury. Am J Physiol Gastrointest Liver Physiol. 2006;291:G1–7. doi: 10.1152/ajpgi.00001.2006. [DOI] [PubMed] [Google Scholar]
- 40.Hassing JM, Rosenberg H, Stohs SJ. Acetaminophen-induced glutathione depletion in diabetic rats. Res Commun Chem Pathol Pharmacol. 1979;25:3–11. [PubMed] [Google Scholar]
- 41.Al-Turk WA, Stohs SJ. Hepatic glutathione content and aryl hydrocarbon hydroxylase activity of acetaminophen-treated mice as a function of age. Drug Chem Toxicol. 1981;4:37–48. doi: 10.3109/01480548109066370. [DOI] [PubMed] [Google Scholar]
- 42.Takakusa H, Masumoto H, Mitsuru A, Okazaki O, Sudo K. Markers of electrophilic stress caused by chemically reactive metabolites in human hepatocytes. Drug Metab Dispos. 2008;36:816–823. doi: 10.1124/dmd.107.018002. [DOI] [PubMed] [Google Scholar]
- 43.Pirmohamed M, Williams D, Madden S, Templeton E, Park BK. Metabolism and bioactivation of clozapine by human liver in vitro. J Pharmacol Exp Ther. 1995;272:984–990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.