HADH mutations are common in consanguineous pedigrees with diazoxide-responsive hyperinsulinaemichypoglycemia; therefore, genetic testing is recommended, even in the absence of abnormal fatty acid oxidation.
Abstract
Context and Objective:
Recessive mutations in the hydroxyacyl-CoA dehydrogenase (HADH) gene encoding the enzyme 3-hydroxyacyl-CoA dehydrogenase are a rare cause of diazoxide-responsive hyperinsulinemic hypoglycemia (HH) with just five probands reported to date. HADH deficiency in the first three identified patients was associated with detectable urinary 3-hydroxyglutarate and raised plasma 3-hydroxybutyryl-carnitine levels, but two recent cases did not have abnormal urine organic acids or acylcarnitines.
Research Design and Methods:
We studied 115 patients with diazoxide-responsive HH in whom the common genetic causes of HH had been excluded. No patients were reported to have abnormal acylcarnitines or urinary organic acids. Homozygosity mapping was undertaken in probands from 13 consanguineous pedigrees to search for regions harboring mutations that are identical by descent.
Results:
HADH sequencing was performed after genome-wide single nucleotide polymorphism analysis revealed a large shared region of homozygosity spanning the HADH locus in six unrelated probands. Homozygous mutations were identified in three of these patients and in a further two probands from consanguineous families. HADH analysis in the remainder of the cohort identified mutations in a further six probands for whom consanguinity was not reported, but who originated from countries with high rates of consanguinity. Six different HADH mutations were identified in 11/115 (10%) patients tested.
Conclusion:
HADH mutations are a relatively common cause of diazoxide-responsive HH with a frequency similar to that of GLUD1 and HNF4A mutations. We recommend that HADH sequence analysis is considered in all patients with diazoxide-responsive HH when recessive inheritance is suspected.
Hyperinsulinemic-hypoglycemia (HH), which is characterized by unregulated secretion of insulin despite a low blood glucose concentration, most commonly presents in the neonatal period with the phenotype ranging from mild to severe medically unresponsive hypoglycemia (1). Diazoxide targets the ATP-sensitive potassium (KATP) channel in the pancreatic β-cell and is often the first line of treatment. Patients who show a poor response to diazoxide therapy are likely to require a pancreatectomy.
Mutations in the ABCC8 and KCNJ11 genes, which encode the SUR1 and Kir6.2 subunits of the KATP channel, most often cause diazoxide-unresponsive HH but rare mutations in these and five other genes (GCK, GLUD1, HADH, HNF4A, and SLC16A1) have been reported in patients with diazoxide-responsive HH (1). Mutations in these genes are often associated with discrete phenotypes, for example GLUD1 mutations also result in hyperammonemia while SLC16A1 mutations cause exercise-induced hyperinsulinism (2, 3). While the clinical characteristics may guide the order of genetic testing, it should be noted that these genotype/phenotype relationships are not absolute. For example, recessive mutations in the hydroxyacyl-CoA dehydrogenase (HADH) gene were first identified in patients with specific fatty acid oxidation defects where urinary 3-hydroxyglutarate was present and plasma 3-hydroxybutyryl-carnitine levels were raised (4–6). However, two patients with homozygous HADH mutations but with normal acylcarnitines and urine organic acids have recently been reported (7, 8).
Recently, we demonstrated that a genetic diagnosis was possible for 27% of cases in our cohort with diazoxide-responsive HH (59/220 patients) (9). Mutations in KCNJ11, ABCC8, GCK, and HNF4A were excluded, but HADH was not sequenced because there was no report of any abnormality in the acylcarnitines and urine organic acids (9). Autozygosity analysis is a useful method for identifying novel genetic etiologies within consanguineous pedigrees through the identification of a genetic region harboring a mutation that is identical by descent (10). In the present study we have undertaken genome-wide single nucleotide polymorphism (SNP) analysis on a subset of unrelated consanguineous probands with diazoxide-responsive HH and no genetic diagnosis.
Materials and Methods
We studied 115 patients with diazoxide-responsive HH without mutations in ABCC8, KCNJ11, GCK, and HNF4A. Mutations in GLUD1 had been excluded in patients with hyperammonemia (n = 7).
Clinical data were provided via a standard request form (www.diabetesgenes.org) with diazoxide-responsiveness defined as the ability to come off intravenous glucose and maintain normoglycemia. No patients had required a pancreatectomy, and patients with evidence of perinatal asphyxia were excluded from the cohort. No cases were reported to have abnormalities in acylcarnitines and urine organic acids or exercise-induced hyperinsulinism. Consanguinity was reported in 18 probands. The study was conducted in accordance with the Declaration of Helsinki with informed parental consent given on behalf of children.
Homozygosity analysis
Genotyping was undertaken on the Affymetrix 6.0 SNP chip by Medical Solutions (Nottingham, UK), ALMAC Diagnostics (Craigavon, Northern Ireland), or Aros Applied Biotechnology (Aarhus, Denmark) for 13/18 consanguineous patients where there was sufficient DNA. Processing of genomic DNA was performed as per the Affymetrix protocol, and the mean SNP call rate was 98.8%. In-house Perl scripts were developed to automatically identify homozygous segments, defined by at least 20 consecutive homozygous SNPs marking a region that exceeded 3cM (11).
HADH analysis
In all 115 patients the 8 exons of HADH (NM_005327.2) were amplified and sequenced as previously described (7). When repeated failure of PCR indicated a homozygous deletion, break points were mapped by sequential PCR and sequencing. Patients with common mutations were further investigated by microsatellite markers (HADH flanking markers D4S2859 and D4S2945).
For patients where conventional sequencing failed to identify a mutation but SNP analysis revealed homozygosity over HADH, the possibility of a partial gene deletion or a mutation in the promoter, 3′ untranslated region (UTR) or alternative exons [as listed on AceView, accessed June 2010 (12)] was investigated. When a novel variant was identified in an alternative exon the presence/absence of the variant transcript in control islets, whole pancreas, liver, and blood was determined by real-time PCR (RT-PCR) and sequencing (primers available on request).
Results
Genetic results
Consanguineous cohort
Genome-wide SNP data were obtained for 13/18 consanguineous probands. Six of the 13 patients shared a region of homozygosity (3.3 Mb) on chromosome 4q25, which contained 21 genes including HADH (http://genome.ucsc.edu/). No further regions of homozygosity shared by four or more individuals were identified.
HADH sequencing identified mutations in 3/6 patients with homozygous regions encompassing HADH. Homozygous mutations were also present in 2/5 patients where insufficient DNA was available for SNP analysis (Table 1). No mutations were identified in the seven patients without homozygous regions spanning HADH. Three different HADH mutations were identified; two novel mutations, Q163X and K136E (each in a single patient), and the previously reported Q236X mutation (8) in three probands. When DNA was available, mutation testing confirmed the carrier status of the unaffected parents. None of the probands had a sibling affected with HH. The K136E mutation is likely to be pathogenic as in silico analysis suggests that it is detrimental to protein function (http://neurocore.charite.de/MutationTaster/), the mutated residue is highly conserved across species, and the variant has not been identified in 362 control chromosomes (http://www.1000genomes.org June 2010). For the three probands with homozygosity over HADH but no coding mutation, dosage analysis, and sequencing of the HADH promoter, 3′UTR and alternative exons was undertaken but no mutations were identified.
Table 1.
Clinical characteristics of patients with HADH mutations
| Patient | Gender | Age at diagnosis | Birth weight (gestation) | Current age (y) | Current diazoxide dose (mg/kg/d) | Reported Consanguinity | Country of origin | Mutation detection method | Mutation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 16 wks | 2.9 kg | 3 | 5 | Yes | Turkey | Homozygosity analysis, then sequencing | K136E/K136E |
| (38 wks) | (c.406A>G) | ||||||||
| 2 | Male | 16 wks | 2.8 kg | 8 | 2 | Yes | Turkey | Homozygosity analysis, then sequencing | Q163X/Q163X |
| (40 wks) | (c.487C>T) | ||||||||
| 3 | Male | 2 wks | 4.1 kg | 2 | 10 | Yes | Turkey | Homozygosity analysis, then sequencing | R236X/R236X |
| (40 wks) | (c.706C>T) | ||||||||
| 4 | Male | 5 days | 4.35 kg | 2 | 10 | Yes | Turkey | Sequence analysis | R236X/R236X |
| (40 wks) | (c.706C>T) | ||||||||
| 5 | Female | 1 wk | 4.0 kg | 7 | 2.5 | No | Turkey | Sequence analysis | R236X/R236X |
| (40 wks) | (c.706C>T) | ||||||||
| 6 | Male | 2 days | 3.2 kg | 1 | 11 | Yes | Pakistan | Sequence analysis | R236X/R236X |
| (39 wks) | (c.706C>T) | ||||||||
| 7 | Female | 12 wks | 3.5 kg | 2 | 15 | No | Iran | Sequence analysis | R236X/R236X |
| (40 wks) | (c.706C>T) | ||||||||
| 8 | Female | 1 day | 3.7 kg | 4 | 8 | No | Iran | Sequence analysis | R236X/R236X |
| (40 wks) | (c.706C>T) | ||||||||
| 9 | Male | 26 wks | 2.95 kg | 2 | 10 | No | India | Sequence analysis | K95SfsX3/IVS6 + 39C>G |
| (40 wks) | (c.283_293delinsT/c.709 + 39C>G) | ||||||||
| 10 | Male | 2 days | 4.0 kg | 2 | 10 | No | India | Long range PCR and sequencing of breakpoints | Ex1del/Ex1del |
| (40 wks) | (c.1-3440_132 + 1943del/c.1-3440_132 + 1943del) | ||||||||
| 11 | Male | 24 wks | 3.8 kg | 3 | 10 | No | India | Long range PCR and sequencing of breakpoints | Ex1del/Ex1del |
| (40 wks) | (c.1-3440_132 + 1943del/c.1-3440_132 + 1943del) |
Mutation nomenclature corresponds to HADH sequence accession NM_005327.2.
Nonconsanguineous cohort
After the identification of mutations in 5/18 (28%) consanguineous patients, HADH sequencing was undertaken in the remainder of the cohort and mutations were identified in 6/97 probands. Three probands were homozygous for the R236X mutation, and a failure to amplify exon 1 by PCR in two probands suggested the presence of a homozygous deletion (Table 1). Mapping of the break points confirmed an identical deletion, which included the minimal promoter and exon 1 (c.1-3440_132 + 1943del). When DNA was available carrier status was confirmed in the unaffected parents. None of the probands had a sibling affected with HH.
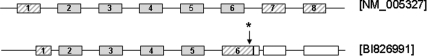
In the sixth patient a maternally inherited frame shift mutation, K95Sfs, was identified. Although a second mutation was not found by analysis of the reference sequence, promoter, 3′UTR, or by dosage studies, a novel paternally inherited variant, c.709 + 39C>G (NM_005327.2), which results in a L254V mutation in exon 6 of an alternative splice variant (cDNA accession number BI826991) (Fig. 1, Table 1), was identified. RT-PCR and sequencing confirmed the presence of this variant transcript in normal pancreas, islets, liver, and blood (data not shown). The c.709 + 39C>G variant was not identified in 362 control chromosomes (http://www.1000genomes.org June 2010).
Fig. 1.
Schematic representation of the full-length HADH gene (Reference sequence NM_005327) and a variant transcript (cDNA accession number B1826991). Exons are represented by boxes, and introns are denoted by lines. Hatched boxes represent coding regions which differ between the two transcripts, and white boxes represent UTRs. An arrow with a star points to the approximate position of the novel variant, c.709 + 39C>G (nomenclature relating to RefSeq NM_005327) identified in patient 9.
Haplotype analysis
Microsatellite analysis suggested the existence of two R236X founder mutations within the Iranian and Turkish/Pakistani populations. The exon 1 deletion was present on a common haplotype, consistent with a common ancestor within the Indian population (Table 1).
Clinical characteristics
The median age at diagnosis of HH for patients with HADH mutations was 7 weeks (range 1 d to 26 weeks) and the median birth weight was 3.6 kg at 40 weeks gestation (range 2.8–4.35 kg) (Table 1). No patients were reported to have defects in fatty acid oxidation, although acylcarnitine profiles and urine organic acid screens were not complete for all patients.
Discussion
HADH sequencing was undertaken after SNP analysis revealed a large shared region of homozygosity spanning the HADH locus in six unrelated consanguineous probands. We identified mutations in three of these patients and in a further two probands from known consanguineous families (28% positive). These results prompted us to sequence HADH in the remainder of our cohort, and homozygous mutations were identified in a further five probands who were not reported as being consanguineous. However, these patients were all referred from countries with high rates of consanguineous marriages. Only one patient with compound heterozygous mutations was identified. In total, HADH mutations were identified in 11/115 (10%) patients with diazoxide-responsive HH of unknown etiology. Our study increases the number of patients reported with HADH HH from five probands (4–8) to 16.
In one patient with a maternally inherited frame shift mutation, extensive studies to search for a second mutation of paternal origin were undertaken and a novel variant was found in intron 6 (corresponding to RefSeq NM_005327.2). In silico splicing prediction programs suggest that this variant will not alter splicing. However, at least eight transcript variants are predicted to exist for HADH, and this substitution results in a missense mutation in exon 6 of a variant transcript (cDNA accession number BI826991) (12) (Fig. 1), which is predicted to encode a protein with an NAD-binding and a C-terminal domain. Although further studies are required to assess the significance of this transcript, its detection in tissues including pancreas and islets suggests that it may be important. While the pathogenicity of the variant is currently unproven, the identification of a frame shift mutation on the opposite allele is consistent with a diagnosis of recessively inherited HH resulting from a HADH mutation.
In keeping with previous reports a range in birth weights and ages at diagnosis of HH were observed in the patients with HADH mutations (5). Interestingly, none of the patients were reported to have abnormalities in plasma acylcarnitines or urine organic acids, a phenotype reported in 3/5 published probands. While the absence of abnormal acylcarnitines and urine organic acids in these patients may be attributable to limitations in laboratory analysis, it is possible that the phenotype is mutation-dependent because none of the mutations identified in this cohort have been found in individuals with abnormal acylcarnitines and urine organic acids. It is unlikely, however, that the disease spectrum reflects the severity of the mutation as the majority of patients with isolated HH have null mutations.
A number of studies have demonstrated that HADH has a pivotal role in regulating insulin secretion (13–16). Most recently a study by Li et al. (15) examined the mechanism of insulin dysregulation in mice with a knock-out of the hadh gene. Pull-down experiments demonstrated protein–protein interactions between HADH and glutamate dehydrogenase (GDH), and studies on isolated islets showed in increased in the affinity of GDH for its substrate α-ketoglutarate. It is therefore likely that HADH mutations cause HH by activation of GDH via loss of inhibitory regulation of GDH by HADH. This finding is of particular interest as activating mutations in GLUD1, which encodes GDH, are a common cause of hyperinsulinism and hyperammonemia (2).
In conclusion we have shown that mutations in HADH account for 10% of cases with diazoxide-responsive HH without a mutation in the known genes. This study takes the number of HADH mutations identified in our cohort to 12 (7), with a prevalence similar to that for HH attributable to HNF4A or GLUD1 (17) mutations. We recommend that analysis of the HADH gene is considered in all patients with diazoxide-responsive HH who originate from known consanguineous pedigrees, isolated populations, or countries where inbreeding is frequent, regardless of whether there is evidence of abnormal fatty acid oxidation.
Acknowledgments
We thank Annet Damhuis and Andrew Parrish for their technical assistance. S.E.F. is the Sir Graham Wilkins Peninsula Medical School research fellow. S.E. is a member of the core staff within the National Institute for Health Research funded Peninsula Clinical Research Facility.
This work was supported by the Wellcome Trust (081188/A/06/Z).
Disclosure Summary: The authors have nothing to declare.
For editorial see page 617
- GDH
- Glutamate dehydrogenase
- HADH
- hydroxyacyl-CoA dehydrogenase
- HH
- hyperinsulinemic-hypoglycemia
- SNP
- single nucleotide polymorphism
- UTR
- untranslated region.
References
- 1. Kapoor RR, Flanagan SE, James C, Shield J, Ellard S, Hussain K. 2009. Hyperinsulinaemic hypoglycaemia. Arch Dis Child 94:450–457 [DOI] [PubMed] [Google Scholar]
- 2. Stanley CA, Lieu YK, Hsu BY, Burlina AB, Greenberg CR, Hopwood NJ, Perlman K, Rich BH, Zammarchi E, Poncz M. 1998. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med 338:1352–1357 [DOI] [PubMed] [Google Scholar]
- 3. Otonkoski T, Jiao H, Kaminen-Ahola N, Tapia-Paez I, Ullah MS, Parton LE, Schuit F, Quintens R, Sipila I, Mayatepek E, Meissner T, Halestrap AP, Rutter GA, Kere J. 2007. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am J Hum Genet 81:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE. 2001. Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest 108:457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Molven A, Matre GE, Duran M, Wanders RJ, Rishaug U, Njolstad PR, Jellum E, Sovik O. 2004. Familial hyperinsulinemic hypoglycemia caused by a defect in the SCHAD enzyme of mitochondrial fatty acid oxidation. Diabetes 53:221–227 [DOI] [PubMed] [Google Scholar]
- 6. Hussain K, Clayton PT, Krywawych S, Chatziandreou I, Mills P, Ginbey DW, Geboers AJ, Berger R, van den Berg IE, Eaton S. 2005. Hyperinsulinism of infancy associated with a novel splice site mutation in the SCHAD gene. J Pediatr 146:706–708 [DOI] [PubMed] [Google Scholar]
- 7. Kapoor RR, James C, Flanagan SE, Ellard S, Eaton S, Hussain K. 2009. 3-Hydroxyacyl-coenzyme A dehydrogenase deficiency and hyperinsulinemic hypoglycemia: characterization of a novel mutation and severe dietary protein sensitivity. J Clin Endocrinol Metab 94:2221–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Candia S, Gessi A, Pepe G, Sogno Valin P, Mangano E, Chiumello G, Gianolli L, Proverbio MC, Mora S. 2009. Identification of a diffuse form of hyperinsulinemic hypoglycemia by 18-fluoro-L-3,4 dihydroxyphenylalanine positron emission tomography/CT in a patient carrying a novel mutation of the HADH gene. Eur J Endocrinol 160:1019–1023 [DOI] [PubMed] [Google Scholar]
- 9. Flanagan SE, Kapoor RR, Mali G, Cody D, Murphy N, Schwahn B, Siahanidou T, Banerjee I, Akcay T, Rubio-Cabezas O, Shield JP, Hussain K, Ellard S. 2010. Diazoxide-responsive hyperinsulinemic hypoglycemia caused by HNF4A gene mutations. Eur J Endocrinol 162:987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lander ES, Botstein D. 1987. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- 11. Woods CG, Cox J, Springell K, Hampshire DJ, Mohamed MD, McKibbin M, Stern R, Raymond FL, Sandford R, Malik Sharif S, Karbani G, Ahmed M, Bond J, Clayton D, Inglehearn CF. 2006. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet 78:889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thierry-Mieg D, Thierry-Mieg J. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7(Suppl 1):S12.1–S12.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martens GA, Vervoort A, Van de Casteele M, Stange G, Hellemans K, Van Thi HV, Schuit F, Pipeleers D. 2007. Specificity in beta cell expression of L-3-hydroxyacyl-CoA dehydrogenase, short chain, and potential role in down-regulating insulin release. J Biol Chem 282:21134–21144 [DOI] [PubMed] [Google Scholar]
- 14. Hardy OT, Hohmeier HE, Becker TC, Manduchi E, Doliba NM, Gupta RK, White P, Stoeckert CJ, Jr, Matschinsky FM, Newgard CB, Kaestner KH. 2007. Functional genomics of the beta-cell: short-chain 3-hydroxyacyl-coenzyme A dehydrogenase regulates insulin secretion independent of K+ currents. Mol Endocrinol 21:765–773 [DOI] [PubMed] [Google Scholar]
- 15. Li C, Chen P, Palladino A, Narayan S, Russell LK, Sayed S, Xiong G, Chen J, Stokes D, Butt YM, Jones PM, Collins HW, Cohen NA, Cohen AS, Nissim I, Smith TJ, Strauss AW, Matschinsky FM, Bennett MJ, Stanley CA. 2010. Mechanism of hyperinsulinism in short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency involves activation of glutamate dehydrogenase. J Biol Chem 285:31806–31818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filling C, Keller B, Hirschberg D, Marschall HU, Jornvall H, Bennett MJ, Oppermann U. 2008. Role of short-chain hydroxyacyl CoA dehydrogenases in SCHAD deficiency. Biochem Biophys Res Commun 368:6–11 [DOI] [PubMed] [Google Scholar]
- 17. Kapoor RR, Flanagan SE, Fulton P, Chakrapani A, Chadefaux B, Ben-Omran T, Banerjee I, Shield JP, Ellard S, Hussain K. 2009. Hyperinsulinism-hyperammonaemia syndrome: novel mutations in the GLUD1 gene and genotype-phenotype correlations. Eur J Endocrinol 161:731–735 [DOI] [PMC free article] [PubMed] [Google Scholar]