Abstract
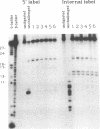
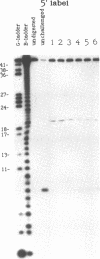
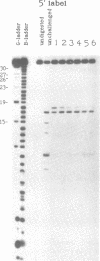
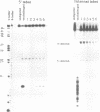
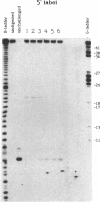
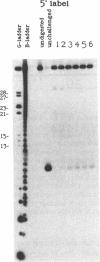
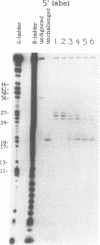
We examined the ribonuclease H (RNase H) specificity of human immunodeficiency virus reverse transcriptase (HIV-RT) using heteropolymeric RNAs hybridized to complementary DNAs. Experiments were performed in the presence of excess challenger polymer (poly(rA)-oligo(dT)) to reveal cleavages resulting from single enzyme binding events. Previous results suggested that initial RNase H directed cleavages were a fixed distance from a DNA primer terminus recessed on an RNA template, i.e. determined by the binding position of the polymerase active site. The influences of recessed RNA termini were not evaluated. In current experiments, RNAs that were 30, 42, or 50 nucleotides long were hybridized to the same 88 nucleotide long complementary DNA, such that the 5' terminal nucleotide of each RNA was hybridized to the 29th nucleotide from the 3' end of the DNA. In all three cases the RNA was initially cleaved between the 19th and 21st nucleotides from its 5' end. Thus, cleavage was not coordinated by the recessed 3' terminus of the RNA. Subsequent cleavages in either direction on the RNA were also observed. An insertion within the RNA that moved the preferred initial cut sequence 10 nucleotides further from the 5' end of the RNA decreased but did not abolish cleavage at the sequence. However, changing the nucleotide sequence in the region of the preferred cleavage either by the insertion experiment or mutagenesis did not significantly alter its capacity for cleavage. These results demonstrated a dominant position preference, plus a sequence priority. In another experiment, a 25 nucleotide long DNA was hybridized such that its 3' terminal nucleotide was 9 nucleotides from the 5' end of a 60 nucleotide complementary RNA. The preferred RNA cleavage sequence discussed above, was 10-14 nucleotides upstream of the 3' end of the DNA. However, initial cleavages occurred 17-20 nucleotides from the DNA 3' end, consistent with cleavage being coordinated by the recessed 3' terminus of the DNA primer.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Artzi H., Zeelon E., Gorecki M., Panet A. Double-stranded RNA-dependent RNase activity associated with human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):927–931. doi: 10.1073/pnas.89.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Gilboa E., Baltimore D. Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J Virol. 1984 Mar;49(3):686–691. doi: 10.1128/jvi.49.3.686-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J. J., Buiser R. G., Mallaber L. M., Bambara R. A., Fay P. J. Human immunodeficiency virus reverse transcriptase displays a partially processive 3' to 5' endonuclease activity. J Biol Chem. 1991 Dec 25;266(36):24295–24301. [PubMed] [Google Scholar]
- DeStefano J. J., Buiser R. G., Mallaber L. M., Myers T. W., Bambara R. A., Fay P. J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991 Apr 25;266(12):7423–7431. [PubMed] [Google Scholar]
- Finston W. I., Champoux J. J. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J Virol. 1984 Jul;51(1):26–33. doi: 10.1128/jvi.51.1.26-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T. B., Taylor J. When retroviral reverse transcriptases reach the end of their RNA templates. J Virol. 1992 Jul;66(7):4271–4278. doi: 10.1128/jvi.66.7.4271-4278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furfine E. S., Reardon J. E. Reverse transcriptase.RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991 Jan 5;266(1):406–412. [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Gopalakrishnan V., Peliska J. A., Benkovic S. J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Moelling K. RNase H activity associated with bacterially expressed reverse transcriptase of human T-cell lymphotropic virus III/lymphadenopathy-associated virus. J Biol Chem. 1987 Sep 15;262(26):12393–12396. [PubMed] [Google Scholar]
- Huber H. E., Richardson C. C. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990 Jun 25;265(18):10565–10573. [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Resnick R., Omer C. A., Faras A. J. Involvement of retrovirus reverse transcriptase-associated RNase H in the initiation of strong-stop (+) DNA synthesis and the generation of the long terminal repeat. J Virol. 1984 Sep;51(3):813–821. doi: 10.1128/jvi.51.3.813-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. K., Cywinski A., Taylor J. M. Initiation of plus-strand DNA synthesis during reverse transcription of an avian retrovirus genome. J Virol. 1984 Jan;49(1):200–204. doi: 10.1128/jvi.49.1.200-204.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. K., Cywinski A., Taylor J. M. Specificity of initiation of plus-strand DNA by Rous sarcoma virus. J Virol. 1984 Nov;52(2):314–319. doi: 10.1128/jvi.52.2.314-319.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnes M. C., Cheng Y. C. Human immunodeficiency virus reverse transcriptase-associated RNase H activity. J Biol Chem. 1989 Apr 25;264(12):7073–7077. [PubMed] [Google Scholar]
- Watson K. F., Schendel P. L., Rosok M. J., Ramsey L. R. Model RNA-directed DNA synthesis by avian myeloblastosis virus DNA polymerase and its associated RNase H. Biochemistry. 1979 Jul 24;18(15):3210–3219. doi: 10.1021/bi00582a004. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990 Nov 6;29(44):10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Volkmann S., Moelling K. Mutations of a conserved residue within HIV-1 ribonuclease H affect its exo- and endonuclease activities. J Mol Biol. 1991 Aug 5;220(3):801–818. doi: 10.1016/0022-2836(91)90119-q. [DOI] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]