Abstract
To compare hairy cell leukemia (HCL) with chronic lymphocytic leukemia (CLL) and normal B cells with respect to their B-cell receptors, somatic hypermutation (SHM) features in HCL were examined in a series of 130 immunoglobulin gene heavy chain rearrangements, including 102 from 100 classic (HCLc) and 28 from 26 variant (HCLv) patients. The frequency of unmutated rearrangements in HCLc was much lower than that in HCLv (17% vs 54%, P < .001) or historically in CLL (17% vs 46%, P < .001), but HCLv and CLL were similar (P = .45). As previously reported for CLL, evidence of canonical SHM was observed in HCLc rearrangements, including: (1) a higher ratio of replacement to silent mutations in the complementarity determining regions than in the framework regions (2.83 vs 1.41, P < .001), (2) higher transition to transversion ratio than would be expected if mutations were random (1.49 vs 0.5, P < .001), and (3) higher than expected concentration of mutations within RGYW hot spots (13.92% vs 3.33%, P < .001). HCLv met these 3 criteria of canonical SHM to a lesser extent. These data suggest that, whereas HCLc cells may recognize antigen-like CLL and normal B cells before malignant transformation, HCLv cells from some patients may originate differently, possibly without undergoing antigen recognition.
Introduction
The status of the B-cell receptor on B-cell leukemias, encoded as on normal B cells by rearranged immunoglobulin genes, is appearing increasingly important with regard to somatic mutations.1–3 For example, in B-cell chronic lymphocytic leukemia (CLL), an unmutated immunoglobulin gene heavy chain variable domain (IGHV), defined as more than 98% homologous to the germline sequence, is a more important prognostic variable than IGHV gene usage.4 In addition to prognostic value, the pattern of somatic hypermutation (SHM) has been studied to explore the development of the B-cell before its malignant transformation. Because normal B cells undergo SHM as a response to antigen, classically within the germinal center, patterns of SHM are thought to provide clues as to the type and likelihood of antigen recognition by the malignant B cell before leukemogenesis. Nonrandom, potentially antigen-driven SHM has been described in B-CLL as well as normal B cells and has been called canonical SHM.5,6 Features of canonical SHM include (1) increased ratio of mutations that change (replace) versus those that do not change (silent) the amino acid sequence (replacement-to-silent [R/S]), particularly in the complementarity determining regions (CDRs) as opposed to framework regions (FRs), (2) increased ratio of transitions (purine to purine or pyrimidine to pyrimidine) to transversions, and (3) more than the expected percentage of mutations in the RGYW hot spots relative to the total number of nucleotides located within these hot spots.7
Hairy cell leukemia (HCL) is also a chronic B-cell leukemia that is composed of only 2% of new cases of leukemia8 but differs greatly from CLL in its morphology, antigen expression, and response to treatment.9 As opposed to the reported CLL studies of up to 1939 patients each,6 studies of mutation status in HCL have contained 5 to 82 patients each10–18 and have included IGHV, IGHD, and IGHJ gene usage and homology to the closest germline sequence, but not characterization of the SHMs. A recent study of 105 untreated HCL patients identified several with multiple light chains.19 In the present study, we were able to study 130 rearrangements in 126 HCL patients, including the 82 patients previously published11,12 where SHMs were not characterized. The 1481 total mutations analyzed allowed us to determine whether SHMs are canonical as in CLL and to detect differences in the pattern of mutations between classic HCL (HCLc) and variant HCL (HCLv), and using historical data, differences between these 2 disorders and CLL.
Methods
Patients and controls
Blood for DNA study was obtained as part of sample acquisition protocols with informed consent approved by the National Cancer Institute Investigator's Review Board in accordance with the Declaration of Helsinki. All samples were retrieved between 2001 and 2010. Of the 130 rearrangements in 126 patients examined, 85 rearrangements in 82 patients were published previously.11,12 All except 12 (10%) of the patients were drawn at the time of relapse from one or more courses of purine analog. Differentiation of HCLc versus HCLv was performed by morphology and flow cytometry as described previously.12,20 Specifically, diagnosis of HCLv was based on the accepted World Health Organization guidelines, which require cytohematologic features (leukocytosis, monocytes, prominent nucleoli, blastic/convoluted nuclei, and/or absence of circumferential shaggy contours), variant immunophenotype (absence of CD25, annexin-A1, or TRAP), and resistance to conventional HCL therapy.21 SHM data for CLL patients discussed herein were reported by other investigators.5,6
IGHVDJ rearrangement sequencing
Total RNA was extracted from peripheral blood drawn into PAXgene tubes (PreAnalytiX) using a PAXgene RNA purification kit (PreAnalytiX) or from CD11c-sorted cells using the MagMax kit (Ambion). First-strand cDNA synthesis was performed by SuperScript III RnaseH-Reverse Transcriptase (Invitrogen) from 1 to 3 μg of total RNA in the reaction mix in the presence of Oligo(dT)20 primer (Invitrogen) and 10mM dNTP mix according to the manufacturer's protocol as described previously.12 Polymerase chain reaction amplification was performed in a single multiplexed reaction consisting of 6 IGHV FR1 primers combined with one IGHJ consensus primer as designed for the BIOMED-2 study.22 All reactions were carried out in 50 μL containing 10 pmol of each primer (InVivoScribe Technologies) using AmpliTaq Gold PCR Master Mix (Applied Biosystems) in the following conditions: denaturation at 95°C for 7 minutes followed by 35 cycles of denaturation at 95°C for 1 minute, annealing at 55°C for 30 seconds, and elongation at 72°C for 1 minute, and finally an elongation step at 72°C for 10 minutes. As reported previously,12 a monoclonal sequence was defined when at least 5 of 10 clones had a completely identical sequence. CD11c sorting was used when the PAXgene tube contained too much contamination from normal B cells to result in a monoclonal peak by CDR3 spectratyping or a monoclonal sequence after cloning.
Cell purification using CD11c sorting
Peripheral blood mononuclear cells were purified from 25 mL of heparin-containing peripheral blood of HCL patients by Ficoll-Paque Plus (GE Healthcare), using the manufacturer's protocol and counted. The peripheral B-cell population was isolated by the Dynal B-cell-negative isolation kit (InVitrogen) and incubated with 20 μL of Fc receptor blocking reagent (Miltenyi Biotec) during 10 minutes at 4°C. A total of 20 μL of CD11c-biotin antibody (Miltenyi Biotec) was added to the sample and incubated at 4°C for 10 minutes. Cells were washed by adding 4 mL of MACS solution (Miltenyi Biotec) and incubated with Streptavidin MicroBeads (Miltenyi Biotec) for 10 minutes at 4°C followed by additional washing by 4 mL of magnetic-activated cell sorting and centrifugation at 300g for 7 minutes at 4°C. The CD11c+ population was captured using 25MS MACS separation columns (Miltenyi Biotec) by the manufacturer's instructions.
Sequence analysis
The closest germline IGHV for each HCL IGHV sequence was assigned using the International ImMunoGeneTics database (www.imgt.org). This database also aligned each sequenced rearrangement in terms of FRs and CDRs. Nucleotide differences between the rearrangement and its closest germline sequence were considered as mutations. For the purposes of counting SHMs, the IGHV region was considered as FR1, CDR1, FR2, CDR2, and FR3.
Statistics
Comparisons between groups of patients were performed by the Fisher exact test or by the Wilcoxon rank-sum test as appropriate.
Results
IGHV repertoire
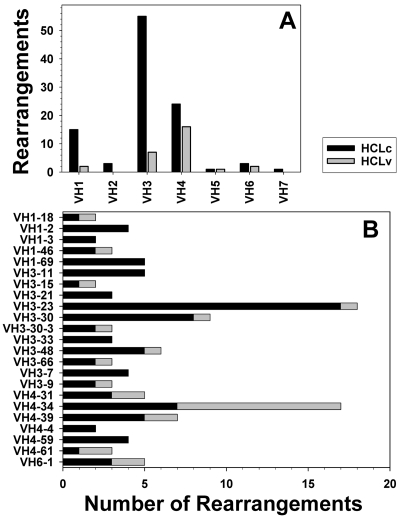
A total of 130 IGHV-D-J sequences were analyzed from 126 patients with HCL, including 100 HCLc and 26 HCLv patients. Two HCLc and 2 HCLv patients each had 2 in-frame rearrangements. As shown in Figure 1A, IGHV3-type variable heavy chains were most commonly used (n = 62) followed by IGHV4 (n = 40) and IGHV1 (n = 17). IGHV3-type variable heavy chains were more common in HCLc than HCLv (54% vs 25%, P = .01), and IGHV4 was more common in HCLv than in HCLc (57% vs 24%, P = .001). A total of 35 different IGHV genes were cloned. More than half (23 of 35) of these occurred in more than 1 patient, and their frequencies are shown in Figure 1B. The most frequently used IGHV genes in HCLc were IGHV3-23 (n = 17, 17%), followed by IGHV3-30 (n = 8, 8%), IGHV4-34 (n = 7, 7%), and IGHV1-69, IGHV3-11, IGHV3-48, and IGHV4-39 (each n = 5, 5%). In HCLv, the most frequently used IGHV gene was IGHV4-34 (n = 10, 36%). Even though IGHV4-34 was the third most common HCLc IGHV gene, the largest difference between HCLc and HCLv rearrangements was observed with IGHV4-34 (7% vs 36%, P < .001).
Figure 1.
IGHV family and gene usage in HCL. Number of IGHV rearrangements in each family (A) and for genes used by more than one patient (B) for HCLc (black) and HCLv (gray).
Structure of HCL V-D-J rearrangement
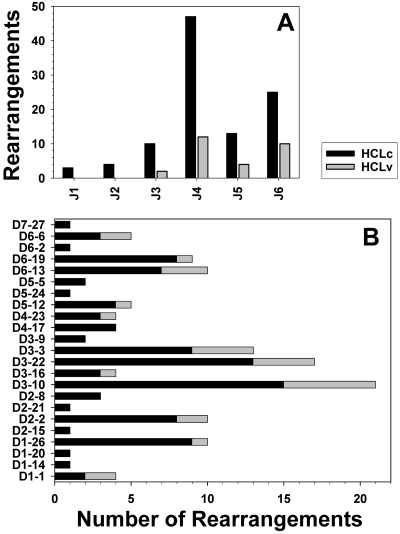
To characterize the CDR3 regions in HCL, the distribution and combination of different IGHJ and IGHD genes in the rearrangements were studied. The IGHJ4 gene followed by IGHJ6 and IGHJ5 were the most commonly used in HCLc and HCLv (Figure 2A). The distribution of IGHJ genes was not significantly different between HCLc and HCLv, or between HCL and normal CD5− B cells.22 As shown in Figure 2B, the most frequently used IGHD gene in both HCLc and HCLv was D3-10, followed by IGHD3-22, IGHD1-26, and IGHD3-3 for HCLc, and then IGHD3-22 and IGHD3-3 for HCLv. No significant differences between HCLv and HCLc were observed with respect to IGHD- or IGHJ-gene usage.
Figure 2.
IGHJ family and gene usage in HCL. IGHJ genes (A) and IGHD gene usage (B) in immunoglobulin heavy chain rearrangements derived from HCLc (black) and HCLv (gray) patients.
IGH mutation status
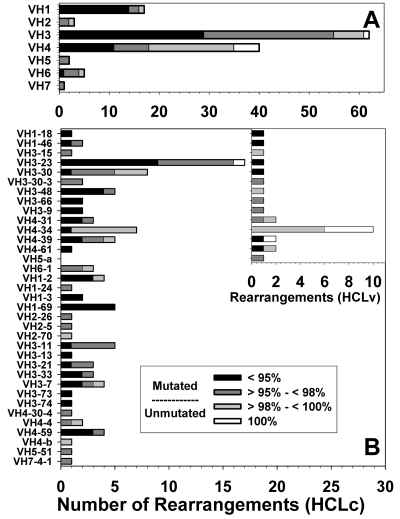
Using the 98% identity cutoff value to make the distinction between “mutated” and “unmutated” HCL rearrangements, 17 (17%) of 102 HCLc and 15 (54%) of 28 HCLv rearrangements were considered unmutated (Table 1; Figure 3). One HCLc and 5 HCLv IGHV sequences were 100% homologous to germline, with no mutations. As shown in Table 1, the 54% of HCLv rearrangements that were unmutated was similar to the 41% to 46% values reported for CLL (P = .45).5,6 However, HCLc rearrangements were more frequently mutated compared with CLL (P < .001). In only 1 of the 126 patients, multiple variants were observed for the cloned rearrangement, indicating that clonal diversity was very rare and that the mutated sequence is essentially fixed before malignant transformation. Moreover, the rearrangement sequence should not be affected whether obtained at the time of diagnosis or at relapse.
Table 1.
Mutated and unmutated rearrangements in HCL
Figure 3.
Mutation status for HCL rearrangements. IGHV family (A) and gene usage (B) are shown for all rearrangements, with the inset in B showing gene usage in HCLv. Homology to germine classified as less than 95% (black), 95% to 98% (dark gray), more than 98% and less than 100% (light gray), and 100% (white).
Unmutated rearrangements with respect to diagnosis and IGHV usage.
As shown in Figure 3A, IGHV families differed markedly with respect to the distribution of unmutated rearrangements. For example, the IGHV4 family was 55% unmutated versus 11% for IGHV3 (P < .001). The IGHV1 family used by 17 rearrangements was only 6% unmutated, also significantly lower than IGHV4 (P < .001). As shown in Figure 3B, unmutated rearrangements were observed in 14 different genes, 6 of which were used by rearrangements from HCLv patients. The IGHV4-34 gene, associated with poor prognosis even in HCLc,12 was most frequently unmutated (94%), and this gene was used by both HCLc (n = 7) and HCLv (n = 10). Subtracting the 17 IGHV4-34 rearrangements, there was no significant difference between HCLc and HCLv (12% vs 28%, P = .13) or between IGHV4 and IGHV3 (26% vs 11%, P = .1) in the frequency of unmutated rearrangements. Interestingly, all unmutated IGHV4-34 gene rearrangements in HCLc (n = 6) and HCLv (n = 10) used IGHV4–34*01 or *02 alleles, whereas the one mutated one was equally homologous to alleles *01, *02, and *08 with 94.56% homology. Although the percentage of IGHV4-34 genes that were unmutated was similar for HCLc and HCLv (83% vs 100%, P = .4), the reported percentage of unmutated IGHV4-34 rearrangements in CLL (21% of 159 rearrangements)6 was much lower than in the present study of HCLc (P = .007) or HCLv (P < .001). IGHV1-69, observed in HCLc (Figure 3B; n = 5) but not in HCLv, was always mutated, in contrast to CLL, where 241 (87%) of 277 rearrangements were unmutated (P < .001)5,6 Of several independent studies of gene usage in HCL rearrangements from a total of 191 specimens obtained before treatment,10,14,15,17,18,23 only one rearrangement expressing IGHV1-69 was reported, and it was also mutated.23 None of the 5 HCLc IGHV1-69 sequences was from allele *01, compared with 214 (83%) of 259 CLL rearrangements that were from allele *01 (P < .001), 177 (83%) of which had 100% homology to germline.6 The IGHV3-23 gene, the most common gene found in HCLc, was unmutated only in one case, very different from IGHV4-34 (P < .001). The IGHV3-23 gene was usually mutated in CLL as well.6 Thus, the unmutated rearrangements were dispersed over a wide variety of genes and between HCLc and HCLv without concentrating within any group except for IGHV4-34. Moreover, differences in the frequency of unmutated rearrangements between HCL and CLL were observed not only with IGHV4-34 but also with IGHV1-69, indicating that the extent of SHM for a particular IGHV rearrangement may be both disease- and allele-specific.
Characteristics of SHM in HCLc and HCLv
The defined characteristics of canonical SHM include: (1) increased R/S ratios in the CDRs, (2) decreased R/S ratios in the FRs, (3) more transitions (A → G, G → A, C → T, and T → C) than transversions, and (4) bias toward mutations in RGYW hot spots.5,24 The 1 HCLc and 5 HCLv sequences with 100% homology to the germline sequences were not evaluable for mutation analysis.
As shown in Table 2, a total of 102 HCLc and 28 HCLv sequences were characterized with respect to number of mutations, R/S ratios in the CDR versus FRs, transition to transversion ratios in the CDR versus FRs, and percentage of the total mutations, which were located in RGYW hot spots. For HCLc, the ratio of total R to S-mutations was 1.79, and higher in the CDR (2.83) than in the FR (1.41) regions (P < .0001). Similarly, for HCLv, total R/S ratio was 1.92, with a trend toward higher R/S ratio in the CDR (2.27) versus FR (1.74) regions (P = .067). There was a trend for higher R/S ratios within the CDR regions for HCLc compared with HCLv (2.83 vs 2.27, P = .07). Although there are 4 possible transitions (A → G, G → A, C → T, and T → C) compared with 8 possible transversions (A → C, C → A, A → T, T → A, G → C, C → G, G → T, and T → G), the transition to transversion ratio was 1.48 for the HCLc sequences and did not differ between the CDR and FRs (Table 2). The transition to transversion ratio was lower for HCLv than HCLc sequences (1.12 vs 1.49, P = .04), and also was not significantly different between the CDR and FRs (1.19 vs 1.00, P = .5). Table 2 shows that 13.92% of the 1214 HCLc mutations were located in RGYW hot spots, significantly higher than the percentage of total HCLc nucleotides within RGYW hot spots (3.33%, P < .001). For HCLv, 8.99% of the 267 mutations were located in RGYW hot spots, significantly higher than the percentage of total HCLv nucleotides within RGYW hot spots (3.54%, P < .001). The percentage of mutations in RGYW hot spots was higher for HCLc than for HCLv (13.92% vs 8.99%, P = .03). Thus, SHMs in HCL displayed all of the established characteristics of canonical SHMs, including higher R/S ratios in the CDR than FRs, more transitions than transversions, and a higher than expected frequency of SHMs in RGYW hot spots. Furthermore, the HCLc sequences met these criteria to a greater extent than did HCLv sequences.
Table 2.
Characteristics of SHMs in HCLc and HCLv
| No. of sequences | No. of mutations | R/S ratio |
Transitions/transversions |
% of mutations within RGYW | % of nucleotides within RGYW | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | CDR | FR | Total | CDR | FR | |||||
| HCLc | 102 | 1214 | 1.79 | 2.83 | 1.41 | 1.49 | 1.49 | 1.49 | 13.92 | 3.33 |
| HCLv | 28 | 267 | 1.92 | 2.27 | 1.74 | 1.12 | 1.19 | 1.00 | 8.99 | 3.54 |
Characteristics of SHM with respect to IGHV family
To determine whether the evidence of canonical SHM was confined to just 1 or 2 IGHV families or was a general characteristic of HCL, the localization and change preferences of the mutations were characterized separately for the 3 most common IGHV families (Table 3). For HCLc, a significant bias toward total R- versus total S-mutations in CDR versus FRs was observed for IGHV1 (P = .007), IGHV3 (P < .001), and IGHV4 (P = .0016). The lower number of HCLv mutations prevented similar analyses of statistical significance for HCLv. There was a trend for a higher R/S ratio in IGHV4 CDRs for HCLc versus HCLv (4.40 vs 1.38, P = .062). The expected transition to transversion ratio, 1:2, was significantly exceeded for HCLc rearrangements of IGHV1 (P < .001), IGHV3 (P < .001), and IGHV4 (P < .001) type. As shown in Table 3, HCLv rearrangements displayed a transition to transversion ratio higher than expected for IGHV4 (P < .001). Only 2 HCLv sequences were obtained for IGHV1, showing only a trend toward higher than expected transition to transversion ratio (P = .088), and 7 HCLv IGHV3 sequences had overall more transversions than transitions, not different from expected (P = .17). The transition to transversion ratio was significantly lower for HCLv IGHV3 compared with HCLc IGHV3 rearrangements (P = .0013). As shown in Table 3, the percentage of mutations in RGYW hot spots was significantly higher than expected based on the percentage of total nucleotides that were located in these hot spots, for HCLc families IGHV1 (P < .001), IGHV3 (P < .001), and IGHV4 (P < .001). Table 3 shows lower percentages of HCLv mutations in hot spots than HCLc, but still significantly more than predicted based on total nucleotides in hot spots for IGHV1 (P = .017) and IGHV3 (P = .019). The number of HCLv IGHV4 mutations was limited and did not display an increase in expected SHMs within RGYW hot spots (P = .33). Thus, characteristics of canonical SHMs in HCLc cannot be accounted for by the mutation pattern in just one specific IGHV family because they are shared among the 3 most common IGHV families. Despite the lower number of HCLv sequences analyzed, evidence of canonical SHM was observed but to a lesser degree than in HCLc. Moreover, in several direct comparisons of HCLc with HCLv, focusing on IGHV3 or IGHV4, evidence of canonical SHM was significantly stronger with HCLc than with HCLv.
Table 3.
SHM characteristics in the most common IGHV families
| No. of sequences | No. of mutations | R/S ratio |
Transitions/transversions |
% of mutations within RGYW | % of nucleotides within RGYW | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | CDR | FR | Total | CDR | FR | |||||
| HCLc | ||||||||||
| IGHV1c | 15 | 242 | 1.66 | 2.95 | 1.3 | 1.57 | 1.76 | 1.26 | 16.94 | 3.96 |
| IGHV3c | 55 | 705 | 1.92 | 2.73 | 1.52 | 1.50 | 1.40 | 1.66 | 13.62 | 3.09 |
| IGHV4c | 24 | 213 | 1.73 | 4.40 | 1.31 | 1.24 | 1.37 | 0.93 | 12.68 | 3.71 |
| HCLv | ||||||||||
| IGHV1v | 2 | 45 | 2 | 3 | 1.64 | 1.14 | 1.42 | 0.78 | 13.33 | 3.72 |
| IGHV3v | 7 | 111 | 1.89 | 2.25 | 1.73 | 0.76 | 0.63 | 1.05 | 8.11 | 3.19 |
| IGHV4v | 16 | 64 | 1.75 | 1.38 | 2 | 2.05 | 2.46 | 1.38 | 6.25 | 3.88 |
Distribution of SHMs along the IGHV sequence
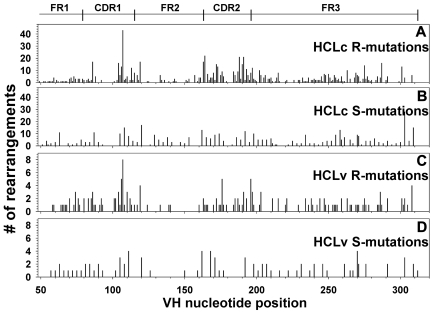
To determine whether the mutations in the IGHV regions occurred randomly within the framework or CDR regions, an analysis was performed to determine the frequency of mutation at each of the 312 nucleotide positions. The positions analyzed included 78 for FR1, 36 for CDR1, 51 for FR2, 30 for CDR2, and 117 for FR3. Sequence information was lacking regarding the first 48 nucleotides of FR1. As shown in Figure 4, S-mutations occurred randomly with relatively even distribution, whereas R-mutations in HCLc and, to a lesser extent, in HCLv rearrangements appeared more localized to specific regions. As shown in Figure 4A, mutations were concentrated to CDR1 and CDR2, and mutations were also observed in the 5′ and 3′ ends of FR2, and to a lesser extent in FR3. The lack of mutations in the middle of CDR1 and CDR2 was simply related to the lack of 2 or 3 codons at these positions in most rearrangements. The HCLv sequences shown in Figure 4C also contained R-mutations more frequently in CDR1, CDR2, and the borders of FR2. Thus, the analysis of mutation frequency at each nucleotide position was consistent with canonical or nonrandom R-mutations, particularly for HCLc, and appeared consistent with a mechanism of SHM, which might lead to matured antibody affinity.
Figure 4.
Distribution of somatic mutations in HCL. Replacement (A,C) and silent (B,C) SHMs are shown for HCLc (A-B) and HCLv (C-D).
Mutations in strong (G-C) versus weak (A-T) nucleotide pairs
To determine whether GC and AT pairs are mutated with approximately equal frequency as they are in normal B cells,25 the types of mutations in the rearrangements were analyzed. As shown in Figure 5A, GC and AT pairs were mutated with similar frequency for both HCLc (593 vs 621) and HCLv (130 vs 137). Thus, the nucleotide positions targeted for SHMs were not related to whether they were connected to the complementary strand with strong or weak hydrogen bonding.
Figure 5.
Nucleotides mutated in HCL. Vertical bars represent the proportion of mutated germline nucleotides, the total of which is listed above each vertical bar, which are mutated to each of 3 other nucleotides. Total HCLc (A) and HCLv (C) mutations are shown, along with just wobble bases for HCLc (B) and HCLv (D).
Mechanism of bias in transitions over transversions
To determine whether the bias toward transitions over transversions was related to selection at the amino acid level, or whether the bias was the result of the mutation machinery independent of the resulting protein sequence, the mutations at wobble bases were studied as previously reported for CLL.5 Wobble bases do not change the amino acid sequence, so bias in wobble mutations would be limited to the mutation machinery at the DNA or possibly transcription level. According to the data shown in Figure 5A, the bias of 4 possible transitions over 8 possible transversions for HCLc (observed 726 vs 488, expected 405 vs 809, P < .001) was also reflected in HCLc wobble mutations depicted in Figure 5B (observed 117 vs 94, expected 70 vs 141, P < .001). In contrast, as shown in Figure 4C-D, the bias of transitions over transversions for HCLv (observed 141 vs 126, expected 89 vs 178, P < .001) was not observed in the HCLv wobble positions (observed 15 vs 24, expected 13 vs 26, P = .8). Compared with HCLv wobble bases, there was a trend for higher transition-transversion ratio for HCLc wobble bases (P = .056). Thus, despite the relatively low frequency of SHMs in HCLv, the canonical analysis suggests that the mechanism of SHM may be different in HCLv compared with HCL.
Stereotyped CDR3 sequences
As in CLL, where groups of patients have been described with similar or “stereotyped” CDR3 SHMs,6,26–30 involving approximately 1% of CLL sequences reported in several studies, several small groups with similar VDJ structure were noted in HCL. Of 7 patients with IGHV-23 rearrangements, 3 with IGHD6-6/IGHJ4 and 4 with IGHD3-10/IGHJ6 were noted. The largest group with similar structure is shown in Table 4, containing IGHV4-34, IGHD3-10, and IGHJ4. Two patients, BH29 and BN15, had identical CDR3 sequences and differed only in the IGHV region. Patient BN38, although expressing the same IGHV, IGHD, and IGHJ genes as the other 4, was too dissimilar to be considered stereotyped.
Table 4.
Stereotyped CDR3 amino acid sequences among 5 patients with IGHV4–34, D3–10, and J4
| AA no. | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | 111.1 | 111.2 | 111.3 | 111.4 | 111.5 | 111.6 | 112.6 | 112.5 | 112.4 | 112.3 | 112.2 | 112.1 | 112 | 113 | 114 | 115 | 116 | 117 | 118 | AA length | CDR3, Daltons | CDR3, pI | % VH homology |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BN38 | C | A | R | G | G | H | F | R | I | G | G | S | G | I | Y | Y | L | S | L | R | T | P | S | L | D | Y | W | 25 | 3046.46 | 9.73 | 100.00 |
| BL18 | C | A | R | R | Y | Y | Y | G | S | — | — | — | — | — | — | — | — | — | — | G | S | Y | Y | I | D | Y | W | 15 | 2186.39 | 8.78 | 100.00 |
| BH29 | C | A | R | E | K | V | Q | G | V | — | — | — | — | — | — | — | — | — | — | M | V | H | Y | F | D | Y | W | 15 | 2131.47 | 7.48 | 100.00 |
| BN15 | C | A | R | E | K | V | Q | G | V | — | — | — | — | — | — | — | — | — | — | M | V | H | Y | F | D | Y | W | 15 | 2131.47 | 7.48 | 99.58 |
| BH32 | C | A | R | E | R | F | R | E | L | — | — | — | — | — | — | — | — | — | — | L | C | N | Y | F | D | Y | W | 15 | 2284.61 | 6.53 | 98.74 |
— indicates no amino acid present at that position.
Discussion
To determine whether the malignant B cells in HCLc and HCLv patients develop using canonical SHM, as was previously reported for normal B cells and CLL,5,6,24 the mutations in a relatively large database of HCL IGHV rearrangements were cloned, sequenced, and analyzed. We found that IGHV genes in HCLc rearrangements were more frequently mutated than in HCLv or CLL. Moreover, HCLc-like CLL exhibited characteristics of canonical SHMs, and to a greater extent than HCLv rearrangements. Analysis of the SHMs affecting wobble bases indicated that for HCLc, as reported for CLL,5 the higher than expected transition to transversion ratio was independent of amino acid selection and might reflect a bias in the mutational machinery. Finally, the lower evidence of canonical mutations in HCLv suggests that, in some patients with HCLv, malignant B cells may develop using a different mechanism or pathway perhaps independent of antigen selection.
Gene usage in HCL with respect to the status of patients tested
Based on previous data regarding IGVH gene usage in predominantly newly diagnosed HCL,23 we found a higher percentage of IGHV4-34+ patients to be unmutated. It should be noted that our study was skewed toward multiply relapsed HCL patients seeking salvage treatments. In comparing our 130 rearrangements with 58 recently reported by Forconi et al from newly diagnosed HCL patients,23 we found 16 of our 17 IGHV4-34 clones to be unmutated versus only 1 of 5 from the reported study (P = .003). We have recently reported that unmutated (but not mutated) IGHV4-34 portends a poor prognosis, particularly for response to single-agent cladribine.12 Consistent with this finding, Forconi et al reported that their one patient with unmutated IGHV4-34 did not respond to cladribine.23 Thus, the usage frequency of certain IGHV genes and the frequency of unmutated rearrangements may depend on the patient population studied. However, the lack of clonal instability makes the analysis of canonical SHM independent of whether patients are newly diagnosed or multiply relapsed.
Canonical SHM as evidence of antigen binding
The presence of canonical SHM in both normal and malignant B cells has been considered as strong evidence of an antigen-driven mutational process.5,6 We and others have previously analyzed SHM in HCL and in other situations in terms of related factors to detect an antigen-driven process, including total R/S ratio, CDR/FR ratio of R-mutations, and p-multinomial and p-binomial statistics.11,14,18,31–37 Our previous study of these statistics in 23 HCL patients identified only 2 patients with strong evidence (P = .001-.003) of antigen-driven SHM.11 Only with a much larger group of patients, as in the present study, was it possible to analyze SHMs for canonical characteristics in HCL. The canonical analysis presented here is more appropriate for comparing populations of patients rather than identifying certain patients with antigen-driven SHMs. With either analysis, it should be emphasized that, in the absence of identification of antigen, firm conclusions cannot be made about antigen dependence for SHMs in HCL cells. Studies of the nonfunctional VDJ rearrangements in normal B cells, which also undergo SHM, indicate that the mutation machinery that is needed for antigen-driven SHM may operate in the absence of selective pressure.38–40 This point is illustrated by the high transition-transversion ratio affecting wobble bases5 (Figure 5B). Conversely, lack of canonical SHM does not exclude antigen-driven SHM, in view of examples where a single framework mutation greatly affects antigen binding.41 Nevertheless, the study of canonical SHM in types of CLL and HCL may be useful and may shed light on their origin and biology.
Evidence of canonical SHM in HCLc versus HCLv
As was reported in CLL,5,6 we found strong evidence of canonical SHM in HCLc but less evidence for HCLv. Despite the limited number of total mutations in HCLv sequences available to analyze, several significant differences were observed in HCLv compared with HCLc, including lower transition to transversion ratio (P = .04), lower fraction of SHMs in RGYW hot spots (P = .03), and trends for lower R/S ratio in CDRs (P = .067) and lower transition-transversion ratio in wobble bases (P = .056). This suggests that, for some patients with HCLv, canonical mutations may not occur and that SHM may proceed randomly or by an alternative mechanism using noncanonical editing of antigen recognition regions. HCLv, at least in some patients, may be derived from B cells within or outside the marginal zone that may mutate via a T-cell independent mechanism that does not use canonical SHM and tends to use different IGHV genes.42,43 Knowing where the B cell becomes malignant may be of clinical importance, particularly when targeting an antigen, such as CD22,44,45 which might be expressed by differentiated HCL cells but not by their malignant progenitors if originated from an earlier antigen-negative B cell.
Acknowledgments
The authors thank Barbara Debrah for data management and our clinical staff, Sonya Duke, Rita Mincemoyer, Linda Ellison, and Elizabeth Maestri, for helping to obtain samples.
This work was supported by the intramural program of the National Cancer Institute, National Institutes of Health.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.A., M.S.-S., and R.J.K. designed research and analyzed and interpreted data; E.A., L.R., J.S., and T.S. performed research; E.A. and R.J.K. wrote the manuscript; and R.J.K. performed statistical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert J. Kreitman, National Institutes of Health, 37/5124b, 37 Convent Dr, MSC 4255, Bethesda, MD 20892; e-mail kreitmar@mail.nih.gov.
References
- 1.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 2.Morabito F, Cutrona G, Gentile M, et al. Definition of progression risk based on combinations of cellular and molecular markers in patients with Binet stage A chronic lymphocytic leukaemia. Br J Haematol. 2009;146(1):44–53. doi: 10.1111/j.1365-2141.2009.07703.x. [DOI] [PubMed] [Google Scholar]
- 3.Lin KI, Tam CS, Keating MJ, et al. Relevance of the immunoglobulin VH somatic mutation status in patients with chronic lymphocytic leukemia treated with fludarabine, cyclophosphamide, and rituximab (FCR) or related chemoimmunotherapy regimens. Blood. 2009;113(14):3168–3171. doi: 10.1182/blood-2008-10-184853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orlandi EM, Zibellini S, Pascutto C, et al. IGHV unmutated status influences outcome more than IGHV1–69 gene usage per se in patients with chronic lymphocytic leukemia. Clin Lymphoma Myeloma. 2009;9(5):390–393. doi: 10.3816/CLM.2009.n.076. [DOI] [PubMed] [Google Scholar]
- 5.Messmer BT, Albesiano E, Messmer D, Chiorazzi N. The pattern and distribution of immunoglobulin VH gene mutations in chronic lymphocytic leukemia B cells are consistent with the canonical somatic hypermutation process. Blood. 2004;103(9):3490–3495. doi: 10.1182/blood-2003-10-3407. [DOI] [PubMed] [Google Scholar]
- 6.Murray F, Darzentas N, Hadzidimitriou A, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111(3):1524–1533. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 7.Neuberger MS, Ehrenstein MR, Klix N, et al. Monitoring and interpreting the intrinsic features of somatic hypermutation. Immunol Rev. 1998;162:107–116. doi: 10.1111/j.1600-065x.1998.tb01434.x. [DOI] [PubMed] [Google Scholar]
- 8.Bouroncle BA, Wiseman BK, Doan CA. Leukemic reticuloendotheliosis. Blood. 1958;13(7):609–630. doi: 10.1182/blood-2016-01-696179. [DOI] [PubMed] [Google Scholar]
- 9.Ravandi F. Hairy cell leukemia. Clin Lymphoma Myeloma. 2009;9(suppl 3):S254–S259. doi: 10.3816/CLM.2009.s.020. [DOI] [PubMed] [Google Scholar]
- 10.Forconi F, Sozzi E, Rossi D, et al. Selective influences in the expressed immunoglobulin heavy and light chain gene repertoire in hairy cell leukemia. Haematologica. 2008;93(5):697–705. doi: 10.3324/haematol.12282. [DOI] [PubMed] [Google Scholar]
- 11.Arons E, Sunshine J, Suntum T, Kreitman RJ. Somatic hypermutation and VH gene usage in hairy cell leukemia. Br J Haematol. 2006;133(5):504–512. doi: 10.1111/j.1365-2141.2006.06066.x. [DOI] [PubMed] [Google Scholar]
- 12.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4–34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114(21):4687–4695. doi: 10.1182/blood-2009-01-201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forconi F, Sahota SS, Raspadori D, Mockridge CI, Lauria F, Stevenson FK. Tumor cells of hairy cell leukemia express multiple clonally related immunoglobulin isotypes via RNA splicing. Blood. 2001;98(4):1174–1181. doi: 10.1182/blood.v98.4.1174. [DOI] [PubMed] [Google Scholar]
- 14.Maloum K, Magnac C, Azgui Z, et al. VH gene expression in hairy cell leukaemia. Br J Haematol. 1998;101(1):171–178. doi: 10.1046/j.1365-2141.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Jimenez P, Garcia-Sanz R, Gonzalez D, et al. Molecular characterization of complete and incomplete immunoglobulin heavy chain gene rearrangements in hairy cell leukemia. Clin Lymphoma Myeloma. 2007;7(9):573–579. doi: 10.3816/clm.2007.n.043. [DOI] [PubMed] [Google Scholar]
- 16.Hockley SL, Giannouli S, Morilla A, et al. Insight into the molecular pathogenesis of hairy cell leukaemia, hairy cell leukaemia variant and splenic marginal zone lymphoma, provided by the analysis of their IGH rearrangements and somatic hypermutation patterns. Br J Haematol. 2010;148(4):666–669. doi: 10.1111/j.1365-2141.2009.07962.x. [DOI] [PubMed] [Google Scholar]
- 17.Thorselius M, Walsh SH, Thunberg U, Hagberg H, Sundstrom C, Rosenquist R. Heterogeneous somatic hypermutation status confounds the cell of origin in hairy cell leukemia. Leuk Res. 2005;29(2):153–158. doi: 10.1016/j.leukres.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Vanhentenrijk V, Tierens A, Wlodarska I, Verhoef G, Wolf-Peeters CD. V(H) gene analysis of hairy cell leukemia reveals a homogeneous mutation status and suggests its marginal zone B-cell origin. Leukemia. 2004;18(10):1729–1732. doi: 10.1038/sj.leu.2403503. [DOI] [PubMed] [Google Scholar]
- 19.Sozzi E, Amato T, Sahota SS, et al. Lack of allelic exclusion by secondary rearrangements of tumour B-cell receptor light chains in hairy cell leukaemia. Hematol Oncol. 2011;29(1):31–37. doi: 10.1002/hon.952. [DOI] [PubMed] [Google Scholar]
- 20.Jasper GA, Arun I, Venzon D, et al. Variables affecting the quantitation of CD22 in neoplastic B cells. Cytometry B Clin Cytom. 2011;80(2):83–90. doi: 10.1002/cyto.b.20567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Vol. 2. Lyon, France: World Health Organization; 2008. [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Forconi F, Sozzi E, Cencini E, et al. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood. 2009;114(21):4696–4702. doi: 10.1182/blood-2009-03-212449. [DOI] [PubMed] [Google Scholar]
- 24.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7(2):248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 25.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 26.Tobin G, Thunberg U, Karlsson K, et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104(9):2879–2885. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 27.Tobin G. The immunoglobulin genes: structure and specificity in chronic lymphocytic leukemia. Leuk Lymphoma. 2007;48(6):1081–1086. doi: 10.1080/10428190701342034. [DOI] [PubMed] [Google Scholar]
- 28.Mauerer K, Zahrieh D, Gorgun G, et al. Immunoglobulin gene segment usage, location and immunogenicity in mutated and unmutated chronic lymphocytic leukaemia. Br J Haematol. 2005;129(4):499–510. doi: 10.1111/j.1365-2141.2005.05480.x. [DOI] [PubMed] [Google Scholar]
- 29.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood. 2004;104(8):2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 30.Forconi F, Potter KN, Wheatley I, et al. The normal IGHV1–69-derived B-cell repertoire contains stereotypic patterns characteristic of unmutated CLL. Blood. 2010;115(1):71–77. doi: 10.1182/blood-2009-06-225813. [DOI] [PubMed] [Google Scholar]
- 31.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15(8):367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lossos IS, Tibshirani R, Narasimhan B, Levy R. The inference of antigen selection on Ig genes. J Immunol. 2000;165(9):5122–5126. doi: 10.4049/jimmunol.165.9.5122. [DOI] [PubMed] [Google Scholar]
- 33.Davi F, Maloum K, Michel A, et al. High frequency of somatic mutations in the VH genes expressed in prolymphocytic leukemia. Blood. 1996;88(10):3953–3961. [PubMed] [Google Scholar]
- 34.Insel RA, Varade WS, Chu YW, Marin E, Fuleihan R, Geha RS. Somatic mutation of human immunoglobulin V genes: bias, rate, and regulation. Ann N Y Acad Sci. 1995;764:158–169. doi: 10.1111/j.1749-6632.1995.tb55820.x. [DOI] [PubMed] [Google Scholar]
- 35.Jukes TH, King JL. Evolutionary nucleotide replacements in DNA. Nature. 1979;281(5732):605–606. doi: 10.1038/281605a0. [DOI] [PubMed] [Google Scholar]
- 36.Varade W, Marin E, Milano M, Insel R, Swerdlow S, Williams M. VH gene repertoire of mantle cell lymphomas. Ann N Y Acad Sci. 1995;764:504–506. doi: 10.1111/j.1749-6632.1995.tb55873.x. [DOI] [PubMed] [Google Scholar]
- 37.Miranda RN, Cousar JB, Hammer RD, Collins RD, Vnencak-Jones CL. Somatic mutation analysis of IgH variable regions reveals that tumor cells of most parafollicular (monocytoid) B-cell lymphoma, splenic marginal zone B-cell lymphoma, and some hairy cell leukemia are composed of memory B lymphocytes. Hum Pathol. 1999;30(3):306–312. doi: 10.1016/s0046-8177(99)90010-2. [DOI] [PubMed] [Google Scholar]
- 38.Dorner T, Foster SJ, Brezinschek HP, Lipsky PE. Analysis of the targeting of the hypermutational machinery and the impact of subsequent selection on the distribution of nucleotide changes in human VHDJH rearrangements. Immunol Rev. 1998;162:161–171. doi: 10.1111/j.1600-065x.1998.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 39.Dorner T, Foster SJ, Farner NL, Lipsky PE. Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur J Immunol. 1998;28(10):3384–3396. doi: 10.1002/(SICI)1521-4141(199810)28:10<3384::AID-IMMU3384>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Delpy L, Sirac C, Le Morvan C, Cogne M. Transcription-dependent somatic hypermutation occurs at similar levels on functional and nonfunctional rearranged IgH alleles. J Immunol. 2004;173(3):1842–1848. doi: 10.4049/jimmunol.173.3.1842. [DOI] [PubMed] [Google Scholar]
- 41.Potter KN, Hobby P, Klijn S, Stevenson FK, Sutton BJ. Evidence for involvement of a hydrophobic patch in framework region 1 of human V4–34-encoded Igs in recognition of the red blood cell I antigen. J Immunol. 2002;169(7):3777–3782. doi: 10.4049/jimmunol.169.7.3777. [DOI] [PubMed] [Google Scholar]
- 42.Weller S, Faili A, Garcia C, et al. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci U S A. 2001;98(3):1166–1170. doi: 10.1073/pnas.98.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297(5589):2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 44.Kreitman RJ, Wilson WH, Bergeron K, et al. Efficacy of the anti-CD22 recombinant immunotoxin BL22 in chemotherapy-resistant hairy-cell leukemia. N Engl J Med. 2001;345(4):241–247. doi: 10.1056/NEJM200107263450402. [DOI] [PubMed] [Google Scholar]
- 45.Kreitman RJ, Stetler-Stevenson M, Margulies I, et al. Phase II trial of recombinant immunotoxin RFB4(dsFv)-PE38 (BL22) in patients with hairy cell leukemia. J Clin Oncol. 2009;27(18):2983–2990. doi: 10.1200/JCO.2008.20.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]