Abstract
A20, a negative regulator of NF-κB, has been implicated as a tumor suppressor gene in multiple types of B-cell lymphoma. AIDS-related lymphomas (ARLs) are high-grade B-cell lymphomas that are frequently associated with EBV infection. We examined a panel of ARLs for A20 alterations. FISH showed A20 deletion in 6 of 33 cases (18%). A20 mutations were found in 3 of 19 cases (16%), including 2 cases with deletions of the comple-mentary allele. Immunohistochemistry showed the absence of A20 protein in 7 of 55 samples (13%). In contrast to reports in Hodgkin lymphoma in which EBV infection and A20 alteration are mutually exclusive, A20 inactivation was observed in both EBV+ and EBV− cases. The EBV latent membrane protein 1, which activates NF-κB, was not expressed in 12 of 13 cases with A20 loss. In ARLs loss of A20 may be an alternative mechanism of NF-κB activation in the absence of latent membrane protein 1 expression.
Introduction
Patients with HIV/AIDS have a significantly increased risk of developing non-Hodgkin lymphoma.1 AIDS-related lymphomas (ARLs) are a heterogeneous group of B-cell lymphomas frequently associated with EBV.2 The mechanism of lymphomagenesis is probably complex and is not because of immune suppression alone. Constitutive NF-κB activation is essential for tumor development in subtypes of lymphoma in HIV-negative persons.3 NF-κB can be activated by upstream signaling initiated by the EBV viral oncogene latent membrane protein 1 (LMP-1)4 or, in some cases, as a consequence of genetic alterations in regulators of NF-κB.5
A20 (TNFAIP3), an ubiquitin-modifying enzyme that inhibits NF-κB,6 has been found to be inactivated in multiple types of B-cell lymphoma, including diffuse large B-cell lymphoma (DLBCL),5,7 MALT lymphoma,7–9 follicular lymphoma,7 marginal zone lymphoma,10 Burkitt lymphoma,7 and Hodgkin lymphoma (HL).9,11,12 A20 is located on chromosome band 6q23, an area frequently deleted in B-cell lymphoma and thought to contain a tumor suppressor gene. Silencing of A20 with small interfering RNA in lymphoblastoid cell lines is associated with increased NF-κB activity, resistance to apoptosis, and enhanced clonogenicity.7 Reconstitution of A20 in cell lines with biallelic A20 deletion leads to apoptosis.5,7 In HL, cases with A20 inactivation are almost exclusively negative for EBV.11,13 In ARLs, the role of A20 and the relationship between A20 and EBV have not been examined.
In this report we describe inactivation of A20 in a significant proportion of ARLs, including EBV+ cases that do not express LMP-1 or express LMP-1 in only a minority of cells. These results suggest that A20 may act as a tumor suppressor gene in ARLs. In EBV-associated ARL, A20 loss may provide an alternate means of NF-κB up-regulation in cells in which LMP-1 is not expressed.
Methods
Patient samples
Sixty-eight formalin-fixed paraffin-embedded samples of ARL were collected from New York Presbyterian Hospital–Weill Cornell, the University of Siena, and the AIDS Malignancy Consortium. Cases were included if > 80% tumor cells were present and the diagnosis of B-cell lymphoma was confirmed. All samples were obtained with the approval of the institutional review boards at both institutions.
Tumor characterization
Immunohistochemistry was performed on tissue microarray to evaluate for the presence of CD10, CD20, CD3, PAX 5, Bcl-6, Bcl-2, BLIMP-1, FOXP-1, CD138, and MUM-1. EBV and Kaposi sarcoma herpes virus status were determined by in situ hybridization for EBV-encoded RNA (EBER) and immunohistochemistry for LMP-1, EBV nuclear antigen 2, and latency-associated nuclear antigen. Tissue diagnosis was made with the use of criteria from the World Health Organization.14 In cases of DLBCL, germinal center B-cell (GCB) versus non-GCB subtype was determined by the Hans algorithm.15,16
Fluorescence in situ hybridization
FISH was performed with the use of the spectrum green-labeled A20 probe (provided by Dr Vundavalli, Columbia University) and the spectrum orange-labeled centromeric probe for chromosome 6 (Vysis/Abbott Molecular), following the standard protocols for paraffin FISH assay.
A20 mutation analysis
Direct sequencing was performed on the coding region and splice sites of A20. Genomic DNA was extracted from paraffin-embedded tissue (QIAGEN). DNA was amplified by PCR with the use of the primers and conditions described in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article. Purified DNA was directly sequenced (Genewiz) and compared with the known germline sequence (NM_006290.2). Samples were included for analysis if sequencing was successfully performed on > 90% of the area of interest. Mutations were screened for known polymorphisms in the National Center for Biotechnology Information SNP (single nucleotide polymorphism) database.17 All mutations were confirmed on independent PCR products in both orientations.
Immunohistochemistry
Immunohistochemical staining of A20 was performed with the use of the Bond Max Autostainer (Leica Microsystems). Formalin-fixed paraffin-embedded tissue sections were baked and deparaffinized, followed by antigen retrieval with the use of the Bond Epitope Retrieval Solution 1 (ER1) at 99°C to 100°C for 30 minutes (Leica Microsystems). Sections were then subjected to sequential incubations with endogenous peroxidase block, primary antibody (Clone EPR2663, 1:50; Epitomics), postprimary (secondary antibody), polymer, diaminobenzidine, and hematoxylin for 5, 25, 15, 25, and 5 minutes, respectively (Bond Polymer Refine Detection Kit; Leica Microsystems). Finally, stained sections were dehydrated and mounted in Cytoseal XYL (Richard-Allan Scientific). Tumors with > 20% positivity were scored as positive.
Results and discussion
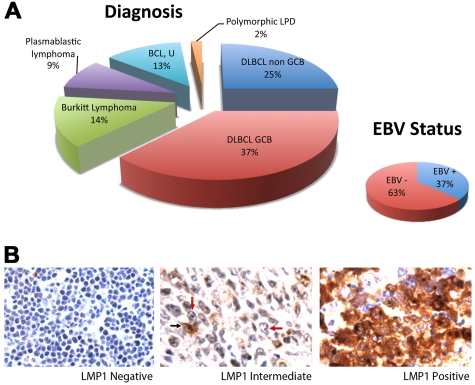
Fifty-six of 68 samples had sufficient tumor and interpretable results. The sample set included DLBCL, GCB (37%) and non-GCB (25%) types; Burkitt lymphoma (14%); plasmablastic lymphoma (9%); B-cell lymphoma, unclassifiable (13%); and polymorphic lymphoproliferative disorder (2%; Figure 1A). EBV was present in 37% of cases. Of 19 EBV/EBER+ cases, 4 were positive for LMP-1 with expression in > 90% of tumor cells. Two additional cases were heterogeneous with intermediate LMP-1 positivity consisting of LMP-1 expression in < 10% of tumor cells. The remaining cases were negative for LMP-1 (Figure 1B).
Figure 1.
ARL Tumor characterization. (A) Distribution of tumor type is shown. Cases included DLBCL of the GCB (n = 21) and non-GCB (n = 14) subtypes, Burkitt lymphoma (n = 8), plasmablastic lymphoma (n = 5), B-cell lymphoma, unclassifiable (BCL, U; n = 7), and polymorphic lymphoproliferative disease (LPD; n = 1). EBV positivity shown was determined by EBER in situ hybridization. (B) LMP-1 expression as determined by immunohistochemistry. Fifty cases lacked LMP-1 in all tumor cells (left). Two cases showed LMP-1 expression in < 10% of tumor cells (middle). A tumor cell expressing LMP-1 is indicated by a black arrow, and representative tumor cells lacking LMP-1 are indicated by red arrows. Four cases expressed LMP-1 in > 90% of tumor cells (right). Original magnification ×600 with 60×/0.80 objective lens. Microscope: Olympus BX 41; camera: Olympus Q-COLOR3; software: QCapture, Version 2.9.8.0 (Quantitative Imaging Corporation).
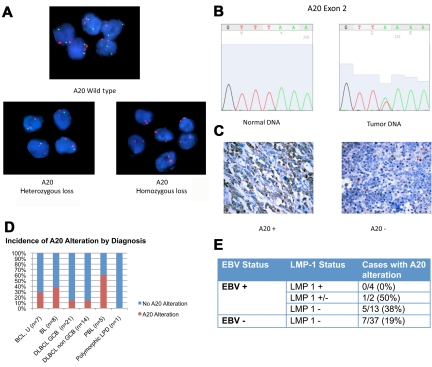
Genetic alterations in A20 were evaluated with the use of FISH and direct sequencing (Figure 2A-B). Monoallelic deletion of A20 was detected by FISH in 5 of 33 cases (15%). Biallelic deletion was detected in one case. Direct sequencing was performed in 19 samples with sufficient DNA. Point mutations were present in 3 of 19 cases (16%; supplemental Table 2). Nonsense mutations coding for a premature stop codon in exon 2 were seen in 2 cases. The third case had a missense mutation in exon 7 resulting in an amino acid change. Two of the 3 cases with A20 mutation also had monoallelic deletion in the complementary allele, indicating biallelic alteration of the A20 gene.
Figure 2.
A20 Alterations in ARL. (A) Dual-color FISH analysis of tumor samples hybridized with A20 probe (green) and chromosome 6 centromeric probe (red). Top case shows wild-type A20. A20 heterozygous loss was shown in 5 cases (bottom left). One case was found to have A20 homozygous loss (bottom right). Magnification: Apochromatic 100× lens with 1.4 aperture; microscope: Nikon Eclipse 80i; camera: Jai CV-A10CL; software: Cytovision Imaging Software (Genetix Corp). (B) Representative chromatogram of normal DNA and tumor sample with A20 nonsense mutation in exon 2. This case had A20 monoallelic deletion, shown by FISH. The normal nucleotide peak in the mutated tumor sample probably represents infiltrating inflammatory cells. Software used was 4Peaks Version 1.7.2. (C) A20 Immunohistochemistry showing tumor cases with presence of A20 (left) and absence of A20 (right). Original magnification ×600 with 60×/0.80 objective lens. Microscope: Olympus BX 41; camera: Olympus Q-COLOR3; software: QCapture, Version 2.9.8.0 (Quantitative Imaging Corporation). (D) Incidence of A20 alteration is shown. (E) Incidence of A20 alteration in LMP-1 positive (+), negative (−), and intermediate (+/−) samples.
Immunohistochemistry for A20 was performed and is described for the first time in this report (Figure 2C). Absence of A20 was shown in 7 of 55 cases (13%). In all cases negative for A20, the infiltrating normal cells stained positive. Included among the cases negative for A20 is the case with biallelic A20 deletion determined by FISH. Cases with A20 mutation or monoallelic deletion or both frequently retained reactivity toward A20. Immunohistochemistry may therefore be useful in identifying cases with complete loss of A20 but not helpful in cases in which A20 is present but altered.
In total 13 of 56 cases (23%) showed evidence of A20 inactivation by FISH, sequencing, immunohistochemistry, or a combination (supplemental Table 3). A20 inactivation was seen in most histologic subtypes, including Burkitt lymphoma (n = 3), DLBCL, GCB (n = 3), and non-GCB (n = 2) subtypes, and B-cell lymphoma, unclassifiable (n = 2). Three of 5 plasmablastic lymphomas (60%) had evidence of A20 loss (Figure 2D), but given the small sample size further investigation is necessary to determine whether A20 inactivation is more common in this lymphoma subtype.
A20 inactivation was seen in both EBV+ and EBV− cases. Interestingly, the EBV viral protein LMP-1, which activates NF-κB, was not expressed in 12 of 13 cases with A20 alteration (Figure 2E). One case with A20 inactivation had evidence of LMP-1 expression however in only 10% of tumor cells (Figure 1B).
This is the first report to show A20 inactivation in EBV-associated lymphoma. This contrasts with previous studies in HL in which A20 inactivation and EBV infection were almost mutually exclusive.11 The EBV gene expression pattern differs in HL and ARL. In HL Reed Sternberg cells, EBV uniformly expresses LMP-1; however, in ARL viral gene expression is more heterogeneous and LMP-1 is frequently absent. Our data indicate that A20 may represent a tumor suppressor gene in a significant subset of ARLs and that A20 inactivation can be present in both EBV+ and EBV− cases. In EBV-related lymphoma the inactivation of A20 may be an alternative mechanism of NF-κB up-regulation in lymphoma cells in which LMP-1 is not expressed.
Supplementary Material
Acknowledgments
We thank Paul A. Chadwick for construction of tissue microarrays; Dr Joshua Nyagol and Dr Emily Rogena from the Department of Human Pathology, University of Nairobi, Nairobi Kenya, for collecting samples via the University of Siena; and the AIDS Malignancy Consortium for contributing samples.
This work was supported in part by the National Institutes of Health (grant R01 CA068939; E.C.) and the AIDS Malignancy Consortium (grant UO1 CA121947). L.G. is supported by the fellowship training program in pediatric hematology/oncology, New York Presbyterian Hospital–Weill Cornell Medical Center and Memorial Sloan-Kettering Cancer Center and the Michael Goldberg Fellowship Fund.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.G. performed sequence analysis, collected and analyzed data, and wrote the paper; S.M. and S.G. performed FISH analysis; G.B. performed sequence analysis and analyzed data; A.C., S.B., G.A., and L.L. collected and characterized samples; Y.F.L. performed immunohistochemistry for A20; W.T. performed the pathology evaluation, including immunohistochemistry; and E.C. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ethel Cesarman, Weill Cornell Medical College, 1300 York Ave C405, New York, NY 10065; e-mail: ecesarm@med.cornell.edu.
References
- 1.Engels EA, Pfeiffer RM, Goedert JJ, et al. Trends in cancer risk among people with AIDS in the United States 1980-2002. AIDS. 2006;20(12):1645–1654. doi: 10.1097/01.aids.0000238411.75324.59. [DOI] [PubMed] [Google Scholar]
- 2.Dal Maso L, Franceschi S. Epidemiology of non-Hodgkin lymphomas and other haemolymphopoietic neoplasms in people with AIDS. Lancet Oncol. 2003;4(2):110–119. doi: 10.1016/s1470-2045(03)00983-5. [DOI] [PubMed] [Google Scholar]
- 3.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80(3):389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 5.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30(8):383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non-Hodgkin lymphomas. Blood. 2009;114(12):2467–2475. doi: 10.1182/blood-2008-12-194852. [DOI] [PubMed] [Google Scholar]
- 8.Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia. 2010;24(2):483–487. doi: 10.1038/leu.2009.234. [DOI] [PubMed] [Google Scholar]
- 9.Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459(7247):712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 10.Novak U, Rinaldi A, Kwee I, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113(20):4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz R, Hansmann ML, Bohle V, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206(5):981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher MA, Schmitz R, Brune V, et al. Mutations in the genes coding for the NF-kappaB regulating factors IkappaBalpha and A20 are uncommon in nodular lymphocyte-predominant Hodgkin's lymphoma. Haematologica. 2010;95(1):153–157. doi: 10.3324/haematol.2009.010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuppers R. Molecular biology of Hodgkin lymphoma. Hematology Am Soc Hematol Educ Program. 2009:491–496. doi: 10.1182/asheducation-2009.1.491. [DOI] [PubMed] [Google Scholar]
- 14.Swerdlow S CE, Harris N. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 15.Hans CP, Weisenburger DD, Vose JM, et al. A significant diffuse component predicts for inferior survival in grade 3 follicular lymphoma, but cytologic subtypes do not predict survival. Blood. 2003;101(6):2363–2367. doi: 10.1182/blood-2002-07-2298. [DOI] [PubMed] [Google Scholar]
- 16.Chadburn A, Chiu A, Lee JY, et al. Immunophenotypic analysis of AIDS-related diffuse large B-cell lymphoma and clinical implications in patients from AIDS Malignancies Consortium clinical trials 010 and 034. J Clin Oncol. 2009;27(30):5039–5048. doi: 10.1200/JCO.2008.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Center for Biotechnology Information. SNP: Single Nucleotide Polymorhism. [Accessed. July 23, 2010]. http://www.ncbi.nlm.nih.gov/snp.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.