Abstract
The mechanisms of the molecular and neuroendocrine responses to stress in the immature rat have been a focus of intense investigation. A principal regulator of the these responses in both mature and developing rat is the neuropeptide corticotropin releasing hormone (CRH), and levels of hypothalamic CRH mRNA are enhanced by stress. In vitro, transcription of the CRH gene is governed by binding of the phosphorylated form of cAMP responsive element binding protein (pCREB) to the promoter. Here we tested the hypothesis that rapid, stress-induced CRH transcription occurred during the first two postnatal weeks, and is associated with pCREB expression. The time-course of induction of unedited, heteronuclear CRH RNA (CRH hnRNA) was examined in hypothalamic paraventricular nucleus (PVN) of immature rats subjected to both modest and strong acute stressors using in situ hybridization; pCREB abundance was determined in individual neurons in specific PVN sub-nuclei using immunocytochemistry and unbiased quantitative analysis. CRH hnRNA signal was negligible in PVN of immature rats sacrificed under stress-free conditions, but was readily detectable within 2 min, and peaked at 15 min, in PVN of stressed animals. Enhanced pCREB immunoreactivity was evident within 2 min of stress onset, and was enhanced specifically in stress-responsive, CRH-expressing medial parvocellular neurons. These data support the notion that, already during early postnatal life, stress induces rapid CREB phosphorylation, interaction of pCREB-containing transcription complexes with the CRE element of the CRH gene promoter, and initiation of CRH hnRNA production in stress-responsive neurons of rat PVN.
Keywords: CREB, CRF, Stress, Gene expression, Heteronuclear RNA
1. Introduction
The ability of the developing rat to mount the full complement of the hypothalamic stress response has been a focus of intense investigation. During the first two postnatal weeks, the immature rat shows an attenuated hormonal response and altered gene regulation in response to stress as compared with the adult [3,30,39,40,44], and the neurohormonal stress response is further modulated by maternal input [33,38]. Recent data suggest that the developing rat may respond to some stressful stimuli [8,12,41,42,44]. However, hypothalamic–pituitary–adrenal activation is diminished compared with the adult [39].
The principal regulator of the neuroendocrine stress response is the neuropeptide corticotropin-releasing hormone (CRH) [17,37]. CRH is released from terminals of parvocellular neurons residing in the medial paraventricular nucleus (PVN) of the hypothalamus to initiate a cascade of hormonal responses mediating the response to a variety of stressors [31,36,44]. Stressful stimuli also result in induction of CRH gene expression in the PVN, resulting in an up-regulation of steady-state CRH mRNA [17,26].
Like most transcriptional processes, RNA transcribed from the CRH gene in the cell nucleus consists of unedited heteronuclear RNA (hnRNA). In the nucleus, the hnRNA is rapidly processed by splicing to remove intronic sequences. After further modification and transport to the cytoplasm, the messenger RNA (mRNA) is utilized for protein translation, resulting in synthesis of the CRH prepropeptide [29]. Steady-state levels of mature mRNA provide a measure of available template for translation, but the half-life of CRH mRNA is rather long [21,22]. Thus, the amount of hnRNA may provide a more direct measure of production of new RNA and of rapid gene activation [19]. Indeed, stress-induced CRH hnRNA has been demonstrated in the mature rat [14,15,20] and, quite recently, during development [8]. However, the mechanisms by which stressful challenges, defined as those activating the neuroendocrine stress response, regulate transcription of the CRH gene in the hypothalamus of the immature rat remain unresolved.
A strong candidate for rapid regulation of CRH transcription is phosphorylated cAMP response element (CRE) binding protein (pCREB). The CRH gene promoter contains a CRE that has been shown to be involved in CRH transcriptional regulation [9,17,32]. Previous studies from the authors’ laboratory, using gel-shift and western blot methods, have shown that acute cold, a powerful stress for the immature rat, increases levels of pCREB and CREB-related DNA binding activity in anterior hypothalamic extracts [11]. However, these hypothalamic extracts included not only the PVN but also additional cell groups, and did not provide the resolution to determine whether CREB phosphorylation was induced by stress in the hypothalamic cell groups that express the CRH gene. Studies of this issue in the mature rat have suggested that total pCREB in the PVN may not increase after stress [5], or that CRH transcription may coincide with CREB phosphorylation [20].
Therefore, the goals of this study were (1) to determine whether the CRH gene was rapidly activated by stress in parvocellular PVN neurons of the immature rat, (2) to analyze the time course for stress-induced CRH hnRNA expression and (3) to determine whether phosphorylation of CREB in CRH-expressing cell groups preceded (and might thus induce) CRH hnRNA production.
2. Materials and methods
2.1. Animals and experimental design
Immature rats were studied during the developmental period considered stress-hyporesponsive. They were evaluated specifically on postnatal days 6 and 9 for the following reasons: (a) induction of CREB phosphorylation in hypothalamus by cold stress has been demonstrated at these ages [11]; (b) quantitative parameters of CRH mRNA expression have been established at these ages [12,44]; (c) the parameters of the cold-challenge paradigm have been standardized for these ages. Rat pups were products of timed-pregnant Sprague–Dawley dams (transported at E10–E15), that were maintained in uncrowded, quiet, NIH-approved animal facility on a 12-h light/dark cycle. Delivery was verified at 12-h intervals and the date of birth was considered day 0. All experiments were approved by the University Animal Care Committee.
On the days of the experiments, rats were treated as follows:
Stress-free controls were sacrificed within 45 s of disturbance (for in situ hybridization histochemistry), or deeply anesthetized with a rapid-acting barbiturate (pentobarbital, 100 mg/kg, into the peritoneal cavity [i.p.]) within the same time frame, for immunocytochemistry. The latter group were then perfused. The adult rat brain, a ‘control’ of the immunocytochemistry, was obtained under the same conditions (rapid anesthesia).
The 2 min stress group animals were carried from the animal facility to the laboratory, and sacrificed or anesthetized at 2 min from their initial disturbance. This group was considered ‘0 min of cold stress’, but was exposed to a 2 min stress of human handling, transportation, novel environment, etc.
Cold stress groups were subjected immediately to a cold challenge modified from a previously described procedure [12,44]. Briefly, immature rats were placed on ice until they showed little response to tactile stimulus. The cold exposure lasted an average of 6 and 12 min for 6- and 9-day-old rats, respectively, and was followed by warming on a euthermic pad. For CRH hnRNA analysis, rats were sacrificed by decapitation at 0, 15, 30 and 60 min from the onset of the cold challenge. For CRH mRNA expression, rats were sacrificed at 4 h after the termination of cold stress, a time point shown previously to coincide with maximal enhancement of CRH mRNA levels by this stress [12,44].
2.2. Hormonal measures
Plasma corticosterone levels were determined by radioimmunoassay [12,13,44] using a commercially available kit (ICN, Irvine, CA). Corticosterone assay sensitivity was 0.05 μg/dl, and interassay variability, determined by three dilutions of adult rat plasma, averaged 15%.
2.3. In situ hybridization histochemistry (ISH)
For ISH, brains were rapidly removed and frozen in powdered dry ice. Frozen coronal sections (20 μm) were collected on gelatin-coated slides and stored at −80°C. Sections were brought to room temperature, air dried and fixed in fresh 4% buffered paraformaldehyde for 20 min, followed by dehydration and rehydration through graded ethanols [2,12,13]. Subsequently, sections were exposed to 0.25% acetic anhydride in 0.1 M triethanolamine (pH 8.0) for 5 min and dehydrated through ethanols. For CRH mRNA, probe synthesis and ISH were carried out as previously described [12,13,44]. For CRH hnRNA, sections were air dried and prehybridized for 1 h at 55°C in a humidity chamber. Prehybridization and hybridization was performed at 55°C in a solution of 50% formamide, 5×SET, 0.2% SDS, 5×Denhardt’s, 0.5 mg/ml salmon sperm DNA, 0.25 mg/ml yeast tRNA, 100 mM DTT, 10% Dextran sulfate. Following prehybridization, sections were hybridized overnight with 1 × 106 cpm of 35S labeled ribonucleotide probe at 55°C. Posthybridization, sections were washed in 2×SSC for 5 min at room temperature, and were RNase-digested (200 μg/ml RNase A) for 30 min at 37°C. Sections were washed successively (at 55°C) in 2×SSC (1×SSC denotes 0.15 M NaCl, 15 mM trisodium citrate buffer, pH 7.0) and 1×SSC for 5 min, 0.25×SSC for 30 min, and in 0.1× and 0.03×SSC for 1 h each. Sections were dehydrated through ethanols containing 0.3 M ammonium acetate followed by 100% ethanol, then apposed to film (Hyperfilm B-Max, Amersham, IL) for 3 weeks.
2.4. Synthesis and labeling of CRH hnRNA probes
A plasmid containing a 530 base pair fragment of the CRH intron was kindly provided by Drs. S.J. Watson and S. Rivest [14,28]. Radioactive antisense cRNA was synthesized by incubating T7 RNA polymerase (30U, Promega, Madison, WI) with 1 μg plasmid linearized with Hind III in 2.5 mM ATP/GTP/UTP, 6 mM [α-35S]–CTP, 10 mM DTT, 40 mM Tris–HCl (pH 7.5), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl and 40 U RNase inhibitor (RNasin, Promega). After 2 h at 37°C, 3 U of RNase-free DNase (RQ1-DNase, Promega) was added for 15 min at 37°C. The probe was subjected to mild alkaline hydrolysis and purified by column chromatography. The specific activity of each probe was 1–3×106 cpm/μg.
2.5. Acquisition and quantitative analysis of ISH signal
Semiquantitative analyses were performed following in situ hybridization. Digitized images of each brain auto-radiogram was acquired using a StudioStar scanner (resolution 1200×1200 dots per inch) and analyzed using the ImageTool software program (University of Texas Health Science Center, version 1.25). For each group a minimum of ten sections from three to four brains were analyzed. Densities were calibrated using C-14 standards [12,13] after correcting for background by subtracting the density of the corpus callosum (white matter background value) from the density of the PVN. The significance of observed quantitative differences among different groups were evaluated using ANOVA or Student’s t-test as appropriate.
2.6. Immunocytochemistry for detection of pCREB
Perfused brains (4% paraformaldehyde in 0.1 M phosphate buffer [PB], pH 7.4) were dissected from the skull, postfixed for 4 h and cryoprotected with 15%, followed by 25% sucrose. Brains were blocked in the coronal plane and cryostat-sectioned at 20 μm. For the hypothalamic region, 1-in-4 sections were stained with cresyl violet. This series was used to ascertain the neuroanatomic boundaries of the various PVN sub-nuclei [7,12,20,35,36], as well as optimal between-group matching of adjacent sections that were subjected to immunocytochemistry.
For pCREB immunocytochemistry, free-floating sections were collected into tissue-culture wells in 0.1 M PB and subjected to standard avidin–biotin complex (ABC) methods [7,43]. Briefly, after several washes with 0.01 M PB-saline (PBS) containing 0.3% Triton X-100 (pH 7.4, PBS-T), sections were treated for 30 min in 0.3% H2O2 in PBS, followed by blockade of non-specific sites with 2% normal goat serum in PBS for 30 min. After a 10-min rinse in PBS, sections were incubated for 48 h at 4°C with rabbit polyclonal IgG directed against phosphorylated CREB (Upstate Biotechnology, Lake Placid, NY) diluted with PBS containing 1% bovine serum albumin and 2% normal goat serum to a concentration of 1:4000. This antiserum is generated against an epitope consisting of the phosphopeptide (including Ser133) portion of pCREB, and does not recognize non-phosphorylated CREB. After three washes in PBS-T, 5 min each, sections were incubated in biotinylated goat–anti-rabbit IgG (1:200, Vector) in PBS for 1 h at room temperature. After washing in PBS-T (3×5 min), sections were incubated in avidin–biotin–peroxidase complex solution (ABC, 1:100, Vector) for 2 h at room temperature. Sections were then rinsed again in PBS-T (3×5 min). The reaction product was visualized by incubating sections for 8–10 min in 0.04% 3,3′-diaminobenzidine (DAB) containing 0.01% H2O2, followed by counter-staining with 0.5% methyl-green to visualize the nuclei.
2.7. Double-labeling CRH/pCREB immunocytochemistry
Free-floating sections were processed for concurrent immunolabeling of CRH and pCREB, using a modification of the dual-chromogen procedure [23]. Briefly, sections were first incubated for 36–48 h at 4°C with rabbit anti-CRH antiserum (1:40 000, a gift from Dr. W.W. Vale) as described previously [43], yielding a diffuse brown DAB reaction product. Subsequently, sections were rinsed with PBS-T, preincubated in 5% normal goat serum for 30 min, then incubated in pCREB antiserum as described above. After these incubations, sections were rinsed in PBS-T for 15 min, and transferred to 0.01 M PB (pH 6.6) for 30–45 min with several changes. Sections were then incubated for 10 min in the same buffer containing 0.025% sodium nitroprusside and 0.01–0.02% benzidine dihydrochloride (BDHC).
Finally, the granular black–blue deposits were visualized by immersing the sections for 3–5 min in fresh incubation solution containing 0.003% H2O2, and the reaction stopped by rinsing in 0.01 M PB containing 0.3% Triton X-100 (pH 6.6).
The CRH antiserum, directed against the C-terminus, does not cross-react with other neuropeptides such as somatostatin, NPY, endorphin, enkephalin, VIP or vasopressin [37]. In this study, the specificity of the primary antiserum was also tested by substituting normal rabbit IgG for the primary antiserum and by pre-adsorbing the antiserum overnight with purified CRH peptide (0.4 mM). No specific signal was found after either procedure [4,7,43]. The specificity of the pCREB immunostaining was addressed by eliminating the primary antiserum during the first incubation, and by using stress-free adult tissue where pCREB levels are low [4].
2.8. Quantitative analysis of pCREB-expressing neurons in PVN
Matching coronal 1-in-4 sections of PVN were examined without knowledge of treatment. The areas analyzed included the anterior parvocellular (ap), medial parvocellular (mp) and posterior magnocellular (pm) sub-nuclei, identified based on cytoarchitectonic relationships, using the 1-in-4, cresyl violet-stained sections [34,35]. The relative density of pCREB-expressing neurons was assessed with a square lattice system [6] over the entire area examined, using light microscopy at 400× magnification.
This system utilizes a grid reticule; 7×10 squares (43 750 μm2) were counted, and the cell density was calculated (see below). pCREB-expressing neurons were included in the count only when more than half of the cell nucleus was labeled (see Fig. 5E and F), and abundance (neuronal density) was expressed as the number of labeled neurons in a 5 ×104 μm2 real area. Because of the size of the PVN in the immature rat (~600 μm), four or five sections were counted per each subnucleus per rat. A similar analysis was conducted for neurons that co-express CRH and pCREB. The denominator in this case was CRH-immunoreactive cells located within the mp sub-nucleus. Student’s t-test was used to assess the differences between the 2-min stress group and matched sections from stress-free controls, with a minimum accepted level of significance of 0.05 (P<0.05).
Fig. 5.
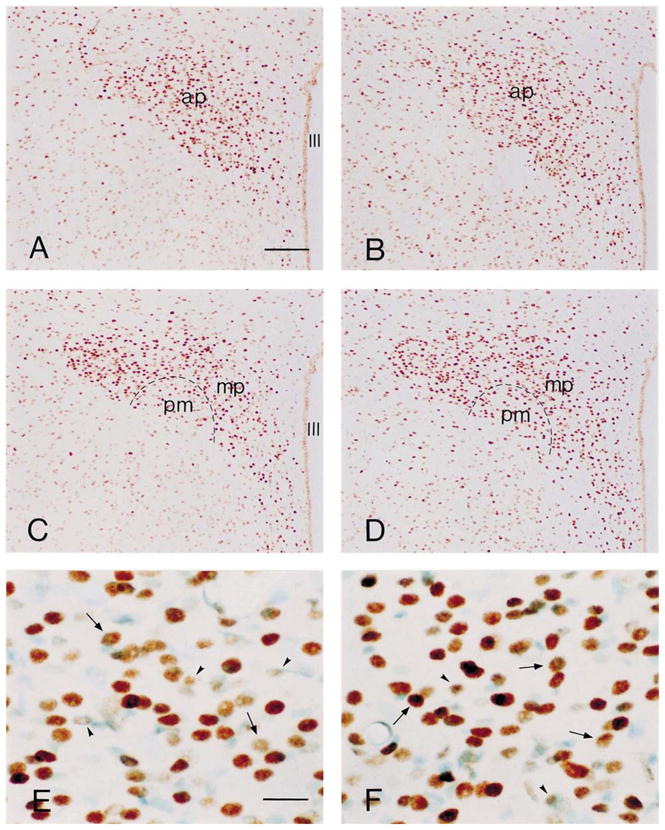
Comparison of immunoreactive labeling for pCREB in coronal sections of the hypothalamic paraventricular nucleus (PVN) from stress-free rats (A, C, E) and those subjected to a 2-min modest stress (B, D, F). Panels A, B show pCREB-immunoreactive neurons in the anterior parvocellular (ap) sub-nucleus of PVN. No significant change was observed in the 2-min stress group compared with stress-free controls. C, D show pCREB-immunoreactive neurons in the medial parvocellular (mp) and posterior magnocellular (pm) sub-nuclei of PVN. Numbers of pCREB-immunoreactive neurons in the mp are increased in section from the stressed group. Panels E, F consist of high magnification of pCREB-labeled cell-nuclei from the mp region of PVN, counterstained with methyl-green. Arrows point to pCREB-immunoreactive neurons with fully labeled cell-nuclei that were counted in the quantitative analysis. Arrowheads indicate partially labeled nuclei that were not counted. III denotes the third ventricle. Scale bars=120 μm in A–D; 20 μm in E, F.
3. Results
3.1. Stress hormone levels are elevated by acute cold challenge in the immature rat
Cold challenge increased plasma corticosterone levels in both groups of immature rats (aged 6 and 9 days; Fig. 1). Peak plasma corticosterone was detected 60 min following the onset of stress, and levels remained significantly elevated for up to 4 h following the cold challenge.
Fig. 1.
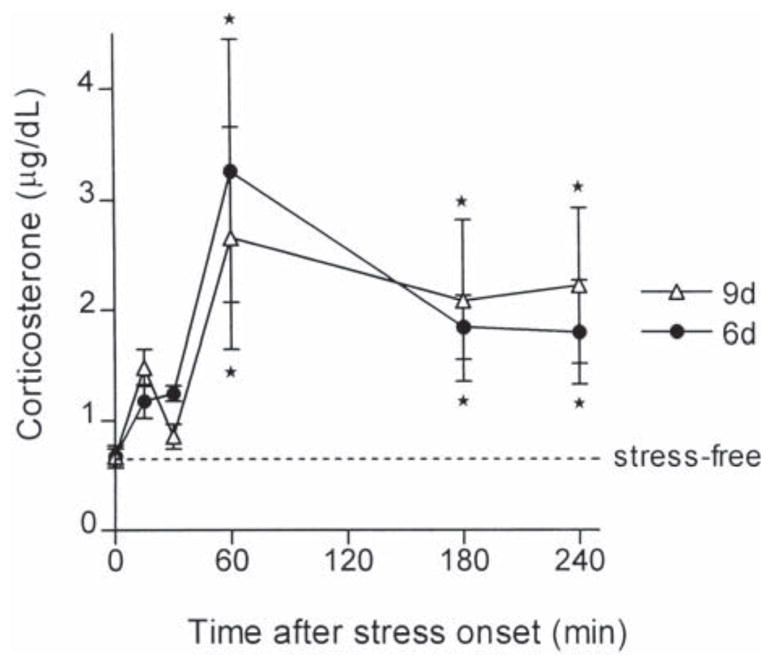
Hormonal response to cold stress in 6- and 9-day-old rats. Immature rats were sacrificed at indicated time-points after the onset of cold stress. Plasma was analyzed for corticosterone levels using radioimmunoassay. Cold-stress resulted in an increase in plasma corticosterone levels in both 6- and 9-day-old rats. Asterisk indicates a significant difference from stress-free controls (P<0.05).
3.2. Transcription of the CRH gene, measured by CRH hnRNA expression, is induced by cold stress in the PVN of the immature rat
The hormonal stress responses (see above) were accompanied by activation of the CRH gene: in situ hybridization performed to detect CRH hnRNA demonstrated little signal above background in the PVN of stress-free immature rats (Fig. 2 left panels). Acute cold challenge induced CRH hnRNA expression within 15 min at both ages studied. Overall levels of unedited mRNA (hnRNA) were low, and better visualized and quantified after transformation to false-color scales (Fig. 2). As shown in the figure, hybridization signal over the PVN was maximal at the 15 min time-point, and persisted at the 30 min time-point. By 60 min from onset of the cold challenge, the CRH hnRNA signal was attenuated and not significantly different from baseline (see below).
Fig. 2.
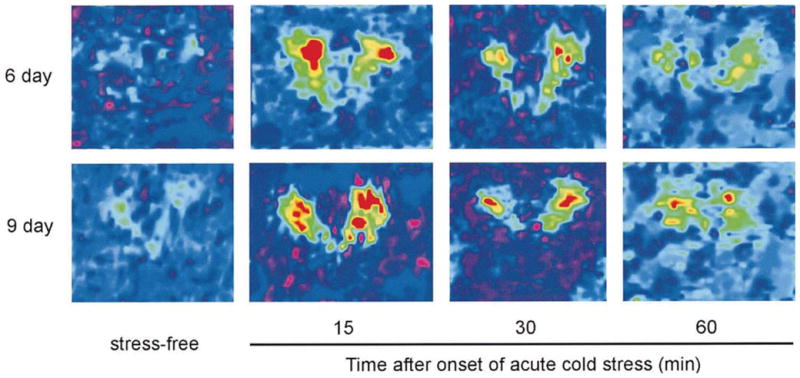
CRH hnRNA in the PVN of 6- and 9-day-old rats following cold-stress. Computer-generated enhanced color-coded quantitative images of in situ hybridization signal of CRH hnRNA in the PVN of representative 6- (top) and 9-day (bottom)-old rats. Rats were sacrificed within 30 s of disturbance (stress-free controls; left panels), or at 15, 30 or 60 min following the onset of cold stress. For both 6- and 9-day-old rats, CRH hnRNA signal in the PVN was barely detectable under stress-free conditions, increased substantially with the onset of stress, peaked at 15 min and was still detected (although at reduced levels) 30 and 60 min following stress.
Semi-quantitative analysis of cold-induced CRH hnRNA expression in the PVN is shown in Fig. 3. In both 6- and 9-day-old animals, low baseline levels of CRH transcription were noted (0.0174±0.002 and 0.0170±0.002 μCi/g, respectively). Peak CRH hnRNA were achieved at 15 min (0.039±0.0032 and 0.0475±0.0049 μCi/g, respectively) with a gradual reduction thereafter. However, the magnitude of induced transcription at 15 min tended to be greater in the 9-day than in the 6-day-old rat (279 vs. 226 percent of stress-free controls, respectively). One-way ANOVA revealed a significant effect of treatment (F=8.53, P<0.0001), and post-hoc Bonferroni tests suggested that the 60-min time-point values did not differ from the stress-free values for either group. Two-way ANOVA demonstrated a strong effect of treatment (F=9.98, P< 0.0001), and a trend towards an effect of age (F=2.81, P=0.096), that did not reach statistical significance.
Fig. 3.
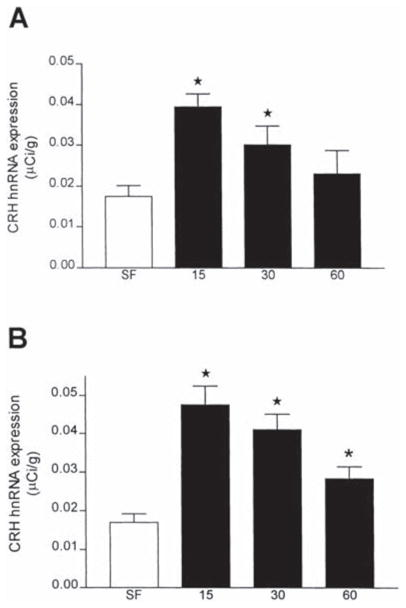
Abundance of CRH hnRNA in the PVN following cold stress. Densitometric analysis was performed on autoradiograms following in situ hybridization for CRH hnRNA in the PVN. Values are expressed as the means±S.E.M. of at least ten values from at least three brains per time-point. At both 6 (A) and 9 days (B) of age, there was greater than a two-fold increase in CRH hnRNA at 15 min after stress onset, and levels decreased at the 30 min time-point. One way ANOVA revealed a significant effect of treatment (F=8.53, P<0.0001). Post-hoc Bonferroni tests suggested that the 60 min time point values did not differ from the stress-free values for either group, although student’s t-test indicated significant (*) elevation in the 9-day-old group. Two way ANOVA demonstrated a strong effect of treatment (F=9.98, P<0.0001), and a trend towards an effect of age (F=2.81, P=0.096) that did not reach statistical significance.
3.3. CRH hnRNA in the PVN is rapidly upregulated also by minimal (handling) stress
Careful analysis of CRH hnRNA induction in the PVN demonstrated rapid gene activation prior to the onset of the cold-stress paradigm (within 2 min of removing the animals from the home cage, Fig. 4). CRH hnRNA levels of these animals were significantly higher than those in stress-free controls (211% of stress-free animals; P<0.05). Thus, whereas a strong challenge such as cold led to a robust and sustained activation of the CRH gene, even the minor perturbation associated with removing the animal to the laboratory sufficed to initiate CRH hnRNA transcription.
Fig. 4.
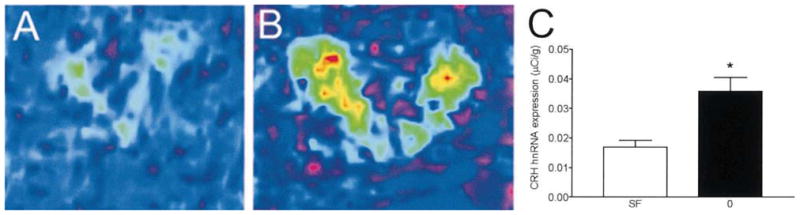
CRH hnRNA is rapidly induced by minimal stress. Nine-day-old rats were sacrificed within 2 min following the initial disturbance of their home cage, namely, after exposure only to human handling-stress. Color-coded quantitative images following in situ hybridization for detection of CRH hnRNA show increased signal over the PVN in animals briefly exposed to human handling and novel environment (B) compared with the stress-free controls (A). Densitometric analysis revealed a significant difference between these groups (P<0.05, C).
3.4. CREB phosphorylation is rapidly enhanced by mild (handling) stress in the medial parvocellular PVN of the immature rat
Because pCREB has been implicated in control of CRH gene transcription [9,16,32] and CRE-DNA binding activity and pCREB have been shown to be enhanced with stress in whole anterior hypothalamus of the immature rat [11], the presence and potential enhancement of pCREB immunoreactivity in parvocellular neurons of the PVN were determined. To consider the possibility that phosphorylated CREB may be involved in the activation of the CRH gene promoter, very early time-points after stress-onset were specifically investigated. Thus, pCREB was evaluated in PVN of ‘stress-free’ rats (deeply anesthetized within 45 s of entry to the animal facility), compared with animals transported to the laboratory, and thus subjected to handling stress (2 min from entry to the animal facility). In addition, medial parvocellular neurons, expressing CRH were distinguished from other, potentially non-stress-responsive PVN neurons. Considering the limitations of quantitative analysis of immunocytochemistry, the intensity or optical density of immunoreactivity signal was not evaluated, and only the number of cells expressing pCREB in sections run together in the immunocytochemistry procedure was evaluated (see Methods for criteria), in carefully matched sections from control and stressed animals.
Fig. 5 demonstrates the distribution of pCREB immunoreactive neurons at two coronal levels of the immature rat PVN. In general, pCREB expression was more abundant than in the adult (Fig. 6D) [20], and more prevalent in PVN compared with surrounding nuclei (Fig. 5A–D). Comparing the stress-free to the 2-min, handling stress group, the number of pCREB immunoreactive cells did not differ in the anterior parvocellular or the posterior magnocellular cell groups (Table 1). However, a significant increase in the number of pCREB immunoreactive neurons in the medial parvocellular PVN of stressed animals was observed, compared with stress-free controls (Table 1, Fig. 5). These data suggest that CREB phosphorylation was upregulated specifically in stress-responsive neurons.
Fig. 6.
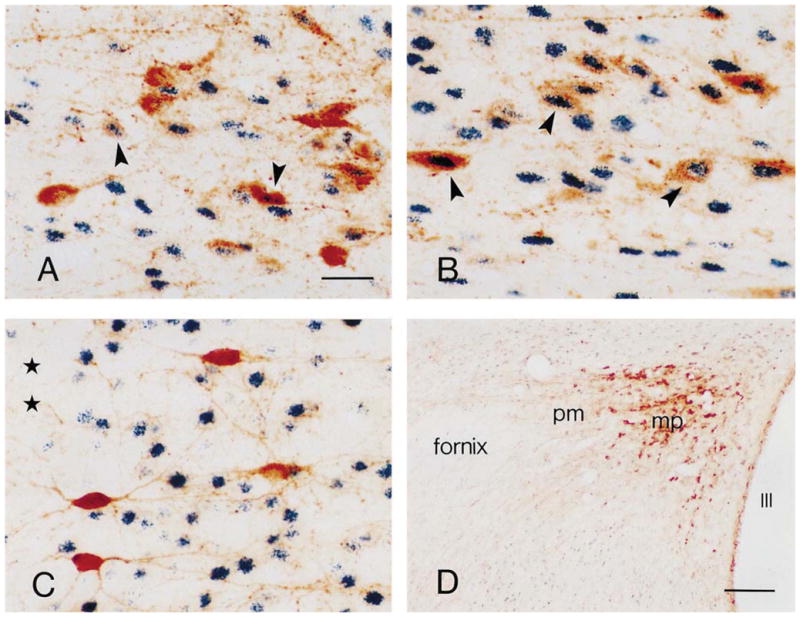
Phosphorylated CREB (pCREB) is primarily localized to CRH-expressing neurons in the paraventricular nucleus (PVN) of the immature rat. Panels A, B show CRH (brown) and pCREB (black–blue) co-expressing neurons (arrowhead) in the medial parvocellular (mp) PVN of stress-free (A) and 2-min stressed (B) immature rats. A higher number of CRH-pCREB co-labeled neurons are observed in the 2-min stress group. C. In contrast to PVN neurons, immature rat cortical CRH-expressing neurons (brown) rarely co-express pCREB (black–blue). D. In the stress-free adult PVN, pCREB immunoreactivity is weak in the posterior magnocellular (pm) sub-nucleus, and rare in the mp sub-nucleus in which CRH is richly expressed. pCREB expression is evident in other hypothalamic region. Asterisks in (C) indicate the cortical pial surface. III in (D) denotes the third ventricle. Scale bars=20 μm in A–C; 150 μm in D.
Table 1.
Quantitative analysis of phosphorylated CREB-immunoreactive cells in three subdivisions of the hypothalamic paraventricular nucleus in stress-free immature rats and 2-min after stress onset
| Regions | Stress-free | 2-min Stress |
|---|---|---|
| Anterior parvocellular (ap) | 197.8±14.7 | 200.6±16.6 |
| Medial parvocellular (mp) | 159.8±3.0 | 186.5±1.6* |
| Posterior magnocellular (pm) | 44.8±0.8 | 44.9±2.7 |
Values are pCREB immunoreactive neurons per 5 × 104 μm2 (ap, mp) or 2 × 104 μm2 (pm) of each PVN subdivision, and are presented as mean±S.E.M. of three rats. See Methods section for details of unbiased counting methods.
Statistically different from stress-free value, P=0.0013.
To evaluate whether the handling stress-induced increased phosphorylation of CREB was associated with CRH-expressing neurons, double-labeling immunocytochemistry was utilized. Within the medial parvocellular PVN, pCREB was frequently co-expressed with CRH (Fig. 6A and B). Comparing the stress-free and 2-min stress groups, a significantly higher number of CRH-pCREB double-labeled neurons was found in the medial parvocellular PVN of the mildly stressed animals (45.5±3.2 vs. 37.8±2.8, P=0.0024). In contrast to the PVN of the stress-free immature rat, no CRH-pCREB co-expressing neurons were observed in the medial parvocellular PVN of the stress-free adult (Fig. 6D). In addition, phosphorylation of CREB was rarely evident in cortical CRH-expressing neurons (Fig. 6C). Thus, rapid CREB phosphorylation in CRH-expressing medial parvocellular neurons of the immature rat was spatially and temporally specific.
3.5. Transcription of the CRH gene leads to significant increase of steady state CRH mRNA levels in the 9-day-old, but not 6-day-old immature rat
While CRH hnRNA production was observed in both groups of immature rats following cold exposure, analysis of CRH mRNA levels in the parvocellular PVN suggested that the magnitude of CRH mRNA production differed in these groups. Fig. 7 demonstrates a robust, 50% increase in steady-state CRH mRNA hybridization signal over the PVN of stressed 9-day-old rats compared with controls, whereas virtually no change of CRH mRNA stores were detected over PVN of 6-day-old stressed pups (P=0.69).
Fig. 7.
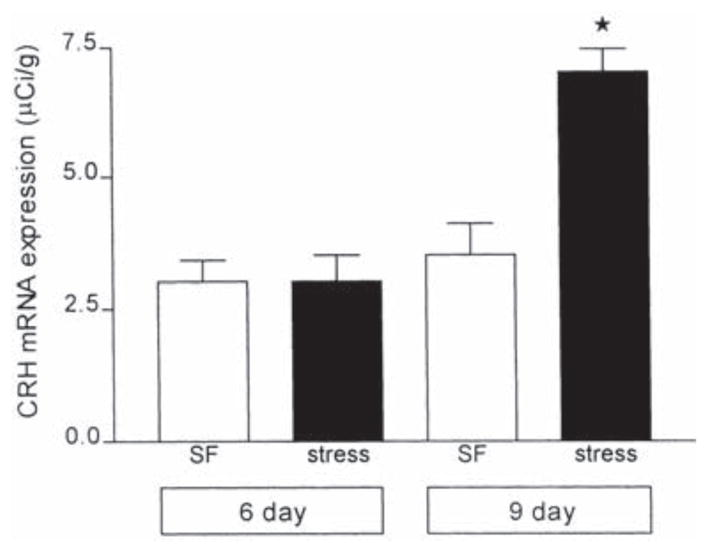
Relative abundance of CRH mRNA in the PVN of 6- and 9-day-old rats under stress-free control conditions and following cold exposure. Values of CRH mRNA levels were obtained from animals sacrificed 4 h after cold exposure (found previously to be optimal for maximal stress-induced CRH expression [12,44]) and are expressed as the mean±S.E.M. of at least five values per group. No significant difference was observed between controls and cold-stressed rats at 6 days of age. However, a significant increase in CRH mRNA (P<0.0005) was observed in 9-day-old rats subjected to cold, compared to age-matched controls.
4. Discussion
The major findings of the current studies are: (1) The CRH gene is strongly activated after cold exposure, a physiological stress in the immature (6- or 9-day-old) rat, and this activation is associated with a significant hormonal response. (2) Even the minimal challenge of human handling and a novel environment results in significant CRH hnRNA induction and CREB phosphorylation, specifically in the medial parvocellular cell groups of the PVN. These results demonstrate that transcription of the CRH gene is induced by mild as well as more rigorous challenges during the first two postnatal weeks, and may be regulated by CREB phosphorylation.
This study demonstrates hormonal and molecular neuroendocrine responses to a strong physiological challenge (cold) in the developing rat. These responses occurred in animals that were maintained in home cages with their mothers until their rapid removal to the laboratory for cold exposure. This finding is remarkable, given that it has generally been accepted that the stress response during the first two postnatal weeks (the stress-hyporesponsive period) is characterized by attenuated hormonal secretion and molecular regulation, and that these parameters are augmented by prolonged maternal deprivation [24,33,38].
The findings shown here are in accordance with a recent study [8] comparing CRH gene expression in maternally deprived and naive animals after saline administration. Similar to the results of the current study, these authors noted augmented CRH transcription in non-deprived rats. Indeed, the levels of CRH hnRNA were higher in that group, compared with maternally deprived cohorts. The data presented here are also in agreement with previous data from the authors’ and other laboratories, showing that the hyporesponsiveness of immature rat is selective rather than absolute [41,42,44]. Thus, exposure to a strong stimulus, such as cold, resulted in robust increase of plasma ACTH and corticosterone throughout the first two postnatal weeks (Fig. 1 [3]), and this hormonal response required CRH release, since it was abolished by both antisera [44] and receptor antagonist [3] of CRH.
The current study also highlights that CRH transcription in the immature rat may be induced by relatively mild challenges – namely, removing a rat pup from its cage to the laboratory. Dent et al. [8] reached a similar conclusion when discussing saline injection to their animals. It may be noted that mere transport of a rat to the laboratory, while involving human handling and a novel environment, is a painless and presumably even less stressful procedure than saline injection. Still, it led to activation of CRH transcription. Whether this procedure would also induce corticosterone secretion is not clear: Dent et al. [8] suggested that saline injection did not elevate plasma corticosterone in their experiments. In the experiments described here, animals were sacrificed within 2 min, a time-frame which may not be sufficient to fully activate adrenal steroid release. It should be noted, however, that the authors have previously demonstrated a significant elevation of plasma corticosterone in 10-day-old laboratory controls, i.e. rats left in the laboratory for 30–60 min, compared with stress-free controls, i.e. rats sacrificed in the animal facility within 45 s of their initial disturbance (2.19±0.56 vs. 0.70±0.08 μg/dl, P=0.01) [13].
The current study provides direct evidence for activation of the CRH gene by mild (handling) and more severe (cold) stress in the 6- and 9-day-old rat. Because expression of CRH hnRNA in stress-free controls was minimal, determination of CRH hnRNA proved a sensitive instrument for detecting induction of CRH gene expression. In addition, the rapid kinetics of CRH hnRNA expression provided a direct and thus more specific means for assessing the effects of stress on this gene.
It should be noted that while CRH gene expression, measured by hnRNA, was provoked by stressful challenges in both the 9- and 6-day-old rat, earlier work [44], as well as the data presented here, demonstrated increased steady state stores of CRH mRNA only in the older age group examined (9 days old). If transcription of the CRH gene is induced by stress both on postnatal days 6 and 9, then why were increased mRNA stores evident only on day 9? The profile of CRH hnRNA expression was similar for rats at 6 and 9 days of age (Figs. 2 and 3). In addition, the kinetics of induction of CRH hnRNA expression were comparable to those previously described in adult rats [15,19,20,27]. It is difficult to compare the magnitude of stress-induced CRH hnRNA increase between developing and adult rats because the stress-paradigms employed are age-specific. For example, acute cold stress is an effective stressor in the young rat but not in the adult [10]. Comparing PVN CRH hnRNA induction by stress as a percentage of the control, a ~300% increase at 15 min following stress was found in the present study and by Dent et al. [8] during the first 2 weeks of life, while an approximate 400% increase at 5 min following either stress [20] was described in adult rat. This indicates that the magnitude of CRH hnRNA induction by stress is comparable between the immature and adult PVN.
However, the correlation between the magnitude or kinetics of CRH hnRNA induction and enhancement of steady-state CRH mRNA levels has proven complex: previous time-course studies of CRH mRNA responses in 6- and 9-day-old rats demonstrated maximal CRH mRNA changes at 4 h [12,44]. In contrast, Dent et al. [8] reported enhanced CRH mRNA levels starting at 15 min after stress, in 6- and 12-day-old rats. The reason for the discrepancy is not clear, but may involve different challenges (saline vs. cold) or different CRH mRNA detection probes: our studies utilized deoxyoligonucleotide probes (60 mer) recognizing solely the regions coding for the 20 C-terminus amino acids of the CRH peptide.
It should be noted that similar discrepancies among groups and paradigms has been described in adult studies: Whereas Kovacs and Sawchenko found a marked induction of CRH hnRNA after ether stress [19], they failed to demonstrate any increase in CRH mRNA levels. In contrast, enhancement of both heteronuclear and messenger RNAs for CRH has been shown after restraint stresses [10,27]. Again, subtle differences in stress paradigms, in CRH hnRNA kinetics or in mRNA stability may account for these discrepancies. Among these, differential effects of age and stress on processing of RNA species provide an attractive mechanism: Age related changes in the rate of mRNA degradation have been noted [18]. This may occur due to differential use of the multiple stop codons in the CRH gene, or different lengths of the poly A+ tail [1,25].
In the present study, increased CRH hnRNA levels were evident already by 2 min from disturbance of rat pups, and levels of this transcript peaked at 15 min. While Dent et al., did not study any earlier time-points, their data indicate a decline in CRH hnRNA signal subsequent to 15 min, as found here. While no formal time course of pCREB expression was performed (and quantitative analysis of immunocytochemistry is undertaken with caution), increased numbers of pCREB immunoreactive cells confined to parvocellular PVN, and predominantly to CRH-expressing neurons, were clearly evident by 2 min from stress onset. This is consistent with phosphorylation of CREB prior to induction of transcription of the CRH gene, and supports the notion that CREB phosphorylation may be part of the mechanism of activation of the CRH gene promoter which contains a CRE.
It may be noted that many more pCREB immunoreactive neurons were apparent in the immature rat material (Fig. 5) compared with adult PVN (Fig. 6D). This is not surprising, based on established and emerging evidence for the roles of CREB in neuronal growth and differentiation [4].
In summary, this study demonstrates that hypothalamic CRH gene expression is rapidly activated by mild disturbance of the environment as well as cold stress during the first 2 weeks of life. The time-course of CRH hnRNA expression and of pCREB phosphorylation in specific, CRH expressing neurons of the parvocellular PVN suggest that pCREB may be involved in stress-induced transcription initiation at the CRH gene promoter of the immature rat. Overall, these findings demonstrate the integrity of the neuroendocrine stress response – including CRH gene activation – to environmental challenges during the first two postnatal weeks in the rat.
Acknowledgments
The authors are grateful to Mariam Eghbal-Ahmadi for technical assistance, and to Michele Hinojosa for excellent editorial support. This work was supported in part by NIH NS 28912 and NS 39307.
References
- 1.Adler GK, Rosen LB, Fiandaca MJ, Majzoub JA. Protein kinase-C activation increases the quantity and poly(A) tail length of corticotropin-releasing hormone messenger RNA in NPLC cells. Mol Endocrinol. 1992;6:476–484. doi: 10.1210/mend.6.3.1350054. [DOI] [PubMed] [Google Scholar]
- 2.Baram TZ, Lerner SP. Ontogeny of corticotropin releasing hormone gene expression in rat hypothalamus – comparison with somatostatin. Int J Dev Neurosci. 1991;9:473–478. doi: 10.1016/0736-5748(91)90033-i. [DOI] [PubMed] [Google Scholar]
- 3.Baram TZ, Yi SJ, Avishai-Eliner S, Schultz L. Development neurobiology of the stress response: multilevel regulation of corticotropin-releasing hormone function. Ann NY Acad Sci. 1997;814:252–265. doi: 10.1111/j.1749-6632.1997.tb46161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender R, Lauterborn J, Gall CM, Cariaga W, Baram TZ. Enhanced CREB phosphorylation in immature dentate gyrus granule cells indicates a specific role for CREB in granule cell differentiation. Eur J Neurosci. 2001;13:679–686. doi: 10.1046/j.1460-9568.2001.01432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilang-Bleuel A, Rech J, Linthorst ACE, Reul JMHM. Long-lasting activation of the cAMP-response element binding protein (CREB) in limbic brain structures following forced swimming. Proc Soc Neurosci. 1999;29:138.7. [Google Scholar]
- 6.Chen YC, Lei JL, Chen QS, Wang SL. Effect of physical training on the age-related changes of acetylcholinesterase-positive fibers in the hippocampal formation and parietal cortex in the C57BL/6J mouse. Mech Ageing Dev. 1998;102:81–93. doi: 10.1016/s0047-6374(98)00026-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Brunson K, Mueller MG, Cariaga W, Baram TZ. Immunocytochemical distribution of corticotropin-releasing hormone receptor type-1 (CRF1)-like immunoreactivity in the mouse brain: light microscopy analysis using an antibody directed against the C-terminus. J Comp Neurol. 2000;420:305–323. doi: 10.1002/(sici)1096-9861(20000508)420:3<305::aid-cne3>3.0.co;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dent GW, Smith MA, Levine S. Rapid induction of corticotropin-releasing hormone gene transcription in the paraventricular nucleus of the developing rat. Endocrinology. 2000;141:1593–1598. doi: 10.1210/endo.141.5.7455. [DOI] [PubMed] [Google Scholar]
- 9.Guardiola-Diaz HM, Boswell C, Seasholtz AF. The cAMP-responsive element in the corticotropin-releasing hormone gene mediates transcriptional regulation by depolarization. J Biol Chem. 1994;269:14784–14791. [PubMed] [Google Scholar]
- 10.Harbuz MS, Lightman SL. Responses of hypothalamic and pituitary mRNA to physical and psychological stress in the rat. J Endocrinol. 1989;122:705–711. doi: 10.1677/joe.0.1220705. [DOI] [PubMed] [Google Scholar]
- 11.Hatalski CG, Baram TZ. Stress-induced transcriptional regulation in the developing rat brain involves increased cyclic adenosine 3′,5′-monophosphate-regulatory element binding activity. Mol Endocrinol. 1997;11:2016–2024. doi: 10.1210/mend.11.13.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatalski CG, Guirguis C, Baram TZ. Corticotropin releasing factor mRNA expression in the hypothalamic paraventricular nucleus and the central nucleus of the amygdala is modulated by repeated acute stress in the immature rat. J Neuroendocrinol. 1998;10:663–669. doi: 10.1046/j.1365-2826.1998.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatalski CG, Brunson KL, Tantayanubutr B, Chen Y, Baram TZ. Neuronal activity and stress differentially regulate hippocampal and hypothalamic corticotropin releasing hormone expression. Neuroscience. 2000;101:571–580. doi: 10.1016/s0306-4522(00)00386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman JP, Schafer MK, Thompson RC, Watson SJ. Rapid regulation of corticotropin-releasing hormone gene transcription in vivo. Mol Endocrinol. 1992;6:1061–1069. doi: 10.1210/mend.6.7.1324419. [DOI] [PubMed] [Google Scholar]
- 15.Imaki T, Shibasaki T, Chikada N, Harada S, Naruse M, Demura H. Different expression of immediate-early genes in the rat paraventricular nucleus induced by stress: relation to corticotropin-releasing factor gene transcription. Endocr J. 1996;43:629–638. doi: 10.1507/endocrj.43.629. [DOI] [PubMed] [Google Scholar]
- 16.Itoi K, Horiba N, Tozawa F, Sakai Y, Sakai K, Abe K, Demura H, Suda T. Major role of 3′,5′-cyclic adenosine monophosphate-dependent protein kinase A pathway in corticotropin-releasing factor gene expression in the rat hypothalamus in vivo. Endocrinology. 1996;137:2389–2396. doi: 10.1210/endo.137.6.8641191. [DOI] [PubMed] [Google Scholar]
- 17.Itoi K, Seasholtz AF, Watson SJ. Cellular and extracellular regulatory mechanisms of hypothalamic corticotropin-releasing hormone neurons. Endocr J. 1998;45:13–33. doi: 10.1507/endocrj.45.13. [DOI] [PubMed] [Google Scholar]
- 18.Keiger CJ, O’Steen WK, Brewer G, Sorci-Thomas M, Zehnder TJ, Rose JC. Cortisol up-regulates corticotropin releasing factor gene expression in the fetal ovine brainstem at 0.70 gestation. Mol Brain Res. 1995;32:75–81. doi: 10.1016/0169-328x(95)00061-v. [DOI] [PubMed] [Google Scholar]
- 19.Kovacs KJ, Sawchenko PE. Regulation of stress-induced transcriptional changes in the hypothalamic neurosecretory neurons. J Mol Neurosci. 1996;7:125–133. doi: 10.1007/BF02736792. [DOI] [PubMed] [Google Scholar]
- 20.Kovacs KJ, Sawchenko PE. Sequence of stress-induced alterations in indices of synaptic and transcriptional activation in parvocellular neurosecretory neurons. J Neurosci. 1996;16:262–273. doi: 10.1523/JNEUROSCI.16-01-00262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak SP, Young EA, Morano I, Watson SJ, Akil H. Diurnal corticotropin-releasing hormone mRNA variation in the hypothalamus exhibits a rhythm distinct from that of plasma corticosterone. Neuroendocrinology. 1992;55:74–83. doi: 10.1159/000126099. [DOI] [PubMed] [Google Scholar]
- 22.Kwak SP, Morano MI, Young EA, Watson SJ, Akil H. Diurnal CRH mRNA rhythm in the hypothalamus: decreased expression in the evening is not dependent on endogenous glucocorticoids. Neuroendocrinology. 1993;57:96–105. doi: 10.1159/000126347. [DOI] [PubMed] [Google Scholar]
- 23.Levey AI, Bolam JP, Rye DB, Hallanger AE, Demuth RM, Mesulam MM, Wainer BH. A light and electron microscopic procedure for sequential double antigen localization using diaminobenzidine and benzidine dihydrochloride. J Histochem Cytochem. 1986;34:1449–1457. doi: 10.1177/34.11.2430010. [DOI] [PubMed] [Google Scholar]
- 24.Levine S. The ontogeny of the hypothalamic–pituitary–adrenal axis: the influence of maternal factors. Ann NY Acad Sci. 1994;746:275–288. doi: 10.1111/j.1749-6632.1994.tb39245.x. [DOI] [PubMed] [Google Scholar]
- 25.Lewis JD, Gunderson SI, Mattaj IW. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J Cell Sci Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 26.Lightman SL, Young WS. Influence of steroids on the hypothalamic corticotropin-releasing factor and preproenkephalin mRNA responses to stress. Proc Natl Acad Sci USA. 1989;86:4306–4310. doi: 10.1073/pnas.86.11.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma XM, Levy A, Lightman SL. Rapid changes in heteronuclear RNA for corticotrophin-releasing hormone and arginine vasopressin in response to acute stress. J Endocrinol. 1997;152:81–89. doi: 10.1677/joe.0.1520081. [DOI] [PubMed] [Google Scholar]
- 28.Rivest S, Laflamme N, Nappi RE. Immune challenge and immobilization stress induce transcription of the gene encoding the CRF receptor in selective nuclei of the rat hypothalamus. J Neurosci. 1995;15:2680–2695. doi: 10.1523/JNEUROSCI.15-04-02680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson BG, D’Angio LA, Pasieka KB, Majzoub JA. Preprocorticotropin releasing hormone: cDNA sequence and in vitro processing. Mol Cell Endocrinol. 1989;61:175–180. doi: 10.1016/0303-7207(89)90128-7. [DOI] [PubMed] [Google Scholar]
- 30.Sapolsky RM, Meaney MJ. Maturation of the adrenocortical stress response: neuroendocrine control mechanisms and the stress hyporesponsive period. Brain Res. 1986;396:64–76. doi: 10.1016/s0006-8993(86)80190-1. [DOI] [PubMed] [Google Scholar]
- 31.Sawchenko PE, Swanson LW. The organization of forebrain afferents to the paraventricular and supraoptic nuclei of the rat. J Comp Neurol. 1983;218:121–144. doi: 10.1002/cne.902180202. [DOI] [PubMed] [Google Scholar]
- 32.Seasholtz AF, Thompson RC, Douglass JO. Identification of a cyclic adenosine monophosphate-responsive element in the rat corticotropin-releasing hormone gene. Mol Endocrinol. 1988;2:1311–1319. doi: 10.1210/mend-2-12-1311. [DOI] [PubMed] [Google Scholar]
- 33.Smith MA, Kim SY, Van Oers H, Levine S. Maternal deprivation and stress induce immediate early genes in the infant rat brain. Endocrinology. 1997;138:4622–4628. doi: 10.1210/endo.138.11.5529. [DOI] [PubMed] [Google Scholar]
- 34.Swanson LW, Kuypers HG. The paraventricular nucleus of the hypothalamus: cytoarchitectonic subdivisions and organization of projections to the pituitary, dorsal vagal complex, and spinal cord as demonstrated by retrograde fluorescence double-labeling methods. J Comp Neurol. 1980;194:555–570. doi: 10.1002/cne.901940306. [DOI] [PubMed] [Google Scholar]
- 35.Swanson LW, Sawchenko PE, Rivier J, Vale WW. Organization of ovine corticotropin-releasing factor immunoreactive cells and fibers in the rat brain: an immunohistochemical study. Neuroendocrinology. 1983;36:165–186. doi: 10.1159/000123454. [DOI] [PubMed] [Google Scholar]
- 36.Swanson L, Simmons DM. Differential steroid hormone and neural influences on peptide mRNA levels in CRH cells of the paraventricular nucleus: a hybridization histochemical study in the rat. J Comp Neurol. 1989;285:413–435. doi: 10.1002/cne.902850402. [DOI] [PubMed] [Google Scholar]
- 37.Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- 38.Van Oers HJJ, de Kloet ER, Whelan T, Levine S. Maternal deprivation effect on the infant’s neural stress markers is reversed by tactile stimulation and feeding but not by suppressing corticosterone. J Neurosci. 1998;18:10171–10179. doi: 10.1523/JNEUROSCI.18-23-10171.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez DM. Stress and the developing limbic–hypothalamic–pituitary–adrenal axis. Psychoneuroendocrinology. 1998;23:663–700. doi: 10.1016/s0306-4530(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 40.Walker CD, Perrin M, Vale W, Rivier C. Ontogeny of the stress response in the rat: role of the pituitary and the hypothalamus. Endocrinology. 1986;118:1445–1451. doi: 10.1210/endo-118-4-1445. [DOI] [PubMed] [Google Scholar]
- 41.Walker CD, Scribner KA, Cascio CS, Dallman MF. The pituitary–adrenocortical system of neonatal rats is responsive to stress throughout development in a time-dependent and stressor-specific fashion. Endocrinology. 1991;128:1385–1395. doi: 10.1210/endo-128-3-1385. [DOI] [PubMed] [Google Scholar]
- 42.Walker CD, Tankosic P, Tilders FJ, Burlet A. Immunotargeted lesions of paraventricular CRF and AVP neurons in developing rats reveal the pattern of maturation of these systems and their functional importance. J Neuroendocrinol. 1997;9:25–41. doi: 10.1046/j.1365-2826.1997.00544.x. [DOI] [PubMed] [Google Scholar]
- 43.Yan XX, Toth Z, Schultz L, Ribak CE, Baram TZ. Corticotropin-releasing hormone (CRH)-containing neurons in the immature rat hippocampal formation: light and electron microscopic features and colocalization with glutamate decarboxylase and parvalbumin. Hippocampus. 1998;8:231–243. doi: 10.1002/(SICI)1098-1063(1998)8:3<231::AID-HIPO6>3.0.CO;2-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi SJ, Baram TZ. Corticotropin-releasing hormone mediates the response to cold stress in the neonatal rat without compensatory enhancement of the peptide’s gene expression. Endocrinology. 1994;135:2364–2368. doi: 10.1210/endo.135.6.7988418. [DOI] [PMC free article] [PubMed] [Google Scholar]


