Abstract
We described an association between a strain of the nematode Caenorhabditis briggsae, i.e. KT0001, and the bacteria Serratia sp. SCBI (South African Caenorhabditis briggsae isolate), which was able to kill the insect Galleria (G. mellonella). Here we show that the Serratia sp. SCBI lines the gut of the nematode, similar to the Heterorhabditis-Photorhabdus complex, indicating that the association is possibly internal. We also expand on the relevance of this tripartite, i.e. insect-nematode-bacteria, interaction in the broader evolutionary context and Caenorhabditis natural history.
Key words: Nematode-bacteria symbiosis, Caenorhabditis ecology, entomopathogenic Caenorhabditis, Caenorhabditis-Serratia complex
From three soil samples collected in three provinces of South Africa we isolated Caenorhabditis briggsae KT0001 and Serratia sp. SCBI through Galleria traps. Our subsequent laboratory experiments demonstrated that Caenorhabditis briggsae KT0001 was able to enter Galleria, overcome the insect's immune response, reproduce, and emerged from the insect cadaver as infective juveniles. Although the nematodes were able to readily enter the insect hemolymph, their ability to kill the insect depended on the bacterial strain they were cultivated on: C. briggsae KT0001 was not able to kill the insect when cured of SCBI and cultivated on E. coli OP50. This demonstrated that Serratia sp. SCBI was the arsenal used to kill the insect host. Further tests on ten wild strains of C. briggsae and four close Caenorhabditis species (C. elegans, C. remanei, Caenorhabditis sp. 5, and C. brenneri) revealed that this ability to enter the insect was not limited to C. briggsae KT0001. Instead all tested species/strains except C. briggsae DR1690 were able to enter Galleria, kill the insect and emerge. Similar to the South African strain these strains were able to kill the insect only when cultivated on Serratia sp. SCBI, indicating the nematode-bacterial association was necessary but non-specific. Such nematode-bacterial associations that result in the death of targeted insect are characteristic of the two archetypical entomopathogenic nematode-bacteria complexes—Steinernema-Xenorhabdus and Heterorhabditis-Photorhabdus.1
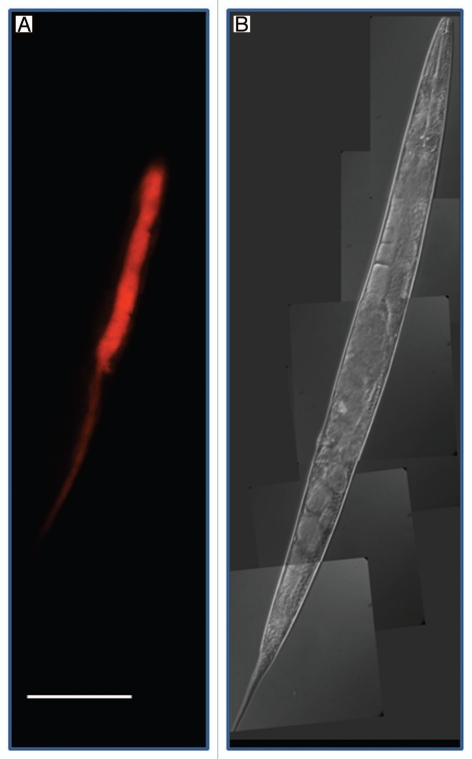
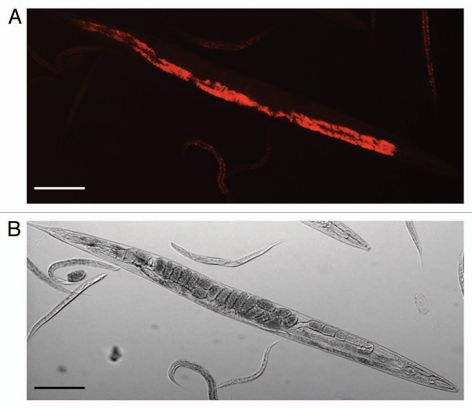
The C. briggsae-Serratia system we described is identical to the archetypical entomopathogenic nematode (EPN) systems in so far as the steps involved in the process and the end product of the interaction—death of the insect and use of its remains as source of nutrient for the nematode and bacteria. The specifics of symbiotic association between C. briggsae KT0001 and Serratia sp. SCBI in terms of whether it is obligatory or facultative is currently not known. Much in the same way as seen in the Photorhabdus-Heterorhabditis association, SCBI is seen lining the gut of the nematode C. briggsae KT0001 (Figs. 1 and 2). We have also observed that juveniles (L1) to carry Serratia sp. SCBI in females undergoing endotokia matricida, i.e. intra-uterine birth causing maternal death (data not shown). Therefore, Serratia sp. SCBI seems to be transferred to the next generation inside the mother. It is, however, only relatively mature juveniles (closer to vulva) that have the SCBI. These observations indicate that Caenorhabditis may use endotokia matricida as a pathway to pass on the bacterial symbionts to the next generation.
Figure 1.
Light microscopic pictures of Caenorhabditis briggsae KT0001. (A) Red Fluorescent Protein (RFP)-tagged Serratia sp. SCBI lining the gut of C. briggsae KT0001. We used plasmid pSPR that harbors the red fluorescent protein gene from DsRedExpress for fluorescent tagging. For a detailed description of the methodology see reference 25. (B) Differential Interference Contrast image of same worm used in (A). (Scale bar = 100µm).
Figure 2.
Light microscopic pictures of Caenorhabditis briggsae KT0001. (A) Red Fluorescent Protein (RFP)-tagged Serratia sp. SCBI lining the gut of C. briggsae KT0001. (B) Bright field image of same worm used in (A) (Scale bar = 100µm).
Our findings that Serratia sp. SCBI works with a number strains/species other than C. briggsae KT0001 in the lab to kill Galleria indicates that the C. briggsae-Serratia sp. SCBI association may not be obligatory, and this is a marked deviation from the well recognized EPN systems.2 Although the association seems to lack specificity, the ability of bacterial associates to colonize more than one worm species and vice versa is not novel to Serratia sp. SCBI.3 The fact that Serratia sp. SCBI causes accelerated death in insect larvae both when injected directly and delivered via nematodes is a clear evidence of its presence as a partner and not a mere staple in the nematode diet.4
It is evident that in order for two species to co-evolve into a co-adapted life three pivotal conditions have to be met:5 (1) Mutual tolerance: if two species can't coexist without killing each other, they can't form symbiotic associations as one would eventually drive the other out of the scenario. (2) Cost-benefit tradeoff: Although it is not always apparent or easy to directly quantify, association always imply costs as well as benefits to the involved parties. In the co-evolution of mutualistic associations, tolerance to one another may be necessary but it is not sufficient to sustain co-adaptive life style. For the co-evolution to persist and lead to co-adapted partners, the participants must remain closely associated and this is best achieved when the cost-benefit tradeoff is positive for both partners. In other words, unless both partners benefit from the association, co-adaptation through co-evolution can't succeed as in the absence of such positive stimulus there is nothing to bind the partners and sooner or later one species may easily defect from the association. (3) Uncompromised reproduction: Continued and unhindered reproduction in the proximity of one another is the third condition that must be met in order to guarantee the continuity of co-adaptation. Unless the partners can reproduce unencumbered in the presence of one another or there would be no continuity in the association.
Seen in light of these considerations, the association between C. briggsae KT0001 and Serratia sp. SCBI falls in an intermediate level in between the highly specific EPN associations and the transient presence of bacteria in bacterivore nematodes. Serratia sp. SCBI, while it has a formidable arsenal with about four to six non-ribosomal peptide synthase complexes, depending on the annotation system used, and around 200 virulence-associated genes and operons6 its toxin complexes are not as redundant as that of Photorhabdus, for example, which contains in excess of twenty such complexes.7
We recognize that where the association between C. briggsae KT0001 and Serratia sp. SCBI falls in the symbiosis continuum is key in its relevance to our understanding of how parasitism evolved.8 It is interesting to note that EPN, as used in the literature, is an exclusive term for those nematodes that use their symbiotic bacteria to kill insects. This definition leaves out all other insect parasitic nematodes such as the Mermithida, a group that as juveniles parasitizes insects but remain free living as adults. Following this narrow definition of EPN, current literature shows that, entomopathogenesis independently arose twice in the phylum Nematoda, i.e. families Steinernematidae and Heterorhabditidae.9 Nematodes and insects are two species-rich and ancient groups,10,11 which in this particular case have a tripartite interaction with an even older and more diverse group: the bacteria. These groups have had plenty of opportunity to co-exist, outcompete and co-evolve with one another. Differences in their biology, structural components, and mechanisms of insect killing point to the fact that the two widely accepted EPN lineages are the result of convergent evolution. But, they are not likely to have evolved these two complex life histories in a single step and there could be more EPN associations and that such associations should be viewed as a spectrum where the specific and highly co-evolved associations are at one end and the transient associations from the other end. The kind of facultative symbiosis we described between C. briggsae KT0001 and Serratia sp. SCBI is probably the transient and non-specific type,12 and this affords flexibility to the relationship. Necromenic association in nematodes, a case where one attaches and waits until its carrier host dies to use its cadaver, was suggested to be the step archetypical EPNs used towards parasitism through symbiosis with insect killing bacteria.8 In light of this, we find it fascinating that insect killing nematode-bacterial associations arose only twice in such a long period of time. Ours is one of a series of reports of insect-killing nematode-bacterial complexes other than Steinernema-Xenorhabdus and Heterorhabditis-Photorhabdus.13–15 However, the association we reported is unique because it involved C. briggsae, which is a congeneric with the widely used animal model species C. elegans.
In contrast to the tremendous scientific infrastructure built around the Caenorhabditis including several complete genome sequences and a rich functional genomics toolbox,16 our knowledge of Caenorhabditis ecology remains almost non-existent and we lack even the most basic understanding of its biology outside the laboratory.17 This represents a serious gap in our knowledge and a plausible reason why a large fraction of the Caenorhabditis genome remains functionally uncharacterized. For example, we know that in Caenorhabditis, the majority of gene knockouts have no discernable laboratory phenotype, and yet those genes are conserved for many millions of years. Also, the natural history of C. elegans still shows critical gaps. The initial assumption that it was a bacterivore, soil inhabiting worm has now given way to the recognition of this species as a colonizer of various microbe-rich habitats, away from the soil environment.17 Now there is overwhelming evidence on the association of Caenorhabditis species, including C. elegans, with invertebrates.18 Nonetheless, none of those reports showed this species to be entomopathogenic. Despite extensive resources and tools, lack of demonstrated ability to parasitize has created a reservation to use C. elegans as a model for parasitism in general.19 In fact some have advocated for the use of Heterorhabditis as the main model system for nematode parasitism because Heterorhabditis is a parasite and, interesting enough, it is phylogenetically closer to C. elegans20 than Steinernema-Xenorhabdus. Our findings provide relevant context to efforts to close some of those gaps that characterized Caenorhabditis natural history17 such that we hope, from now on, its association with insects will be explored in a different context, and the Caenorhabditis-Serratia system may serve as a model for understanding basic host-pathogen interactions and parasitic biology.21
Some Serratia kill C. elegans,16,22 but the interaction of various C. elegans strains with various Serratia strains may be more specific, suggesting genetic variation in host susceptibility and parasitic virulence.23 Under laboratory conditions it has been shown that C. elegans avoids some strains of Serratia when provided as food.24 Serratia sp. SCBI on the other hand was harmless when provided as food to the various Caenorhabditis species we tested. When culturing these nematodes on Serratia sp. SCBI it is very clear that the behavior of the nematodes is very different and they do not avoid Serratia sp. SCBI but in fact appear to be preferentially attracted to Serratia sp. SCBI.
Future Direction
We recognize a need to define EPN interactions with the emphasis that the primary co-evolution has to happen between the bacteria and the nematode. We want to use genetic techniques to identify mechanisms of interaction between Caenorhabditis and Serratia sp. SCBI. Questions we would like to address include: What pathways are critical to the relationship with Serratia sp. SCBI and not other bacteria? What nematode genes are critical to these interactions and how does the bacterium regulate mechanisms of killing to avoid killing the nematode partner? Considering the fact that there are strains of C. briggsae that do not reproduce in Galleria, and given the abundant genetic tools that are available for both Caenorhabditis and the partner bacteria, it is feasible to test the effect of every gene of the bacteria on the survival, reproduction and overall fitness of the nematode as an EPN. Also, we plan to devise ways of engineering the system to target specific insect pests or vectors of diseases or to conduct experimental evolution by subjecting the duo to facilitated coevolution under selective pressure to learn more about key events in the establishment of permanent and specific EPN partnership.
Acknowledgements
Eyualem Abebe was supported in part by the US Army Research Laboratory and US Army Research Office under Contract number W911N7-08-1-0402 and US National Science Foundation Award Number 0808632.
References
- 1.Adams BJ, Fodor A, Koppenhöfer HS, Stackebrandt E, Stock SP, Klein MG. Biodiversity and systematics of nematode-bacterium entomopathogens. Biol Control. 2006;37:32–49. [Google Scholar]
- 2.Stock SP. Insect-parasitic nematodes: From lab curiosities to model organisms. J Invert Pathol. 2005;89:57–66. doi: 10.1016/j.jip.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Chapuis E, Emelianoff V, Paulmier V, Le Brun N, Pagès S, Sicard M, et al. Manifold aspects of specificity in a nematode bacterium mutualism. J Evol Biol. 2009;22:2104–2117. doi: 10.1111/j.1420-9101.2009.01829.x. [DOI] [PubMed] [Google Scholar]
- 4.Abebe E, Jumba M, Bonner K, Gray V, Morris K, Thomas WK. An entomopathogenic Caenorhabditis briggsae. J Exp Biol. 2010;213:3223–3229. doi: 10.1242/jeb.043109. [DOI] [PubMed] [Google Scholar]
- 5.Roughgarden J. The theory of coevolution. In: Futuyama DJ, Slatkin M, editors. Coevolution. Sunderland, MA: Sinauer Associates Inc. Publishers; 1983. pp. 33–64. [Google Scholar]
- 6.Abebe-Akele F, Cooper V, Tisa L, Abebe E, Thomas WK. Complete genome sequence of an entomopathogenic Serratia isolated from Caenorhabditis briggsae in the wild and its comparative genomic analysis with other members of the genus Serratia and known entomopathogens of the genera Photorhabdus and Xenorhabdus. Genome Res. 2011 Submitted. [Google Scholar]
- 7.Duchaud E, Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 8.Sudhaus W. Die mittels symbiontischer Bakterien entomopathogenen Nematoden Gattungen Heterorhabditis and Steinernema sind keine Schwestertaxa. Verh Deutsch Zool Ges. 1993;86:146. (Ger). [Google Scholar]
- 9.Dorris M, De Ley P, Blaxter ML. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol Today. 1999;15:188–193. doi: 10.1016/s0169-4758(99)01439-8. [DOI] [PubMed] [Google Scholar]
- 10.Meldal BHM, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol. 2007;42:622–636. doi: 10.1016/j.ympev.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 11.Bologna MA, Oliverio M, Pitzalis M, Mariottini P. Phylogeny and evolutionary history of the blister beetles (Coleoptera, Meloidae) Mol Phyl Evol. 2008;48:679–693. doi: 10.1016/j.ympev.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 12.Perru O. Cooperation strategies, signals and symbiosis. C R Biologies. 2006;329:928–937. doi: 10.1016/j.crvi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Young-Keun Y, Park HW, Shrestha S, Seo J, Kim YO, Shin C, et al. Identification of two entomopathogenic bacteria from a nematode pathogenic to the oriental beetle, Blitopertha orientals. J Microbiol Biotechnol. 2007;17:968–978. [PubMed] [Google Scholar]
- 14.Zhang C-X, Young S-Y, Xu M-X, Sun J, Liu H, Liu J-R, et al. Serratia nematodiphila sp. nov., associated symbiotically with the entomopathogenic nematode Heterorhabditidoides chongmingensis (Rhabditida: Rhabditidae) Int J Syst Evol Microbiol. 2009;59:1603–1608. doi: 10.1099/ijs.0.65718-0. [DOI] [PubMed] [Google Scholar]
- 15.Ye W, Torres-Barragan A, Cardoza YJ. Oscheius carolinensis n. sp. (Nematoda: Rhabditidae), a potential entomopathogenic nematode from vermicompost. Nematology. 2010;12:121–135. [Google Scholar]
- 16.WormBase. http://www.wormbase.org/
- 17.Félix M-A, Braendle C. The natural history of Caenorhabditis elegans. Curr Biol. 2010;20:965–969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 18.Caswell-Chen EP, Chen J, Lewis EE, Douhan GW, Nadler SA, Carey JR. Revising the standard wisdom of C. elegans natural history: ecology of longevity. Sci Aging Knowl Environ. 2005;40:pe30. doi: 10.1126/sageke.2005.40.pe30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geary TG, Thompson DP. Caenorhabditis elegans: how good a model for veterinary parasites? Vet Parasitol. 2001;101:371–386. doi: 10.1016/s0304-4017(01)00562-3. [DOI] [PubMed] [Google Scholar]
- 20.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol. 2007;17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 21.Lewis EE, Campbell J, GriYn C, Kaya H, Peters A. Behavioral ecology of entomopathogenic nematodes. Biol Contr. 2006;38:66–79. [Google Scholar]
- 22.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 23.Schulenburg H, Ewbank JJ. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol Biol. 2004;4:49. doi: 10.1186/1471-2148-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradel E, Zhang Y, Matsuyama T, Bargmann CI, Ewbank J. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poltak SR, Cooper SV. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5:369–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]