Abstract
Retinoic acid inducible gene I (RIG-I) is a pattern recognition receptor (PRR) responsible for detection of nucleic acids from pathogens in the cytoplasm of infected cells and induction of type I interferon (IFN). RIG-I-specific pathogen associated molecular patterns (PAMPs) are characterized by RNA molecules with a 5′-triphosphate (5′-ppp) group and partial double-stranded composition. Although many RNA molecules capable of activating RIG-I have been described, the exact nature of viral RNAs that are responsible for triggering RIG-I activity during the course of an infection has not been extensively explored and the specificity of RIG-I for various viral RNA molecules remains largely unknown. By examining endogenous RIG-I/RNA complexes in influenza virus- and Sendai virus-infected cells we were able to identify viral RNA molecules that specifically associated with RIG-I during infection. We showed that in Sendai virus-infected cells, RIG-I specifically and preferentially associated with the copy-back defective interfering (DI) particle RNA and not with the full-length Sendai virus genome or Sendai virus encoded mRNAs. In influenza virus-infected cells RIG-I also preferentially associated with DI RNAs as well as with the shorter genomic segments.
Key words: RIG-I, Sendai, influenza, PAMP, PRR
RIG-I is a cytoplasmic pattern recognition receptor that plays an essential role in intracellular innate immunity to RNA virus infections.1,2 Located at the top of the signaling cascade that leads to production of IFN and other proinflamatory cytokines, RIG-I is critical for detection of replicating viruses and initiation of cellular antiviral responses. The physiological importance of this antiviral sensor is supported by the increased susceptibility of RIG-I knockout mice to infections with numerous RNA viruses.3 The molecular mechanism of RIG-I activation has been the subject of intense research since the discovery of this pivotal sensor. Both biochemical and structural studies have identified a 5′-triphosphate (5′-ppp) group on an RNA molecule as a unique and specific ligand for RIG-I.4–7 The presence of the 5′-ppp appears to be very important for RIG-I activation, as dephosphorylated viral RNA will no longer induce a RIG-I mediated IFN response.5 Indeed some viruses have been shown to enzymatically remove their 5′-ppp in order to avoid recognition by RIG-I.8 Other viruses are thought to hide this important PAMP by either tight encapsidation of viral genomic RNA or replication in privileged compartments inaccessible to RIG-I. In addition to the 5′-ppp, a bluntended double-stranded RNA component directly adjacent to the 5′-ppp also appears to be important for RIG-I activation, as short single-stranded RNAs even in the presence of a 5′-ppp are not able to induce RIG-I signaling.9,10 Importantly, single-stranded viral RNAs such as genomes of many viruses, as well as leader and trailer RNAs of the Mononegavirales family, have not been formally ruled out as activators of RIG-I and it is possible that some partial base-paired RNA composition within these molecules could function as a RIG-I PAMP. In addition to the 5′-triphosphate group and base-paired RNA composition, RNA sequence has also been implicated in having a role in RIG-I activation. Specifically 5′-ppp-containing RNA, rich in U residues, has been shown to be an especially potent stimulator of RIG-I responses; however, this activation seems to be very context-dependent as some U-rich RNAs do not act as a strong RIG-I inducers.11,12 Double-stranded RNA molecules that do not possess a 5′-ppp have also been shown to activate RIG-I, although it appears that the length of the RNA plays an important role in the magnitude of the response, as RNAs that are too short or too long fail to trigger the sensor.5,6,13,14 Interestingly proteolytic cleavage analysis of polyI:C (a double-stranded RNA mimic) bound RIG-I revealed that RIG-I undergoes a different type of conformational change when bound to poly I:C in comparison to 5′-ppp RNA, suggesting that the mechanism of activation by these two types of ligands might be fundamentally different.4 Despite vast information as to what types of synthetic RNAs can activate RIG-I in cells and in vitro, the question of which viral RNAs are responsible for activating this sensor during an infection has not been thoroughly explored.
In our work we wanted to address this important question by analyzing endogenous RIG-I/RNA complexes generated during a virus infection.15 We chose to study Sendai Cantell virus and influenza PR8 ΔNS1 virus based on their very different life cycles and known ability to induce high amounts of IFN through the RIG-I pathway.3,16–18 Sendai virus is a prototypical member of the Paramyxoviridae family: a single-stranded, non-segmented, negative sense RNA virus with a cytoplasmic replication cycle. The Cantell strain of Sendai virus is a very well characterized potent inducer of the IFN response due to its tendency to accumulate large amounts of defective interfering particles (DIs).17 DI particles are subgenomic viral RNA species generated through mistakes in virus replication. DI RNAs have a replication advantage over full length genomes and therefore inhibit virus replication through sequesteration of viral polymerase complexes.19
In order to identify which RNA molecules activate RIG-I in the course of a Sendai virus infection we infected A549 human lung carcinoma cells with Sendai virus and isolated RIG-I/RNA complexes from infected cells by immunoprecipitation. Examination of RIG-I associated RNA by deep sequencing analysis revealed that only the copy-back DI RNA specifically associated with RIG-I at both early (4 hpi) and late (24 hpi) time-points in infection. Interestingly, we could not detect specific binding of full-length Sendai genome to RIG-I at either time during infection. In a recent study Rehwinkel et al. reported that RIG-I associates with the genome of Sendai virus.20 However since the approaches used to identify Sendai RNA such as primer extension and size fractionation would have picked up both full-length virus genome as well as DI RNA it is possible that the data from this study agrees with our findings. Since Sendai genomic RNA and DI RNA share identical sequence in the 5′ end of the genome it can be difficult to tell the two species apart based on conventional assays illustrating the advantage of an unbiased approach such as deep sequencing. However it is also possible that differences in virus stocks, cells and experimental approaches might account for the differing observations in the two studies. The question of whether a full-length genome of a paramyxovirus can and does function as a RIG-I PAMP during infection remains to be answered. The reason for preferential DI binding by RIG-I is not clear, although it is possible that the DI RNA (a molecule structurally distinct from the full-length genome) presents a much better ligand for RIG-I and may simply outcompete the full-length genome for binding. Unlike the Sendai virus genome, the DI genome has a 92 nt base-paired region adjacent to the 5′-triphosphate group. Since length and dsRNA composition directly adjacent to the 5′-ppp have both been shown to play an important role in activation of RIG-I, perhaps it is not surprising that the DI molecule presents a better activator of this sensor than the full-length genome of Sendai virus. It is also possible that the full-length genome of a paramyxovirus does not function as a RIG-I PAMP even in the absence of DIs, based on its length, structure, or encapsidation. It will be very interesting to see what types of RNA molecules (if any) will interact with and activate RIG-I during infection with a virus that does not contain or produce high amounts of DI RNAs. Our observation that Sendai DI RNA functions as a specific RIG-I ligand might partially explain the known association between accumulation of DI molecules and induction of an IFN response.17
Examination of RIG-I associated RNA from influenza virus-infected cells using the same approach revealed that all influenza virus RNA segments specifically interacted with RIG-I. This is consistent with experiments using exogenously expressed RIG-I.20 However in our study we observed differences between the relative extent of this association among the different influenza segments. The NS segment and M segments, as well as subgenomic DI RNAs generated from the PB1 and PA segments bound RIG-I to a greater extent than other influenza RNAs. It is interesting that again, in the context of a very different virus we found DI RNA species to preferentially interact with RIGI. Unlike the Sendai DI RNAs, influenza virus DI genomes are generated by internal deletions of genomic RNA. Therefore influenza DI RNAs have identical ends to the full-length genome, but are simply shorter in length. Since the terminal end structure of all influenza segments should be fairly similar, we concluded that one common factor to all RNAs that preferentially interacted with RIG-I was their relatively short length (between 500 and 1000 nt). It is likely that other factors present during an infection are important determinants of which molecules will interact with RIG-I; these factors might include localization and accessibility of RNA to RIG-I. Since influenza virus replicates in the nucleus and RIG-I recognition takes place in the cytoplasm, the localization of various influenza RNAs at different time points in the replication cycle could play an important role in which viral RNA species will act as PAMPs for RIG-I.
The results of our experiments clearly illustrate that there is a large difference in RIG-I binding between virally encoded RNAs present during an infection cycle (Fig. 1). It is very intriguing that subgenomic DI RNAs were found to specifically and preferentially associate with RIG-I in the context of infections with two different RNA viruses. Although DI molecules, especially those of the Mononegavirales species have long been known to be associated with a strong IFN response, this was assumed to be due to a shift in balance between viral PAMPs and virally encoded IFN antagonist proteins. The conclusions of our work indicate that viral DI RNAs might function as more potent PAMPs than other viral RNA species such as the full-length genomes of Mononegavirales. Interestingly both the NS1 segment of influenza virus and the 5′ end of Sendai virus genome (sequence corresponding to DI molecule) were predicted to be potent activators of RIG-I based on their rich U composition.11 Although we did not find specific U enrichment in other RNAs that preferentially associated with RIG-I it is still possible that sequence composition might play a role in RIG-I induction. Our work also raises a possibility that mutant viruses known to induce high amounts of IFN do so partially because of generation of aberrant PAMPs not present in wildtype virus infections. The identification of viral DI RNAs as preferential ligands for RIG-I leads to an interesting question of what role the generation of these molecules plays in IFN induction in vivo during a natural virus infection.
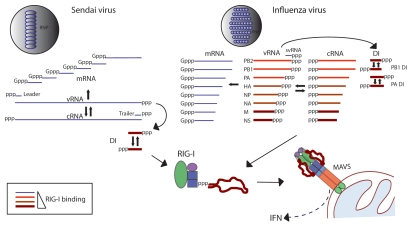
Figure 1.
Association of RIG-I with influenza and Sendai virus RNAs in infected cells. Viral RNAs produced in the course of infection by Sendai and influenza viruses are shown. These RNAs include genome (vRNA), antigenome (cRNA), mRNAs, DI RNAs, small viral RNAs (svRNA) of influenza virus24–26 as well as leader and trailer RNAs produced by Sendai virus. RNAs that we have found to associate with RIG-I during infection are depicted in red, with darker color and thicker lines representing greater extent of RIG-I association.
Since the discovery of 5′-ppp RNA as a RIG-I specific PAMP, the potential use of this RNA as a vaccine adjuvant has begun to be explored.21,22 In a recent report Easton et al. showed that DI containing influenza virus is able to provide heterologous protection against pneumovirus infection in mice in an IFN-dependent manner further supporting the possible clinical use of influenza DIs as an IFN inducing agent.23 Triphosphate-modified siRNAs were also shown to be potent RIG-I inducers in tumor cells leading to apoptosis of these cells in vivo, highlighting a potential role of these molecules as anti-cancer therapeutics.24 The identification of natural viral DI molecules as preferential RIG-I ligands now allows for more rational design of RIG-I-specific agonists for use as vaccine adjuvants or modulators of innate immune responses. Further characterization of which structural components of DI molecules are responsible for preferential RIG-I binding and stimulation will lead to a better understanding of how this sensor is induced in vivo and allow for more effective therapeutic design of RIG-I specific activators and inhibitors.
Acknowledgements
Work in the laboratory of AG-S is being supported by NIH grants R01AI46954, P01AI58113, P01AI82325, U01AI70469, U19AI62623, U19AI83025, the Northeast Biodefense Center U54-AI057158-Lipkin and by CRIP (Center for Research in Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Research and Surveillance, HHSN266200700010C. A.B. is supported by National Institute of Allergy and Infectious Diseases Training Program in Mechanisms of Virus-Host Interactions (2T32AI007647-11).
References
- 1.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 2.Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 4.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–1272. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, et al. Recognition of 5′ triphosphate by RIG-I Helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 15.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 17.Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, et al. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 19.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547–555. doi: 10.1016/0092-8674(76)90223-3. [DOI] [PubMed] [Google Scholar]
- 20.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarthy KV, Bonoiu AC, Davis WG, Ranjan P, Ding H, Hu R, et al. Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc Natl Acad Sci USA. 107:10172–10177. doi: 10.1073/pnas.0914561107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjan P, Jayashankar L, Deyde V, Zeng H, Davis WG, Pearce MB, Bowzard JB, et al. 5′PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol J. 7:102. doi: 10.1186/1743-422X-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easton AJ, Scott PD, Edworthy NL, Meng B, Marriott AC, Dimmock NJ. A novel broad-spectrum treatment for respiratory virus infections: Influenza-based defective interfering virus provides protection against pneumovirus infection in vivo. Vaccine. 2011;29:2777–2784. doi: 10.1016/j.vaccine.2011.01.102. [DOI] [PubMed] [Google Scholar]
- 24.Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 25.Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. 107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. MBio. 1:e00204–e00210. doi: 10.1128/mBio.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]