Abstract
Heterogeneous DNA methylation leads to difficulties in accurate detection and quantification of methylation. Methylation-sensitive high resolution melting (MS-HRM) is unique among regularly used methods for DNA methylation analysis in that heterogeneous methylation can be readily identified, although not quantified, by inspection of the melting curves. Bisulfite pyrosequencing has been used to estimate the level of heterogeneous methylation by quantifying methylation levels present at individual CpG dinucleotides. Sequentially combining the two methodologies using MS-HRM to screen the amplification products prior to bisulfite pyrosequencing would be advantageous. This would not only replace the quality control step using agarose gel analysis prior to the pyrosequencing step but would also provide important qualitative information in its own right. We chose to analyze DAPK1 as it is an important tumor suppressor gene frequently heterogeneously methylated in a number of malignancies, including chronic lymphocytic leukemia (CLL). A region of the DAPK1 promoter was analyzed in ten CLL samples by MS-HRM. By using a biotinylated primer, bisulfite pyrosequencing could be used to directly analyze the samples. MS-HRM revealed the presence of various extents of heterogeneous DAPK1 methylation in all CLL samples. Further analysis of the biotinylated MS-HRM products by bisulfite pyrosequencing provided quantitative information for each CpG dinucleotide analyzed, and confirmed the presence of heterogeneous DNA methylation. Whereas each method could be used individually, MS-HRM and bisulfite pyrosequencing provided complementary information for the assessment of heterogeneous methylation.
Key words: MS-HRM, pyrosequencing, digital PCR, heterogeneous DNA methylation, DAPK1, chronic lymphocytic leukemia
Introduction
DNA methylation changes in cancer have been considered to have great potential as a source of locus-specific biomarkers that can be used in early detection and monitoring of cancer.1,2 This promise is largely yet to be fulfilled as DNA methylation, particularly in the cancer context, is often heterogeneous in nature [e.g., CDKN2B (p15),3 DAPK1,4 MGMT,5 PROX1,6 and PTCH7]. This means that for a given region, multiple epialleles, each with a different pattern of methylated and unmethylated CpG dinucleotides, can co-exist. Quantification of heterogeneous methylation remains challenging as each methodology has its strengths and limitations (reviewed in Mikeska et al.8). Many techniques used to investigate DNA methylation, especially those that are based on methylation-specific PCR (MSP), assume that DNA methylation is homogeneous, making data obtained from heterogeneously methylated regions difficult to interpret.
Methylation-sensitive high resolution melting (MS-HRM),9 is capable of analyzing homogeneous methylation in a semi-quantitative manner. This is not the case for heterogeneous methylation, due to the complex population consisting predominantly of heteroduplexes formed between complementary strands differing only at a few CpG positions. This results in a broadened melting profile that only allows a qualitative interpretation.10 However, whereas heterogeneous methylation is not quantifiable, it is nevertheless readily recognizable by its characteristic melting profiles. Thus, MS-HRM can be used either in its own right to score for methylation or as a screening step to select those amplification products requiring more quantitative investigation, usually by sequencing methodologies.
Bisulfite pyrosequencing is a sensitive and quantitative technique that assesses the average methylation at each individual CpG dinucleotide within a given amplicon.11–13 Bisulfite pyrosequencing has thus been used for the investigation of heterogeneously methylated loci.14–16
Here we present a combined MS-HRM and bisulfite pyrosequencing approach that identifies heterogeneous methylation in the MS-HRM step and then quantifies the levels of methylation detected at the pyrosequencing step. As the combined approach conforms to assay design criteria for both methodologies,17,18 the PCR amplicons generated from MS-HRM can be used directly for quantitative DNA methylation analysis by bisulfite pyrosequencing without the need for the design of a new PCR assay.
We chose to analyze the death associated protein kinase 1 (DAPK1) promoter in chronic lymphocytic leukemia (CLL) samples, as DAPK1 is an important gene whose promoter is often heterogeneously methylated in cancer.19 It is frequently methylated in a variety of solid tumors20–22 and hematological malignancies.19,20,23 DAPK1 is often methylated in sporadic CLL, where the estimates of the proportion of patients showing promoter methylation (e.g., Katzenellenbogen et al. (1/11),24 Seeliger et al. (16/32),25 Raval et al. (64/65),19 Rossi et al. (8/30)26) and the estimate of the degree of methylation vary widely due to the different methodologies used. DNA methylation analysis using methods that take heterogeneous methylation into account is more likely to provide an accurate picture of the methylation at this locus.
Results
The effect of 5′-end biotinylation of the PCR primers on melting patterns.
Biotinylation of one of the PCR primers is necessary for pyrosequencing analysis. This allows the same PCR product to be sequentially analyzed by MS-HRM and bisulfite pyrosequencing. Biotinylation of either the forward or reverse PCR primers was shown to have essentially no effect on the melting behavior of the panel of DNA methylation standards compared to the case where neither primer is biotinylated (Fig. 1).
Figure 1.
The effect on melting behavior of 5′-end biotin labeling of PCR primers. DNA methylation standards (100, 50, 25, 5 and 0% (WGA) methylation) were run in duplicate using primers (1) without biotin labeling (black curves), (2) with a biotin labeled forward primer (red curves) and (3) with a biotin labeled reverse primer (green curves). There does not appear to be any significant difference in melting behavior introduced by either biotin label. Replicates are grouped into a single line for clarity.
Methylation-sensitive high resolution melting (MS-HRM).
MS-HRM for DAPK1 was performed on the mononuclear cell fraction from ten chronic lymphocytic leukemia (CLL) patients. The mononuclear fraction was not enriched and thus contained both CLL cells and normal cells. The MS-HRM assay used analyzed nine CpG dinucleotides within a 106 bp amplicon. The analyzed region is congruent with that generally studied in the literature and first analyzed by Katzenellenbogen et al.24
All ten CLL samples showed heterogeneous DNA methylation as can be seen by their melting profiles which do not conform to any of the methylation controls (Fig. 2A and B). These altered melting profiles are caused by heteroduplex formation between closely related single complementary DNA strands.10 Because of the enormous possible variation in the patterns and copy number of heterogeneously methylated templates, and the earlier melting temperature of the heteroduplexes, the resulting melting profile can only be interpreted in a qualitative manner.
Figure 2.
MS-HRM of the DAPK1 promoter for 10 CLL samples. (A) Normalized HRM curves. The DNA methylation standards of 0 (WGA), 25, 50 and 100% methylation are indicated. All amplicons from the CLL samples begin melting before the unmethylated control as can be seen by the earlier drop in fluorescence. Only samples 2 and 8 continue to melt after the unmethylated control has finished melting. (B) Tm plot (negative first derivative of the HRM curves). The broader peaks in the CLL samples that begin before the unmethylated control correspond to the earlier melting seen in (A), and result from heteroduplex formation. Only samples 2 and 8 encroach into the area under the peaks corresponding to methylation indicating the presence of more highly methylated templates.
The melting curve of sample 3 does not cross over into the methylated area but shows an earlier melt compared to the unmethylated control (Fig. 2B). This is typically caused by the formation of heteroduplexes between sequences that have comparatively low levels of DNA methylation.
Sample 2 contains a fraction of PCR products with the highest melting temperature (Fig. 2B). We would thus expect this sample to contain the most heavily methylated epialleles. Sample 8 has a similar late melting profile (Fig. 2B), but as the amplicons finish melting just before those of sample 2, we expect the most heavily methylated epialleles to not be as methylated as those in sample 2.
The other samples have melting profiles that finish melting shortly after the unmethylated control (Fig. 2B). This is consistent with low to moderate levels of DNA methylation that is also heterogeneous.
To complement the qualitative information from the melting profiles, bisulfite pyrosequencing was used to assess the quantitative DNA methylation information.
Bisulfite pyrosequencing.
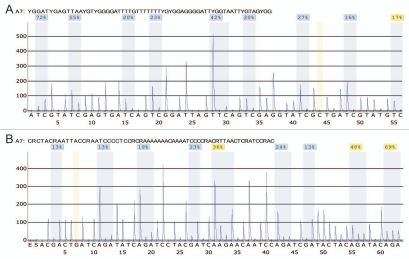
The DNA methylation values obtained from each pyrogram for each CpG position analyzed were very similar for both pyrosequencing directions (Figs. 3 and 4). The mean difference in methylation for the forward and reverse directions at each CpG position across the 10 CLL samples was in the range of 2–5% with a maximum deviation of 9%. Pyrosequencing of the biotinylated MS-HRM products confirmed the presence of heterogeneous DNA methylation patterns within the populations of all of the MS-HRM amplicons generated during PCR amplification (Fig. 3).
Figure 3.
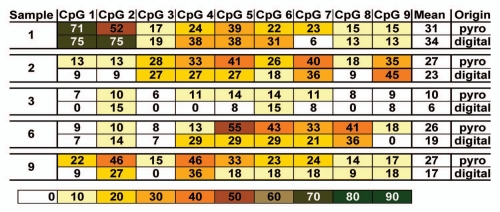
Bisulfite pyrosequencing data. The MS-HRM products of the ten CLL samples were analyzed by bisulfite pyrosequencing. Pyrosequencing was performed on both the forward and reverse strands, following MS-HRM using one or the other of the biotin-labeled primers. The methylation percentage measured for each CpG dinucleotide obtained in each sequencing direction, the calculated means for each CpG dinucleotide of both sequencing reactions and the overall average calculated from all nine CpG positions of a given sample are shown. The individual methylation ratios as measured in both sequencing directions for each CpG position are similar. The heterogeneous methylation at an overall population level is revealed, with the average methylation level at each CpG dinucleotide given.
Figure 4.
Representative pyrograms. DNA methylation level measured by bisulfite pyrosequencing of sample one in the forward (A) and reverse direction (B), respectively. The peaks used to determine the methylation ratio at each CpG dinucleotide are highlighted by light blue shading. The calculated methylation percentage for each CpG position is provided above the highlighted area. Nucleotides 44 and 7 are highlighted by light yellow shading in (A and B), respectively, and indicate control peaks to estimate incomplete bisulfite conversion. In (A), the CpG dinucleotides are analyzed from CpG 1 to CpG 9, whereas in (B), the CpG dinucleotides are analyzed from CpG 9 to CpG 1 because of the reverse sequencing of the upper strand of the PCR product.
Sample 3 shows the lowest level of average methylation (mean value of 10% across all CpG dinucleotides analyzed) (Fig. 3). The low amount of methylation measured for each CpG position can be explained either by the presence of very few heavily methylated epialleles in a large background of unmethylated alleles or by the presence of epialleles which have only a smaller number of CpG dinucleotides methylated across the entire amplicon. The melting profile from the MS-HRM data indicates the latter is the case. Since the levels detected are quite low, it is currently unclear as to whether any of this methylation is above the background level.
Samples 1, 2, 4, 6, 7, 8 and 9 show similar mean DNA methylation levels (values in the range of 22% to 31% across all CpG dinucleotides analyzed) (Fig. 3). However, this is where the similarities end. The methylation levels obtained for each of the nine CpG dinucleotides interrogated reveal differences across all the samples, and while there is a hint of a pattern of CpG positions that seem more prone to methylation, this is not consistent across all of the samples. There does appear to be a minimum consensus region containing CpG positions 5, 6 and 7 (Fig. 3). However, increased methylation levels are not restricted to certain CpG dinucleotides. For example, sample 1 shows high methylation at CpG dinucleotides 1 and 2, whereas sample 2 shows elevated methylation levels for CpG positions 3, 4, 5, 6, 7 and 9. This result is consistent with the MS-HRM melting profile of sample 2, where a fraction of the templates amplified showed a considerable amount of CpG dinucleotides being methylated.
Validation of the results by Sanger sequencing of digitally obtained clones.
Five CLL samples (1, 2, 3, 6 and 9) that represented the range of diversity observed in both the MS-HRM and the bisulfite pyrosequencing assays were further studied using Sanger sequencing of digital MS-HRM (dMS-HRM) products, to assess the validity of the combined approach presented. Figure 5 shows each of the epialleles obtained for each of the CLL samples. Figure 6 shows the mean DNA methylation level per CpG position obtained by pyrosequencing and sequencing of the dMS-HRM “clones.” Overall, the Sanger sequencing data validates the bisulfite pyrosequencing data.
Figure 5.

Bisulfite sequencing of digitally obtained “clonal” PCR products. Five CLL samples representing the range of diversity of DNA methylation as determined by both the MS-HRM and the bisulfite pyrosequencing assays underwent bisulfite sequencing of digitally obtained “clonal” PCR products. Each horizontal line belongs to an epiallele. The CpG dinucleotides are represented by circles, where open and filled circles correspond to unmethylated and methylated CpG positions, respectively. The epiallelic resolution allows the determination of the DNA methylation patterns of each sample analyzed. The dMS-HRM amplicons encompass 16 CpG dinucleotides. The last nine CpG dinucleotides correspond to CpG positions 1 to 9 which were analyzed by the combined MS-HRM and bisulfite pyrosequencing approach. For clarity, sequencing data for just the last nine CpG dinucleotides is shown in this figure.
Figure 6.
Comparison of bisulfite pyrosequencing with digital sequencing data. The mean DNA methylation values obtained for each CpG dinucleotide analyzed either by bisulfite pyrosequencing (from Fig. 3) or digital Sanger sequencing (calculated from the individual epialleles of a given sample from Fig. 5) are comparable, validating the combined MS-HRM and bisulfite pyrosequencing approach.
In sample 1, bisulfite sequencing of the digital clones confirmed the bisulfite pyrosequencing data that CpG positions 1, 2 and then 4 to 6 are the most densely methylated. Examination of the individual clones confirms that CpG positions 1, 2 and 5, 6 in particular are often methylated together (Fig. 5). Aside from this, the occurrence of methylated CpG dinucleotides appears to be at random.
Another set of CpG dinucleotides (3, 4, 5, 7 and 9) are most often methylated together in sample 2 (Fig. 5) and confirms the MS-HRM interpretation that there is a subset of heavily methylated templates and the bisulfite pyrosequencing results. The remainder (if any) of the low-level sporadic methylation seems to be below the detection threshold of bisulfite pyrosequencing and hence not detected.
Sample 3 shows the lowest level of DNA methylation across the samples analyzed (Figs. 5 and 6) and the results are consistent with the results obtained by MS-HRM and bisulfite pyrosequencing. Of the 13 clones analyzed, the majority were unmethylated. The apparent differences between the pyrosequencing and dMS-HRM sequencing mean values can be explained by the relatively few methylated epialleles contributing to the data.
There are four CpG dinucleotides (5 to 8) that seem to be significantly methylated in sample 6 according to the bisulfite pyrosequencing results, but the digital sequencing results show that not all of these CpG dinucleotides are methylated on the same epiallele (Figs. 5 and 6). Only CpG positions 5 to 7 appear to be methylated together.
In sample 9, in which the epialleles show quite low amounts of methylation like sample 3, none of the CpG dinucleotides appear to be methylated with any other CpG dinucleotide (Fig. 5).
Interestingly, despite considerable intra-sample heterogeneity, each sample seems to have a distinct family of methylated epialleles that resemble each other more than they resemble the other samples. This can only be seen from the analysis of the individual dMS-HRM clones. However, the bisulfite pyrosequencing data suggests that there appears to be a consensus pattern including CpG positions 5, 6 and 7. This highlights the limitations of any methodology that examines non-clonal PCR products.
Discussion
MS-HRM is a useful method for rapidly assessing the presence of DNA methylation and distinguishing homogeneous methylation from heterogeneous methylation patterns. MS-HRM can semi-quantitatively estimate the amount of homogeneous methylation. However, it cannot estimate the amount of heterogeneous methylation due to the formation of heteroduplexes between the different epialleles in heterogeneously methylated samples. In homogeneously methylated samples, the sequence differences between methylated and unmethylated epialleles are generally too great for heteroduplexes to form. Even though in some cases the melting differences in methylation between heterogeneously methylated samples and unmethylated samples can be subtle (Fig. 2), they are nevertheless obvious to the experienced observer.
In this investigation, we have chosen cases in which DAPK1 is likely to be methylated in nearly every sample and it could be argued that pyrosequencing is all that is necessary. However, in many investigations, only a fraction of samples show any methylation and in this case MS-HRM will be invaluable in identifying those unmethylated samples that do not require further analysis by pyrosequencing thereby minimizing analysis costs.
Bisulfite pyrosequencing has emerged as another of the few methodologies that allows the recognition and analysis of heterogeneous DNA methylation patterns. The results presented here show that MS-HRM and bisulfite pyrosequencing are complementary techniques that facilitate DNA methylation analysis. MS-HRM can be used as a pre-screening tool to identify samples of interest that warrant further investigation, and the appropriate MS-HRM products can be directly analyzed using a bisulfite pyrosequencing approach.
The utility of the presented workflow is the direct use of PCR products from MS-HRM pre-screening in the pyrosequencing system without further sample manipulation. This allows the omission of the agarose gel analysis quality control step prior to pyrosequencing. The only considerations to take into account are the use of a biotinylated MS-HRM primer during the MS-HRM setup and additional pyrosequencing primer(s) for bisulfite pyrosequencing (if required).
Pyrosequencing assays usually require short read lengths of up to 100 bp,14,27 which complements the use of shorter MS-HRM products that are typically used in methylation analysis and are applicable to analyzing degraded material, e.g., those derived from formalin-fixed paraffin-embedded (FFPE) specimens,28 or other archival specimens.29
If longer amplicons generated from better quality DNA are being analyzed, the requirement for short pyrosequencing assays may be overcome by using a tandem assay approach.30 While there are more limitations in designing a bisulfite pyrosequencing assay compared to Sanger sequencing (e.g., the avoidance of secondary structure formations or long homopolymeric stretches), the results obtained are clearer and a greater sensitivity is achievable (10–20% vs. approximately 5%8).
Bisulfite pyrosequencing does not reveal all of the desirable DNA methylation information. The data can be misleading for heterogeneous methylation as it only gives quantitative information for each CpG position across the entire population of amplified templates. For the four samples that are more heavily methylated, the bisulfite pyrosequencing data indicates that there is a consensus region for DNA methylation at CpG dinucleotides 5, 6 and 7. However, when examining the digital sequencing data, there are DNA methylation patterns within particular samples, but not across samples. This highlights the aforementioned shortcoming of the bisulfite pyrosequencing approach: there is no information given as to the epiallelic DNA methylation patterns. It is dangerous to assume that any given CpG dinucleotides that are more methylated in a given sample are often methylated together at the epiallelic level.
The most accurate method for quantification of DNA methylation is the sequencing of amplicons obtained from digital PCR as this avoids potential PCR amplification and cloning biases31,32 and reveals the DNA methylation status of each individual CpG dinucleotide on each epiallele analyzed. Nevertheless, this approach is both cost- and labor-intensive. In most non-research applications, detailed epiallelic resolution is not necessary, and the combination of MS-HRM and pyrosequencing would be appropriate.
Materials and Methods
DNA samples.
Ten CLL patients presenting to the Peter MacCallum Cancer Centre were selected for inclusion in this study. The use of these patients' blood in this study was approved by the institutional ethics committee (Approval number: 02/70). Mononuclear cells were isolated using Histopaque-1077 (Sigma Aldrich, Cat. No. 10771-6 × 100 ML), according to the manufacturer's instructions. DNA was extracted using the QIAamp DNA Blood mini kit (Qiagen, Cat. No. 51106) following the manufacturer's directions, except that the samples were treated with 20 µL proteinase K (20 mg/mL) (Worthington Biochemical Corporation, Cat. No. PROK) for up to three days.
Fully methylated control DNA was obtained commercially (Millipore, Cat. No. S7821). Unmethylated control DNA was generated from DNA extracted from the peripheral blood of a normal individual by performing whole genome amplification (WGA) twice, as described previously in reference 33.
Bisulfite treatment.
The unmethylated DNA control and DNA derived from CLL samples were quantified using the ND-2000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific). One µg of the fully methylated and unmethylated controls and 200 ng of the CLL DNAs were modified using the Methyl Easy Xceed kit (Human Genetic Signatures, Cat. No. ME002) according to the manufacturer's instructions and eluted twice in a final volume of 100 µL or 20 µL respectively, to give a theoretical concentration of 10 ng/µL (theoretical amount, assuming no loss of DNA during bisulfite conversion).
Methylation-sensitive high resolution melting (MS-HRM).
The fully methylated and unmethylated DNA controls were compared for amplifiable bisulfite modified DNA using a DNA input control assay designed within the COL2A1 gene in a region lacking CpG dinucleotides.34 The concentration of DNA for each was then adjusted accordingly, allowing the accurate preparation of 50, 25 and 5% DNA methylation standards.
MS-HRM was performed on a Rotorgene 6000 (Corbett, Sydney, Australia). Each sample was run in duplicate. The primer sequences for the analysis of the DAPK1 promoter region are: 5′-TTG TTT CGG AGT GTG AGG AGG ATA GT-3′ (GeneWorks, Adelaide, Australia) and 5′-biotin-GCC GAC CCC AAA CCC TAC C-3′ (Sigma-Aldrich) or 5′-biotin-TTG TTT CGG AGT GTG AGG AGG ATA GT-3′ (Sigma-Aldrich) and 5′-GCC GAC CCC AAA CCC TAC C-3′ (GeneWorks). The amplified region corresponds to GenBank accession number AL161787, nucleotides 47,000 to 47,105. Our primers overlap the binding regions for the PCR primers used extensively in the literature and first used by Katzenellenbogen et al.24
PCR was performed in 0.1 mL tubes with a final reaction volume of 20 µL containing 200 nmol/L of each primer, 200 µmol/L of each dNTP, 5 µmol/L SYTO 9 (Invitrogen, Life Technologies, Cat. No. S-34854), 2.5 mmol/L MgCl2, 0.5 U HotStarTaq DNA Polymerase in its supplied buffer (1x) (Qiagen, Cat. No. 203209) and 10 ng bisulfite modified DNA (theoretical amount, assuming no loss of DNA during bisulfite conversion). PCR was performed as follows: 1 cycle of 95°C for 15 min, 50 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 20 sec. This was immediately followed by a hold at 95°C for 1 min, 72°C for 1.5 min and a HRM step from 72 to 95°C rising at 0.2°C per second and holding for 1 sec after each stepwise increment.
Increasing the annealing temperature during the PCR stage of MS-HRM is correlated with increasing bias towards amplification of methylated templates. The temperature at which this bias is minimized is preferable, so that the pyrosequencing results are a true reflection of the methylation in a given sample. We tested multiple annealing temperatures with mixtures of methylated controls to find the least biased conditions.
Bisulfite pyrosequencing.
15 µL of the MS-HRM PCR products were taken for bisulfite pyrosequencing. In the case where the reverse MS-HRM primer was biotinylated, the pyrosequencing primer used for primer extension was 5′-AGT GTG AGG AGG ATA GT-3′ (GeneWorks) which interrogates the sequence: 5′-YGG ATY GAG TTA AYG TYG GGG ATT TTG TTT TTT TYG YGG AGG GGA TTY GGT AAT TYG TAG YGG-3′ (Dispensation order: 5′-ATC GTA TCG AGT GAT CAG TCG GAT TAG TTC AGT CGA GGT ATC GCT GAT CGT ATG TC-3′). In the case where the forward MS-HRM primer was biotinylated, the pyrosequencing primer used for primer extension was 5′-CGA CCC CAA ACC CTA C-3′ (GeneWorks) which interrogates the sequence: 5′-CRC TAC RAA TTA CCR AAT CCC CTC CRC RAA AAA AAC AAA ATC CCC RAC RTT AAC TCR ATC CRA C-3′ (Dispensation order: 5′-ACG ACT GAT CAG ATA TCA GAT CCT ACG ATC AAG AAC AAT CCA GAT CGA TAC TAC AGA TAC AGA-3′). The underlined nucleotides in each dispensation order above are control nucleotides, which allow the detection of significant incomplete bisulfite conversion.
The pyrosequencing reaction was performed on a PyroMark Q24 instrument (Qiagen) using the Pyro Gold Q24 Reagents (Qiagen, Cat. No. 970802). Purification and subsequent processing of the biotinylated single-stranded DNA was performed according to the manufacturer's recommendations. The pyrosequencing primers were used in a final concentration of 0.3 µmol/L. Resulting data were analyzed and quantified with the PyroMark Q24 software version 2.0.6 (Qiagen). MS-HRM products were pyrosequenced once for each sequencing direction.
Digital methylation-sensitive high resolution melting (dMS-HRM).
PCR was performed as described for MS-HRM using the primers 5′-GTT AGT TCG TTT GTA GGG TTT TTA TTG GT-3′ and 5′-GCC GAC CCC AAA CCC TAC C-3′ (GeneWorks). The amplified region corresponds to GenBank accession number AL161787, nucleotides 46,924 to 47,105. Serial dilutions of the bisulfite modified DNA from the CLL samples were made. The dilution where approximately two thirds of the replicates amplified was identified. According to the Poisson distribution, this would indicate an average concentration of one copy of the template per well, and around half of the successful amplifications will have arisen from a single template copy. Once the appropriate dilution was established, each sample was run with 66 replicates, presuming that around 22 of the expected 44 positive wells would contain “clonal” PCR products. HRM was used to identify wells that had a single amplified template by the presence of one clean peak in the Tm plot (negative first derivative of the HRM curves).
Sanger sequencing of dMS-HRM amplicons.
PCR products that arose from a single template (“clones”) were selected for sequencing. The PCR primers for MS-HRM were used as sequencing primers (both forward and reverse reactions). Three and a half microliters of a 1:35 dilution of the clonal PCR product was used in a sequencing reaction using the Big Dye Terminator v3.1 chemistry (Applied Biosystems, Life Technologies, Cat. No. 4337457), according to the manufacturer's instructions. The cycling conditions were 95°C for 10 min, followed by 30 cycles of 95°C for 10 sec, 59°C for 30 sec and 72°C for 3 min. The products were cleaned up using ethanol precipitation prior to sequencing. The sequencing data for the dMS-HRM products were analyzed and visualized using the BiQ Analyzer software35 (Max-Planck-Institut für Informatik, Saarbrücken, Germany).
Conclusions
MS-HRM can readily be incorporated as an effective “front-end” for bisulfite pyrosequencing as the PCR product that is used in MS-HRM analysis can be directly pyrosequenced and this may eliminate the necessity for gel analysis. The use of MS-HRM does not make any significant change to the conventional bisulfite pyrosequencing work-flow apart from the addition of a fluorescent dye and the programming of an extra melt step at the end of the PCR. MS-HRM also gives information about DNA methylation in its own right and pyrosequencing can be used to further investigate selected MS-HRM results. In situations where complex DNA methylation patterns require detailed analysis without the expense of sequencing of multiple clones, a combination of MS-HRM and bisulfite pyrosequencing is an effective solution that will yield considerable DNA methylation information.
Acknowledgements
AD received grant support from the CLL Global Research Foundation, Victorian Cancer Agency, the National Breast Cancer Foundation of Australia, and the Cancer Council of Victoria. ILMC is supported by the Fay Marles scholarship from The University of Melbourne. The authors thank Giada Zapparoli for critical reading of the manuscript.
References
- 1.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 2.Levenson VV. Biomarkers for early detection of breast cancer: what, when and where? Biochim Biophys Acta. 2007;1770:847–856. doi: 10.1016/j.bbagen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Dodge JE, List AF, Futscher BW. Selective variegated methylation of the p15 CpG island in acute myeloid leukemia. Int J Cancer. 1998;78:561–567. doi: 10.1002/(sici)1097-0215(19981123)78:5<561::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 4.Aggerholm A, Hokland P. DAP-kinase CpG island methylation in acute myeloid leukemia: methodology versus biology? Blood. 2000;95:2997–2999. [PubMed] [Google Scholar]
- 5.Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- 6.Versmold B, Felsberg J, Mikeska T, Ehrentraut D, Kohler J, Hampl JA. Epigenetic silencing of the candidate tumor suppressor gene PROX1 in sporadic breast cancer. Int J Cancer. 2007;121:547–554. doi: 10.1002/ijc.22705. [DOI] [PubMed] [Google Scholar]
- 7.Wolf I, Bose S, Desmond JC, Lin BT, Williamson EA, Karlan BY. Unmasking of epigenetically silenced genes reveals DNA promoter methylation and reduced expression of PTCH in breast cancer. Breast Cancer Res Tr. 2007;105:139–155. doi: 10.1007/s10549-006-9440-4. [DOI] [PubMed] [Google Scholar]
- 8.Mikeska T, Candiloro ILM, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics. 2010;2:561–573. doi: 10.2217/epi.10.32. [DOI] [PubMed] [Google Scholar]
- 9.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candiloro IL, Mikeska T, Hokland P, Dobrovic A. Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenetics Chromatin. 2008;1:7. doi: 10.1186/1756-8935-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. BioTechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 12.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. BioTechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 13.Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- 14.Brakensiek K, Wingen LU, Langer F, Kreipe H, Lehmann U. Quantitative high-resolution CpG island mapping with Pyrosequencing reveals disease-specific methylation patterns of the CDKN2B gene in myelodysplastic syndrome and myeloid leukemia. Clin Chem. 2007;53:17–23. doi: 10.1373/clinchem.2007.072629. [DOI] [PubMed] [Google Scholar]
- 15.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikeska T, Bock C, El-Maarri O, Hubner A, Ehrentraut D, Schramm J. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 18.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903–1908. doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 19.Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi K, Oda Y, Saito T, Yamamoto H, Takahira T, Tamiya S. Death-associated protein kinase (DAP kinase) alteration in soft tissue leiomyosarcoma: Promoter methylation or homozygous deletion is associated with a loss of DAP kinase expression. Hum Pathol. 2004;35:1266–1271. doi: 10.1016/j.humpath.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–942. [PubMed] [Google Scholar]
- 22.Satoh A, Toyota M, Itoh F, Kikuchi T, Obata T, Sasaki Y. DNA methylation and histone deacetylation associated with silencing DAP kinase gene expression in colorectal and gastric cancers. Br J Cancer. 2002;86:1817–1823. doi: 10.1038/sj.bjc.6600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong TS, Chang HW, Tang KC, Wei WI, Kwong DL, Sham JS. High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients. Clin Cancer Res. 2002;8:433–437. [PubMed] [Google Scholar]
- 24.Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- 25.Seeliger B, Wilop S, Osieka R, Galm O, Jost E. CpG island methylation patterns in chronic lymphocytic leukemia. Leukemia Lymphoma. 2009;50:419–426. doi: 10.1080/10428190902756594. [DOI] [PubMed] [Google Scholar]
- 26.Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli M, Bhatia K. Aberrant promoter methylation of multiple genes throughout the clinico-pathologic spectrum of B-cell neoplasia. Haematologica. 2004;89:154–164. [PubMed] [Google Scholar]
- 27.Gharizadeh B, Nordstrom T, Ahmadian A, Ronaghi M, Nyren P. Long-read pyrosequencing using pure 2′-deoxyadenosine-5′-O′-(1-thiotriphosphate) Sp-isomer. Anal Biochem. 2002;301:82–90. doi: 10.1006/abio.2001.5494. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen LS, Wojdacz TK, Thestrup BB, Wiuf C, Hager H, Hansen LL. Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer. 2009;9:453. doi: 10.1186/1471-2407-9-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong N, Morley R, Saffery R, Craig J. Archived Guthrie blood spots as a novel source for quantitative DNA methylation analysis. BioTechniques. 2008;45:423–424. doi: 10.2144/000112945. [DOI] [PubMed] [Google Scholar]
- 30.Tost J, El abdalaoui H, Gut IG. Serial pyrosequencing for quantitative DNA methylation analysis. BioTechniques. 2006;40:721–722. doi: 10.2144/000112190. [DOI] [PubMed] [Google Scholar]
- 31.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 33.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive Melting Analysis after Real Time-Methylation Specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res. 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- 35.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]