Abstract
An understanding of the number and types of progeny produced by progenitor cells during development provides a foundation for studies of when and where cell fate determination takes place. Lineal relationships can be revealed by the identification of descendents of cells that express a recombinase, such as Cre or Flp. This method provides data concerning gene expression history, but does not provide clonal resolution among the descendents. An alternative method employs retroviral labeling, which permits the identification of clones, but does not allow for the tracking of gene expression history. Here we report a combination of these methods to circumvent each method’s limitations. By employing the specificity of Cre expression, and by selecting only a subset of cells with a Cre history for retroviral infection, clones with a gene expression history can be labeled. The method utilizes a conditional allele of the avian tumor virus receptor A (TVA), which allows infection of mouse cells following Cre activity, with mammalian retroviral vectors pseudotyped with the ASLV-A envelope glycoprotein (EnvA). We quantified the efficiency and specificity of this system in vivo and in vitro. We also generated a series of retroviral vectors encoding a variety of histochemical and fluorescent reporter genes that enable the tracking of mixtures of clones, thus enabling better resolution of clonal boundaries. This method and new vectors can be used to further our understanding of the gene expression patterns of progenitor cells that make particular daughter cells, as well as provide a platform for manipulating identified subsets of developing cells.
Keywords: TVA, lineage analysis, retrovirus, conditional TVA
Background
One of the challenges to understanding the development of multicellular organisms is determining the relationship between gene expression and cell fate decisions. To begin to address this challenge, one can determine the progenitor-progeny relationships, i.e. perform lineage analyses of developing cells. This descriptive exercise can provide insights into when and where decisions are being made concerning cell fates. The link to gene expression patterns can be provided by marking cells that are expressing a particular gene, or marking cells with a particular gene expression history. Engineered expression of the recombinases, Cre and Flp, has provided a powerful set of tools for this latter purpose, and has provided a means to perform tissue-specific loss or gain of function analyses (Kim et al., 2009). In most cases of engineered recombinase expression, the regulatory elements employed to direct recombinase expression lead to the labeling of a fairly broad spectrum of daughter cell types. Labeled descendents typically comprise multiple cell types, with unclear clonal relationships among the labeled cells. One approach to improve the resolution of this method has been the intersectional method, in which Cre and Flp mouse lines are crossed to a Cre- and Flp-sensitive reporter line. Only cells with an expression history of both Cre and Flp express the reporter gene, permitting the labeling of a more restricted group of cells which share a history of expression of two genes (Farago et al., 2006). An additional limitation in recombinase fate mapping is due to the lack of control of the timing of recombinase action. Unless one uses a regulated recombinase, such as an estrogen receptor fusion of Cre (Metzger et al., 1995), one cannot choose the time to initiate labeling. The lack of timing control can create an ambiguity regarding whether a labeled cell is a descendent of a Cre-expressing progenitor cell, or whether a labeled cell became marked following Cre expression later in its development.
Another approach that can be used to obtain specificity in the marking of descendents is the EnvA/TVA system. Mice are not infectable with viruses carrying the EnvA glycoprotein from avian retroviral vectors (Weiss et al., 1985). However, since the discovery of the avian TVA, which encodes the receptor for EnvA, mouse strains have been created that allow infection by EnvA vectors through expression of TVA in murine tissues (Federspiel et al., 1994; Lewis et al., 2001; Young et al., 1993). This method has been productively employed previously for several applications (Doetsch et al., 1999; Federspiel et al., 1994; Fisher et al., 1999; Holland et al., 1998;). For each study, independent mouse lines have been generated to express TVA under the control of different promoters. This approach is both time and labor-intensive, as each transgenic line needs to be generated and characterized for each set of experiments. Recently, Seidler et al. reported the production of a mouse permitting conditional expression of TVA (Seidler et al., 2008). We have independently generated three mouse lines conditionally expressing TVA and have demonstrated their utility in tracking clones with a particular gene expression history. This method combines the strengths of the recombinase fate mapping strategy and the clonal resolution afforded by retroviral lineage analysis. In addition, the fact that the viral vectors used for lineage analysis can only infect mitotic cells provides a guarantee that cells that are marked came from a progenitor cell with a history of Cre expression, and were not labeled by expression of Cre at a later point in their development.
We have performed quantitative analyses of the efficacy and specificity of this method. These studies are crucial, as infectability must be restricted to Cre-expressing/TVA expressing cells, or gene expression cannot be correlated with clonal labeling. We report that infectability is indeed restricted to cells with a Cre expression history in vivo. Interestingly, we also discovered a “hitchhiking” phenomenon whereby mixtures of virions with different envelope proteins can use each other’s envelope proteins to gain access to host cells. Finally, we report the development of several retroviral vectors encoding discrete histochemical and fluorescent markers for distinctive labeling of multiple clones, to aid in the definition of clonal boundaries, and to give flexibility in terms of labeling protocols.
Materials and Methods
Viruses and Virus Production
All EnvA viruses were produced in 293T cells (Cepko and Pear, 2001; Pear et al., 1993). Viruses encoding genes for histochemical analysis include LIA, which encodes human placental alkaline phosphatase (PLAP) (Bao et al., 1997), BAG, which encodes β-galactosidase (lacZ) (Price et al., 1987), and NinII, which is similar to LIA but encodes a nuclear lacZ gene in place of PLAP. All viruses encoding fluorescent reporters were derived from the pQCXIX vector (Clontech). To produce the EnvA viruses, on Day 1, at 70% confluency, a 10 cm plate was transfected with a pool of plasmids containing 6 μg viral genomic plasmid, 3 μg pMD gagpol plasmid (Ory et al., 1996), and 1 μg EnvA envelope plasmid (Landau et al., 1992). Media was changed the following day and replaced with 5 mL of fresh media. On days 3 and 4, the supernatant was collected and frozen. Supernatants were thawed and viruses concentrated by ultracentrifugation (Cepko and Pear, 2001). The method of production of VSV-G enveloped viruses was the same, except they were produced in the platE cell line, which expresses gagpol and the ecotropic envelope gene (Morita et al., 2000; Naldini et al., 1996). In this case, 1 μg of pCL VSV (gift from Richard Mulligan, Harvard Medical School) was transfected into platE cells together with 6 μg of genomic plasmid.
Cell culture
For cell culture experiments, NIH 3T3 cells were grown to confluency on a 10 cm plate, and split onto 6-well plates at a 1 : 20 split ratio. Each condition was done in triplicate. The next day, the cells were transfected with 5 μg of total DNA using FUGENE6 (Roche) according to the manufacturer’s instructions. In order to maintain a constant FUGENE : DNA ratio, for the cTVA and CAG-Cre:GFP (Matsuda et al., 2007) combination, 2.5 μg of each plasmid was co-transfected, whereas for CAG-TVA, UbC-cTVA (cTVA), and CAG-Cre:GFP, 2.5 μg of plasmid was co-transfected with 2.5 μg of pBluescript II KS(−) (genbank accession number X52329). The day after transfection, each well was infected with NinII containing the EnvA protein on its envelope (NinII(EnvA)) or QCXIX encoding a nuclear form of tdTomato cloned 5′ of the Ires (QC NLS-tdTomato IX (EnvA)). Xgal assays were performed as described (Price et al., 1987), and tdTomato fluorescence was observed by fluorescent microscopy. Transfection efficiency of NIH 3T3 cells was measured by transfecting a nuclear localized green fluorescent protein (CAG-Cre:GFP) and, three days following transfection, comparing GFP positive nuclei to the 4′,6-diamidino-2-phenylindole (DAPI)-positive nuclei. Co-transfection ratios were calculated by co-transfecting CAG-GFP:Cre along with pCALNL DsRed (a reporter plasmid for Cre activity encoding DsRed, Matsuda and Cepko, 2007) and comparing cells that were both green and red (express genes from both plasmids) to those that were only green (expressing GFP:Cre).
For MEF cultures, 10 cm confluent tissue culture plates were split 1 : 6 onto 10 cm plates, and infected 8 hours after splitting with NinII(EnvA). Cells were glutaraldehyde fixed 3 days later and Xgal stained, as detailed below.
Mice
Each of the three cTVA transgenic lines was created by pronuclear injection. Each line was maintained as heterozygotes. The construct used for injection is as shown in Figure 1. The human ubiquitin-C (UbC) promoter is a broadly active promoter which should drive expression of a gene (in this case, destabilized GFP) in all or most cells in the mouse (Lois et al., 2002; Schorpp et al., 1996). The TVA gene should only be expressed after Cre has recombined out the upstream GFP. A transcriptional stop prevents transcription of the TVA gene until Cre has acted. GFP expression in mice with cTVA in the absence of Cre should provide evidence of where and when the construct is expressed. However, we were unable to detect GFP in any of the lines, presumably due to the use of the destabilized GFP allele and/or low level expression. For genotyping, primers were designed to the UbC promoter, as follows: 5′-GAGCGGAACAGGCGAGGA AAAGTA-3′ and 5′-CGCCCCCGCAACCAGACGACAA-3′. The amplicon is 554 bp. The Chx10-Cre (Rowan et al., 2004), hβactin-Cre (Lewandoski and Martin, 1997), and Wnt1-Cre (Danielian et al., 1998) mice were maintained as heterozygotes. Genotyping primers for each Cre line are as follows: 5′-GCAGAACCTGAAGATGTTCGC-3′ and 5′-ACACCAGAGACGGAAATCC-3′. The amplicon is 494 bp.
Figure 1.
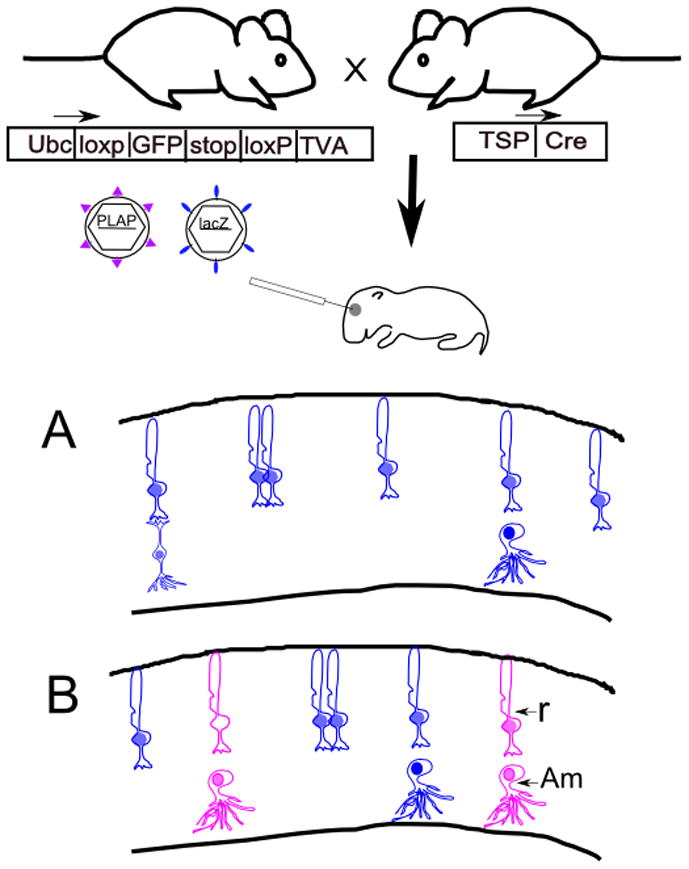
Experimental strategy for use of the cTVA allele for lineage analysis. A mouse with conditional expression of TVA can be crossed to a Cre line of choice, with expression of Cre directed by a tissue-specific promoter (TSP). TVA, a receptor for viruses with the EnvA glycoprotein, will be expressed in cells with a history of Cre expression. Viruses pseudotyped with the EnvA glycoprotein are able to infect TVA-expressing cells, and are not able to infect non-TVA expressing mouse cells. Viruses pseudotyped with the VSV-G glycoprotein can infect any mitotic cell. Two viruses, one pseudotyped with EnvA, encoding PLAP, and one pseudotyped with VSV-G, encoding lacZ, can be coinjected into a developing tissue, e.g. the eye. The drawings in A and B depict retinal sections following histochemical processing for PLAP (purple) and β-galactosidase (blue). (A). If a control, TVA-negative retina is injected with a mixture of the two viruses, only blue clones will result. (B) If a TVA expressing retina is injected with this mixture of viruses, both types of viruses will infect mitotic cells, giving rise to blue and purple clones. In this example, the cells with a Cre history generate clones of rod photoreceptors (R) and amacrine interneurons (Am).
Quail TVA (pg800, (Bates et al., 1993)) was generated by RT-PCR using the primers 5′-AAGAATTCTGAGCCGCCACCATGGCGCGGCTGCTGCCCGCGCTG-3′ and 5′-ATATGCGGCCGCTCACTCAGTCCCATCTCACCAGCTCACAGCAA-3′ and RNA isolated from quail fibrosarcoma QT6 cells (ATCC #. CRL-1708). The PCR amplified fragment was digested with EcoRI/NotI and cloned into pCAGEN ((Matsuda and Cepko, 2007), addgene #11160) digested with EcoRI/NotI to make pCAG-TVA.
LoxP-GFPd2 was generated by PCR using the primers 5′-ATGCACGCGTATAACTTCGTATAGCATACATTATACGAAGTTATGGATCCACCGGT CGCCACCAT-3′ and 5′-GAGTCGCGGCCGCGCATCTACACATTGATCC-3′ and pCAG-GFPd2 ((Matsuda and Cepko, 2007), addgene #14760) as a template. SV40polyA-loxP was generated by PCR using the primers 5′-ATGCGCGGCCGGGACTCTAGATCATAATCAGCCATACCAC-3′ and 5′-TTCTCGAGATATAACTTCGTATAATGTATGCTATACGAAGTTATCGCCTTAAGATAC ATTGATGAG-3′ and pCALNL-GFP ((Matsuda and Cepko, 2007), addgene #13770) as a template. To make CAG-loxP-GFPd2-SV40polyA-loxP-GFP (pCALGd2L-GFP), these two PCR fragments were digested with MluI/EagI and EagI/XhoI, respectively, and simultaneously ligated into pCALNL-GFP digested with MluI/XhoI. To make human UbC promoter-loxP-GFPd2-SV40polyA-loxP-GFP (pUbCLGd2L-GFP), loxP-GFPd2-SV40polyA-loxP-GFP was excised from pCALd2L-GFP with MluI (blunt ended) and NotI, and ligated into pUbC-GFP ((Matsuda et al., 2004), addgene #11155) digested with SacI (blunt ended) and NotI. To make pUbCLGd2L-TVA, the coding region of TVA excised from pCAG-TVA with EcoRI and NotI (blunt ended) was ligated into pUbCLGd2L-GFP digested with EcoRI and Sau36I (blunt ended). The expression cassette UbCLGd2L-TVA was isolated as a SspI (blunt ended)/SfiI fragment, and inserted into the XhoI site (blunt ended) of pBS-Ins4s obtained from Dr. David Rowitch (UCSF) to flank UbCLGd2L-TVA with the chicken β-globin HS4 insulators (1.2kb HS4, (Chung et al., 1993)). The insulated UbCLGd2L-TVA was isolated as a NotI fragment, and injected into the pronuclei of fertilized SJL-BL/6 F1 blastocysts and backcrossed into the C57BL/6 strain. All experiments involving mice were approved by the Longwood Medical Area Institutional Animal Care and Use Committee.
Retinal injections and dissections
Mouse pups were injected subretinally at P0 and sacrificed at P21 as described previously (Turner et al., 1987). For quantification of EnvA : VSV-G efficiency in the retina, 500 nL of a mixture of 1×107 cfu/mL LIA(EnvA) and 5×106 cfu/mL BAG(VSV-G) were used. Dilutions of 1:5 and 1:25 of this mixture were used for sparser retinal infection.
Retinae were processed according to the staining or viral detection protocol required for the marker gene. β-galactosidase and PLAP histochemical staining protocols were conducted as previously described (Fields-Berry et al., 1992; also see http://genetics.med.harvard.edu/~cepko/protocol/xgalplap-stain.htm). Whole eyeballs were fixed in 0.5% glutaraldehyde for 20 minutes for Xgal stain, and 4% formaldehyde for 1 hour for PLAP or fluorescent analysis. The retinal pigmented epithelium (RPE) and lens were removed after fixation. Retinae were post-fixed for 15 minutes, then rinsed three times for 15′ each in PBS. For fluorescent analysis, antibody staining was conducted as previously described (Punzo and Cepko, 2008).
For confocal and clone counting analyses, retinae were flat mounted between cover slips. After histochemical or immunohistochemical processing, retinae were rinsed 3 times in PBS for 15 minutes, then cut 4 times so as to lay flat. Retinae were mounted with fluoromount (Southern Biotech) for β-galactosidase and PLAP detection or Prolong Gold for fluorescence detection.
For experiments on mouse retinal explants, retinae were taken from P0 mice, and cultured on filters as previously described (Altshuler et al 1993). Retinae were immediately infected with either 1 μL of 2.7 × 104 infectious BAG(EnvA) virions or 1 μL of a mixture of 2.7×104 BAG(EnvA) and 1.35 × 104 LIA(VSV-G) virions. Retinae were maintained for three days in vitro, fixed, and stained for β-galactosidase only.
For postnatal brain injections of cTVA x hβactin-Cre animals, P2 mice were injected into the midbrain with 1 μL of a mixture of 5×10^8 cfu/mL LIA(EnvA) and 1×10^8 BAG(EnvA). Mice were perfused with 4% PFA at P28, and brains were stained histochemically for both β-galactosidase and PLAP activities. For prenatal brain injections of cTVA x Wnt1-Cre animals, pups were injected in utero into the junction of the third and fourth ventricles at embryonic day 13.5 using ultrasound-guided injection (Punzo and Cepko, 2008). Two different viral solutions were injected, a 200 nL mixture of 1×109 cfu/mL LIA(EnvA) and 5×108 cfu/mL NinII(EnvA), or 200 nL of 1×1010 cfu/mL QC NLS-GFP IX(EnvA). Mice were sacrificed at P0, fixed with 4% PFA, and examined for PLAP enzyme activity, β-galactosidase enzyme activity, or immunohistochemically detectable GFP.
Antibodies
| Primary Antibody | Dilution | Source |
|---|---|---|
| Primary Antibodies | ||
| Chicken anti-GFP (GFP) (ab13970) | 1:2000 | Abcam |
| Rabbit anti-DsRed (tdTomato) (632496) | 1:1000 | Clontech |
| Goat anti- β-galactosidase (4600-1409) | 1:400 | AbD Serotec |
| Secondary Antibodies | ||
| Goat anti-rabbit Cy3 (tdTomato) (111-165-003) | 1:250 | Jackson Immunoresearch |
| Goat anti-chicken A488 (GFP) (103-485-155) | 1:250 | Jackson Immunoresearch |
| Donkey anti-rabbit Cy3 (tdTomato, mCherry) (711-165-152) | 1:250 | Jackson Immunoresearch |
| Donkey anti-goat Cy5 (β-galactosidase) (705-495-003) | 1:250 | Jackson Immunoresearch |
| Donkey anti-goat A488 (β-galactosidase) (705-485-147) | 1:250 | Jackson Immunoresearch |
Results
Test of infectivity of murine cells with EnvA pseudotyped retroviruses
Mammalian cells have been shown to be uninfectable with retroviruses carrying the EnvA protein in the viral envelope (Federspiel et al., 1994; Weiss et al., 1985; Young et al., 1993). However, mouse cells can be made infectable by introduction of the TVA receptor. In order to confirm these findings and quantify the specificity and efficiency of use of the transgenic cTVA mice that we constructed, we first performed control experiments. Two different TVA expression vectors were created using two different broadly active promoters: CAG-TVA and hUbC-loxP-GFP-stop-loxP-TVA (cTVA). The CAG-TVA expresses the TVA receptor ubiquitously using the CAG promoter (Niwa et al., 1991), while cTVA uses the ubiquitous human ubiquitin C promoter (Lois et al., 2002; Schorpp et al., 1996), and is expressed only after a Cre-mediated recombination event.
In order to test for any leakiness in this system, murine NIH 3T3 cells were examined for infectivity with an EnvA pseudotyped murine retrovirus following transfection of several types of plasmids. For a positive control, they were transfected with CAG-TVA. To test for activity of cTVA, they were transfected with both cTVA and CAG-GFP:Cre. Two negative controls were used, one with cTVA in the absence of GFP:Cre, and one with GFP:Cre in the absence of cTVA. Transfected cells were inoculated a day later with the NinII(EnvA) murine retrovirus, which encodes nuclear β-galactosidase, or QC NLS-tdTomato IX(EnvA). Infected cells were scored for expression of β-galactosidase by Xgal histochemistry, or for tdTomato by its intrinsic fluorescence. Clones were identified as a coherent cluster of labeled cells surrounded by unlabeled cells. More clones were observed when CAG-TVA alone was transfected than when cTVA + CAG-GFP:Cre were co-transfected (Figure 2). Many more clones were observed following infection of cells transfected with cTVA + GFP:Cre than cTVA or GFP:Cre alone. The efficiency of infection for each transfection was calculated as the number of clones relative to infectious particles, which had been measured on a 293 cell line, TVA800, constitutively expressing TVA (Narayan et al., 2003). The efficiency for CAG-TVA was 6.08×10−2 and for cTVA + GFP:Cre was 1.27×10−2. The number of infections of cells transfected with cTVA alone or CAG-Cre alone was not zero. These efficiencies were 3.28×10−4 for cTVA alone and 2.60×10−4 for GFP:Cre alone.
Figure 2.
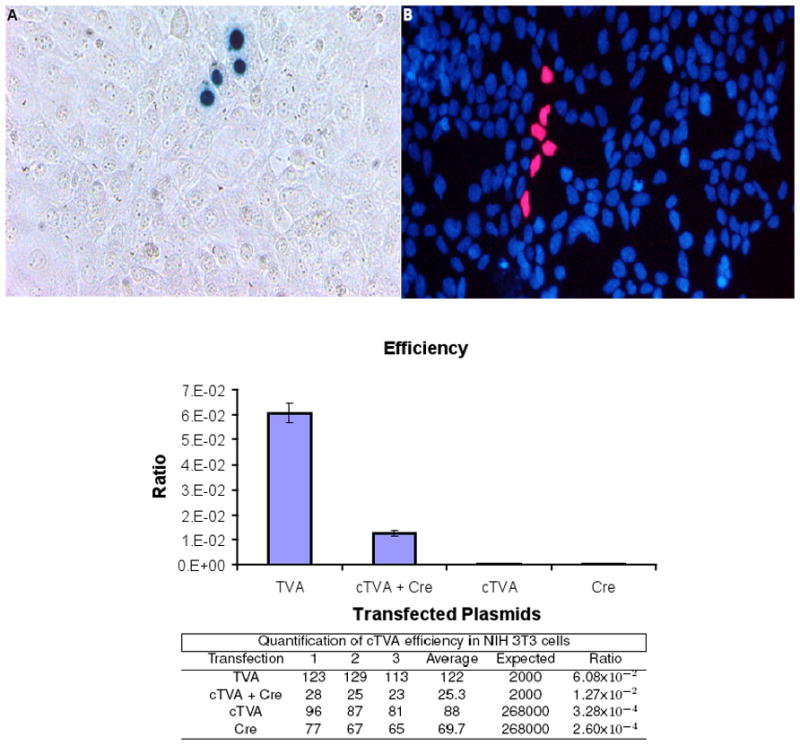
Infection of murine NIH 3T3 cells with EnvA pseudotyped retroviruses. Mouse NIH 3T3 cells were transfected with either pCAG-TVA, pUbC-cTVA + pCAG-GFP:Cre, pUbC-cTVA, or CAG-GFP:Cre. The next day, they were infected with MMLV retroviruses pseudotyped with the EnvA glycoprotein. (A) An image of a single NinII-infected clone detected by Xgal histochemistry. (B) An image of a single fluorescent NLS-tdTomato-infected clone (red) with DAPI-labeled nuclei (blue). The infection ratios, which are the number of clones observed on the transfected cells vs. the number of infectious particles observed on 293T TVA 800 cells (which constitutively express TVA, (Narayan et al., 2003)) are plotted with corresponding values shown in the table below. The number of infections in the cTVA alone or CAG-GFP:Cre alone conditions were not significantly different from each other (t-test).
It should be noted that all measured efficiencies not only reflect the efficiency of the viral events, but also the efficiency of transfection of 3T3 cells, which was observed to be approximately 1% (Supplemental Figure 1). It was possible that the lower efficiency of infection of the cTVA + GFP:Cre transfected cells was due to lack of recombination of the cTVA construct. To measure how well Cre from this construct was able to recombine a plasmid template, and to measure the co-transfection rates, another experiment was performed. pCAG-GFP:Cre was co-transfected with pCALNL-DsRed, which expresses DsRed only in cells having a Cre expression history (Matsuda et al., 2007). The cells expressing Cre-GFP were then examined for expression of DsRed. It was observed that 230/230 GFP:Cre expressing-cells also expressed DsRed (Supplementary Figure 1), indicating that Cre was active in all cells expressing GFP:Cre. Low efficiency of Cre activity would thus not explain the difference between infectivity of CAG-TVA transfected cells and cTVA transfected cells.
Characterization of transgenic mouse lines with conditional TVA expression
For use in a developmental context, transgenic mouse lines were generated to allow for expression of TVA following a Cre-mediated recombination event. The same cTVA plasmid that was tested in NIH 3T3 cells was used to establish three separate cTVA transgenic lines by pronuclear injection, designated L1, L2, and L3.
To test the effectiveness and fidelity of the conditional allele in the three mouse lines, each line was crossed to a constitutive Cre expressing line, hβactin-Cre (Lewandoski and Martin, 1997). E14.5 embryos from each cross were used to produce mouse embryonic fibroblast cultures (MEFs). The same MMLV virus preparations used to test for infectivity of NIH 3T3 cells were used to infect the MEFs. Two questions were addressed: the efficiency of EnvA/TVA-mediated infection, and the specificity of viral infection, e.g. if any viral infection occurred when TVA should not be expressed.
MEFS from each cTVA line were infected with NinII(EnvA). Each culture of the genotype cTVA+/Cre+ was infectable with these viruses, while all other cultures of genotypes cTVA+/Cre−, cTVA−/Cre+, and cTVA−/Cre− were not (Figure 3). These data indicate that the conditional allele was working as designed. All three lines had similar infectivity ratios and showed no background infectivity in the absence of Cre (data not shown).
Figure 3.
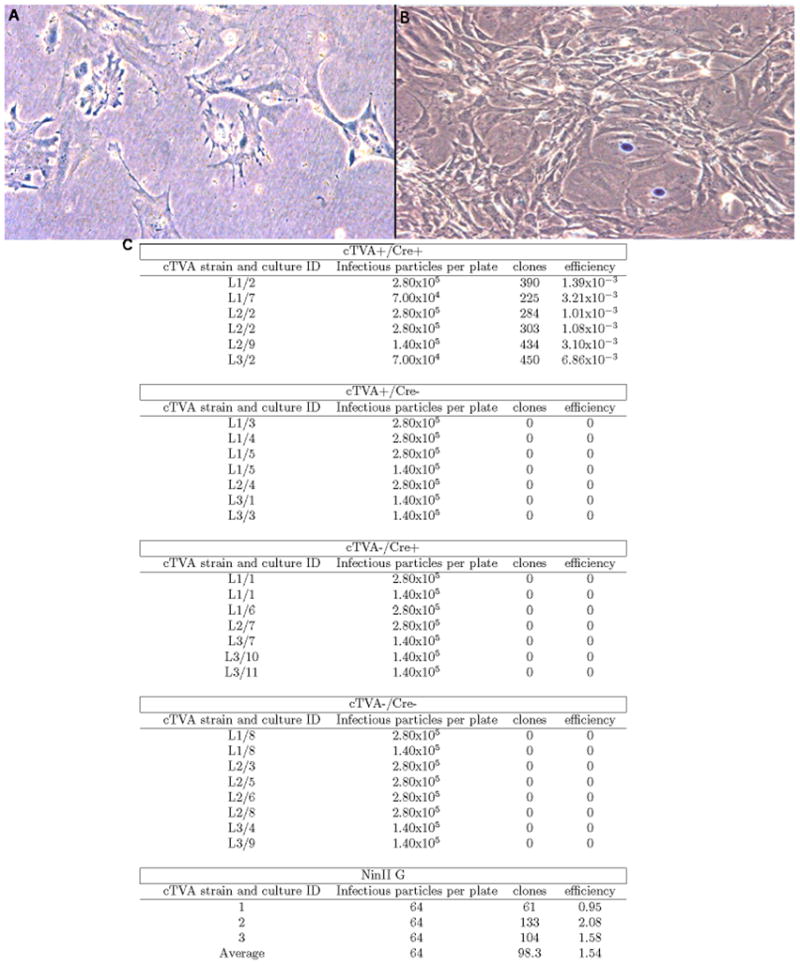
Infection of cTVA MEFs with EnvA pseudotyped retroviruses. MEFS from three different cTVA mouse lines (L1, L2, and L3) were infected with NinII(EnvA). (A) An image of a dish from a TVA- Cre- culture. (B) An image from a dish of a TVA+/Cre+ culture. Both cultures were infected with the same number of NinII infectious particles. (C) Quantitative data from the infection of MEFs by NinII. Column 1 lists the culture ID number, which refers to an individual MEF culture, derived from a unique embryo. Column 2 indicates the number of infectious viral particles applied to the dish. Column 3 indicates the number of clones observed. Column 4 is the ratio of clones observed (column 3) divided by the infectious virions applied (column 2). The number of infectious virions was measured by infection of 293 TVA800 cells, which constitutively express TVA.
To test how efficiently retroviruses with a highly promiscuous envelope protein could infect these cells, and thus provide a comparison for EnvA pseudotyped retroviruses, VSV-G pseudotyped retroviruses were used. VSV-G mediates infection into a wide variety of species and is commonly used as the glycoprotein for retroviral infections (Emi et al., 1991; Kwon et al., 2003; Yoshida et al., 1997). NinII(VSV-G) virus was incubated with the MEF cultures and, as expected, a greater efficiency was observed relative to that of the EnvA virions. The highest infectivity of an EnvA retrovirus relative to a VSV-G enveloped retrovirus in the MEFs was about 1 : 554. Though reduced, this efficiency may not affect utility in the mouse, as only a small number of infection events are desired for clonal lineage analysis.
Efficiency and specificity of EnvA/cTVA in the retina
To use the cTVA mouse lines for in vivo lineage analysis, it was useful to perform an in vivo characterization in a well-defined system. The retina was chosen, as extensive lineage analyses have been performed in the retina by our laboratory in the past, providing a robust set of data for comparison (Fields-Berry et al., 1992; Morrow et al., 1999; Turner and Cepko, 1987; Turner et al., 1990). The genesis of retinal neurons begins at e11 and continues until P11 (Young, 1985a), and the birthdating of these cells has been well established (Young, 1985a; Young, 1985b).
In order to test the efficiency of TVA/EnvA-mediated viral entry, we first normalized for injection efficiency. The efficiency was examined by co-injecting an EnvA pseudotyped murine vector encoding alkaline phosphatase (LIA) with a VSV-G enveloped murine vector encoding β-galactosidase (BAG). LIA(EnvA) and BAG(VSV-G) were injected into the mouse retinae at P0. At P21, after development was complete, the number of clones arising from each virus was scored and analyzed. Clones were identified as radial clusters of cells, separated from other clusters of labeled cells. Previous studies in our laboratory, in which a dilution series of viruses (Turner and Cepko, 1987), or mixtures of viruses encoding distinct markers (Fekete et al, 1994; Fields-Berry et al. 1992; Rompani and Cepko, 2008) were used, have established these radial clusters as clones. As MEFs derived from mouse lines L1, L2, and L3 all showed similar infectivity, L1 was used for all further analysis in the mouse. A schematic diagram of this experiment is shown in Figure 1.
Similar EnvA/VSV-G ratios were observed from counts made on whole mount retinal preparations or cryosections. Additional viral cocktails were examined and included mixtures of nuclear/cytoplasmic β-galactosidase, as well as NLS-tdTomato/β-galactosidase viruses. All ratios of infection efficiency appeared to be consistent independent of the viral cocktail used. Two different Cre lines were used to activate TVA expression – hβactin-Cre (Lewandoski and Martin, 1997), which is ubiquitously expressed, and Chx10-Cre, which is expressed in the majority of retinal progenitor cells during development (Rowan and Cepko, 2004). These two Cre-expressing lines crossed to the cTVA mouse showed similar ratios of EnvA : VSV-G infection, a finding that is expected, given that almost all retinal cells should have a history of expression of Cre.
The ratio of EnvA/VSV-G mediated infection events in retinae of mice with the cTVA and Cre alleles was roughly 80 : 1000, or 1 : 12.5 (Figure 4, Table 1). Since the cocktails were each injected with an EnvA/VSV-G ratio of 2 : 1, this indicates that the EnvA/TVA-mediated viral entry was about 25 times less efficient than VSV-G-mediated entry in the P0 mouse retina. Interestingly, retinae from the cross cTVA +/− x hβactin-Cre +/− that should not express TVA (cTVA+/Cre−; cTVA−/Cre+, and cTVA−/Cre−) showed low infectivity from the EnvA virus. This is in contrast to the results obtained in MEFs from the same cTVA expressing lines. However, the results indicate that there were no more infection events in the cTVA+/Cre− condition than in the cTVA−/Cre+ and cTVA−/Cre− conditions, demonstrating that the infection events were independent of the presence of Cre or TVA.
Figure 4.
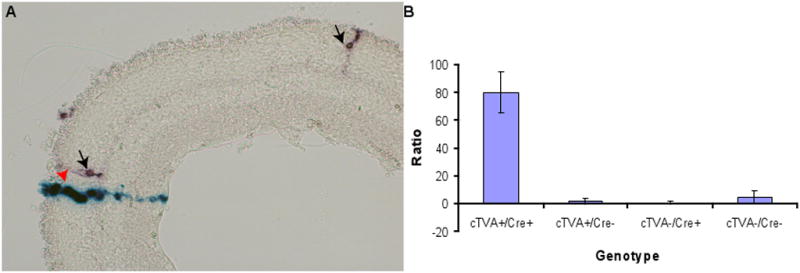
Efficiency of infection of virus pseudotyped with EnvA vs. VSV-G. Retroviruses (LIA(EnvA) and BAG (VSV-G)) were mixed and injected into the subretinal space of P0 mice in vivo. Retinas were analyzed for clones following the completion of development at P21, either by scoring infections on retinal whole mount preparations, or by scoring clones on cryosections. (A) A cryosection of a retina exhibiting clones from LIA(EnvA) (arrows) and BAG (VSV-G) (arrowhead) are shown. (B) The ratio of VSV-G to EnvA events was calculated for the four resulting genotypes of the cTVA+/− x hβactin-Cre +/− cross, as shown. The values for these calculations are listed in Table 1.
Table 1.
Quantification of clones from retinae infected with LIA(EnvA) and BAG(VSV-G). While all four genotypes showed infection with EnvA pseudotyped virus, the cTVA+ Cre+ condition showed a twenty-three-fold enhancement of infection. Employing the t-test with Bonferroni corrections, the number of clones observed on TVA+/Cre−, TVA−/Cre+, and TVA−/Cre− were not significantly different from one another.
| cTVA+/Cre+
| ||||||
|---|---|---|---|---|---|---|
| Mouse | Retina | lacZ (G) | PLAP (A) | Total | G/A ratio | A clones/1000 G clones |
| 11767 | 1 | 374 | 33 | 407 | 11.3 | 88.2 |
| 11767 | 2 | 885 | 56 | 941 | 15.8 | 63.2 |
| 11769 | 1 | 642 | 58 | 700 | 11.1 | 90.3 |
| 11770 | 1 | 1253 | 96 | 1349 | 13.1 | 76.6 |
| 11770 | 2 | 1515 | 156 | 1671 | 9.71 | 103 |
| 11741 | 1 | 884 | 55 | 939 | 16.1 | 62.2 |
| 11774 | 1 | 641 | 49 | 690 | 13.1 | 76.4 |
|
| ||||||
| Total | 6194 | 503 | 6697 | 12.3 | 81.2 | |
| cTVA+/Cre−
| ||||||
|---|---|---|---|---|---|---|
| Mouse | Retina | lacZ (G) | PLAP (A) | Total | G/A ratio | A clones/1000 G clones |
| 11740 | 1 | 320 | 0 | 320 | - | 0 |
| 11740 | 2 | 398 | 2 | 400 | 199 | 5.03 |
| 11745 | 1 | 1603 | 6 | 1609 | 267 | 3.74 |
| 11771 | 1 | 199 | 0 | 199 | - | 0 |
| 11771 | 2 | 38 | 0 | 38 | - | 0 |
| 11772 | 1 | 1696 | 4 | 1700 | 424 | 2.36 |
| 11772 | 2 | 11 | 0 | 11 | - | 0 |
| 11753 | 1 | 1892 | 3 | 1895 | 631 | 1.59 |
| 11753 | 2 | 18 | 0 | 18 | - | 0 |
|
| ||||||
| Total | 6175 | 15 | 6190 | 412 | 2.43 | |
| cTVA−/Cre+
| ||||||
|---|---|---|---|---|---|---|
| Mouse | Retina | lacZ (G) | PLAP (A) | Total | G/A ratio | A clones/1000 G clones |
| 11754 | 1 | 81 | 0 | 81 | - | 0 |
| 11754 | 2 | 1394 | 3 | 1397 | 465 | 2.15 |
| 11773 | 1 | 103 | 0 | 103 | - | 0 |
| 11773 | 2 | 10 | 0 | 10 | - | 0 |
|
| ||||||
| Total | 1588 | 3 | 1591 | 529 | 1.89 | |
| cTVA−/Cre−
| ||||||
|---|---|---|---|---|---|---|
| Mouse | Retina | lacZ (G) | PLAP (A) | Total | G/A ratio | A clones/1000 G clones |
| 11768 | 1 | 500 | 1 | 501 | 500 | 2 |
| 11768 | 2 | 1250 | 15 | 1265 | 83.3 | 12 |
| 11749 | 1 | 644 | 4 | 648 | 161 | 6.21 |
| 11749 | 2 | 2268 | 24 | 2292 | 94.5 | 10.6 |
| 11750 | 1 | 719 | 1 | 720 | 719 | 1.39 |
| 11750 | 2 | 4552 | 28 | 4580 | 163 | 6.15 |
| 11751 | 1 | 4733 | 24 | 4757 | 197 | 5.07 |
| 11751 | 2 | 3121 | 13 | 3134 | 240 | 4.17 |
| 11754 | 1 | 70 | 0 | 70 | - | 0 |
| 11754 | 2 | 43 | 0 | 43 | - | 0 |
|
| ||||||
| Total | 17900 | 110 | 18010 | 163 | 6.15 | |
A non-zero background of EnvA infection in the absence of TVA has been indicated previously (Wickersham et al., 2010). However, the number of infection events observed here was over 103 higher than previously reported. Since the previous study did not employ co-infection with VSV-G pseudotyped virus, it seemed that the coinfection strategy used here might be responsible for the higher rate of EnvA virus entry in the absence of TVA. To test this idea, retinae from P0 mice were explanted and infected with 2.7 × 104 infectious EnvA pseudotyped BAG virions. The retinae were kept for 3 days and subsequently processed for β-galactosidase activity. Analysis of 26 retinae from 13 mice that were cTVA+/Cre−, cTVA−/Cre+, or cTVA−/Cre− showed no Xgal staining, indicating that the virus did not infect the retinae. Subsequently, LIA (VSV-G) was added to the same tube of BAG(EnvA) virus used in the experiments above. A mixture of the same 2 : 1 EnvA/VSV-G ratio as was used previously in vivo was used to for the infection of the explants, which was 1 μL (2.7×104 BAG(EnvA) and 1.35 × 104 LIA(VSV-G) virions) per infection of a P0 retinal explant. The presence of BAG viral infection was detected in all 11 retinae scored that were not cTVA+/Cre+ and therefore did not express TVA (Figure 5). This demonstrates that, in the presence of VSV-G enveloped retroviruses, EnvA retroviruses can infect cells in the absence of TVA expression.
Figure 5.

Test for effect of a virus with one glycoprotein on the infectivity of a virus with a different glycoprotein. EnvA retroviruses were used to infect retinal explants with and without co-infection with VSV-G retroviruses. Retinal explants were infected with BAG(EnvA) at P0. The retina in (A) was TVA+/Cre+ and was infected with BAG(EnvA) retrovirus only, while that in (B) was TVA+/Cre− and was infected with the same amount of infectious BAG(EnvA) virus. The retina in (C) was also TVA+/Cre−, but was infected with a mixture of BAG(EnvA) and LIA(VSV-G). All retinae were stained for β-galactosidase only.
Viruses with EnvA can successfully infect several tissues in the developing cTVA mouse
As recombination rates and expression levels in transgenic mice can be variable across tissues, we wished to explore whether the cTVA mice would be useful for analyses in multiple tissues. To this end, infection of two other developing tissues was carried out. The progeny of hβactin-Cre x cTVA mice were injected in the midbrain at P2 with a cocktail of β-galactosidase and PLAP-expressing EnvA pseudotyped retroviruses with the aim of labeling postnatally born glial cells. In this case, no attempt was made to perform clonal analysis, as this was done in the past with this combination of viruses injected into this site, albeit with virions with the MMLV ecotropic envelope protein (Levison and Goldman, 1993). The experiment done here was to address only whether the cTVA would recombine and express at a sufficient level for infection with EnvA-pseudotyped viruses. Mice were allowed to develop for 3 weeks after inoculation to permit clonal expansion and cell differentiation. Thereafter, brains were analyzed for the presence of viral infection. Both astrocytes and oligodendrocytes labeled for either β-galactosidase or PLAP activity were observed in cTVA+/Cre+ (6/7) (Figure 6A, B) but not cTVA+/Cre− (0/4) mice. Astrocytes and oligodendrocytes were the predicted progeny based upon classical birthdating studies and the previous retroviral lineage studies (Altman, 1966; Levison and Goldman, 1993).
Figure 6.
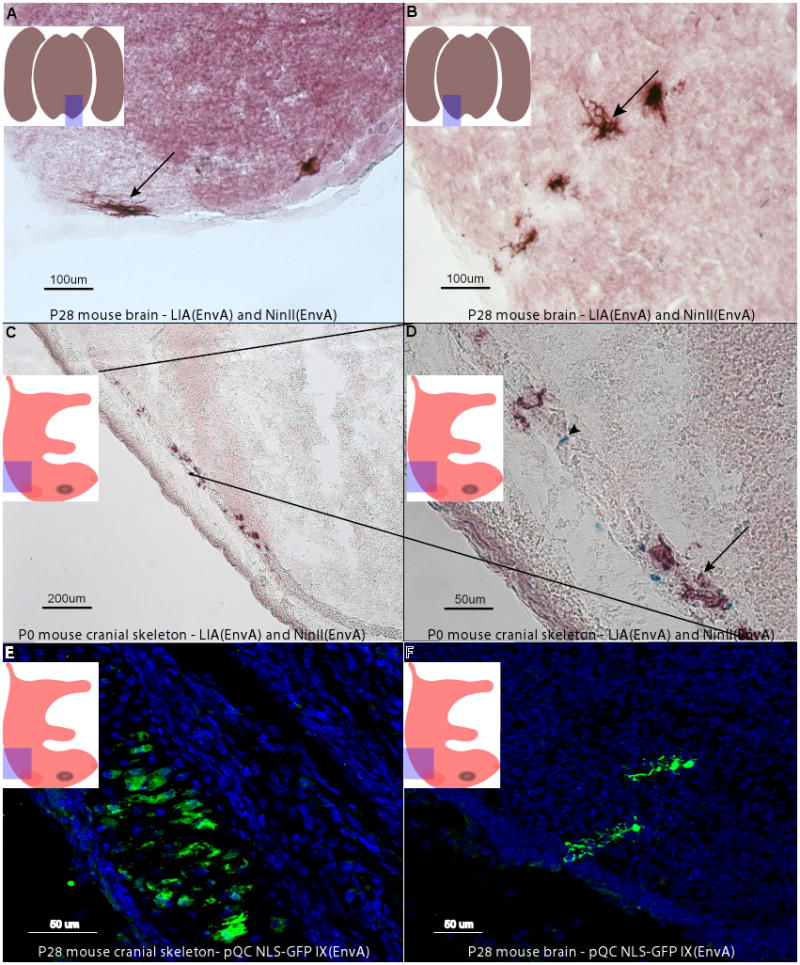
Infection of mouse brains with LIA and NinII pseudotyped with EnvA. P2 mouse pups of the cross cTVA+/− x hβactin-Cre +/− were injected in the subventricular zone of the midbrain and sacrificed at P28. Coronal sections were made and stained histochemically for PLAP and β-galactosidase activity. The area of the sections is indicated in the highlighted area of the embedded cartoon. Both viruses labeled oligodendrocytes (A, arrow) and astrocytes (B, arrow), glial types known to be produced postnatally. Viral labeling was only seen in animals with TVA and Cre; all other genotypes of this cross were negative. Wnt1-Cre +/− mice were crossed to cTVA +/− mice, and the embryos were injected in utero into the third ventricle with LIA(EnvA) and NinII(EnvA), or QC NLS-GFP IX(EnvA) (C–F). The embedded cartoons show the area of the sections that are shown. Cells in the cranial skeleton that are descended from Wnt1 expressing progenitor cells, infected by LIA(EnvA) (arrow in D), NinII(EnvA) (arrowhead in D), or QC NLS-GFP IX(EnvA) (E) in cTVA+/Cre+ mice, are shown. Hindbrain neurons also were labeled, as demonstrated by a labelled Purkinje cell (F), generated by infection with QC NLS-GFP IX(EnvA). Injections into TVA− or Cre− animals were negative.
To investigate the utility of this tool in a line expressing Cre in a more restricted cell population, a Wnt1-Cre mouse was crossed to a cTVA L1 mouse. Wnt1-Cre is expressed embryonically, starting at approximately e8.5 in the neural tube (McMahon and Bradley, 1990). Areas shown to express Wnt1 include the embryonic mesencephalon, dorsal myelencephalon, and spinal cord, and later also the rhombic lip and caudal diencephalon (Dymecki and Tomasiewicz, 1998). The early expression domain of Wnt1-Cre ultimately gives rise to cells in the skeleton of the head, as well as neurons in the cerebellum, midbrain, spinal cord, and dorsal root ganglia (Dymecki and Tomasiewicz, 1998; Ikeya et al., 1997; McMahon and Bradley, 1990; Shackleford and Varmus, 1987). As progenitor cells with a history of Wnt1-Cre expression (and therefore TVA-expressing) were accessible by ventricular injection at early developmental stages, viral injections were targeted to the cycling progenitors lining the ventricular space at e13.5. Pups were sacrificed at birth and tissues scored for virally-labeled cells. In a single litter, 4/4 cTVA+/Wnt1-Cre+ embryos had cells with evidence of LIA infection, and other cells showing evidence of NinII infection (Figure 6C–E). No embryos (0/3) that should not express TVA exhibited virally-labeled cells. The labeled cells observed in the cTVA+/Wnt1-Cre+ positive embryos were in the cranial skeleton, which is derived from the cranial neural crest. As well, neurons in the hindbrain, including cerebellar Purkinje cells, which are known to have a Wnt1-Cre history in this transgenic line, were labeled (Figure 6F). The productive infection of TVA+ progenitor cells, and the lack of infection in mice with no TVA in these two additional areas, demonstrates that cTVA mice should be useful for applications in which EnvA-pseudotyped viruses are employed, either for lineage studies, or simply introduction of genes into specific cells with a Cre history.
Multiple types of reporter viruses for marking infected cells
Coinfection with viruses that encode distinct marker genes allowed for the comparison described above regarding the efficiencies of infection with viruses with EnvA and VSV-G (Figure 4). This same strategy of mixing together viruses with distinct marker genes can also be employed using viruses with the same glycoprotein in order to enable definition of clonal boundaries. This is sufficient when there is a small or moderate amount of mixing of clones, and allows for a more efficient way to follow lineages, as one can examine multiple clones following infection of the same animal (Cepko et al., 1993; Fields-Berry et al., 1992; Halliday et al., 1992; Leber et al., 1995; Peters et al., 2002). When an extreme amount of migration causes clones to be completely intermingled, infection with a few types or markers is not sufficient, and the use of viral libraries with, e.g. >102 genetic tags, is required (Cepko et al., 1998; Golden et al., 1995; Walsh et al., 1992). Beyond enabling clonal assignments, viruses with distinct histochemical or fluorescent reporters enable different types of assays for detection of infected cells, and for co-labeling of infected cells with cell type-specific antibodies. To this end, a number of replication-incompetent retroviruses were created and tested in several combinations.
A nuclear, cytoplasmic, and membrane version of fluorescent reporters, including, CFP, GFP, YFP, Kusabira Orange, tdTomato, and mCherry were made and tested (Shaner et al., 2005). In addition to viruses encoding fluorescent reporters, the viruses encoding histochemically detectable enzymes, such as nuclear or cytoplasmically localized β-galactosidase, and membrane-bound PLAP, can be combined with each other and/or with the fluorescent reporter viruses. For examples, we tested several combinations, as shown in Figure 7. It is worth noting that the combinations shown in Figure 7 were not meant to highlight clonal boundaries: high concentrations of viruses were used so that a comparison of cells infected with different viruses could be seen in the same micrograph. For the establishment of clonal boundaries, one would use many fewer viral particles, and for added confidence, a dilution series would be run (e.g. see Turner and Cepko, 1987).
Figure 7.
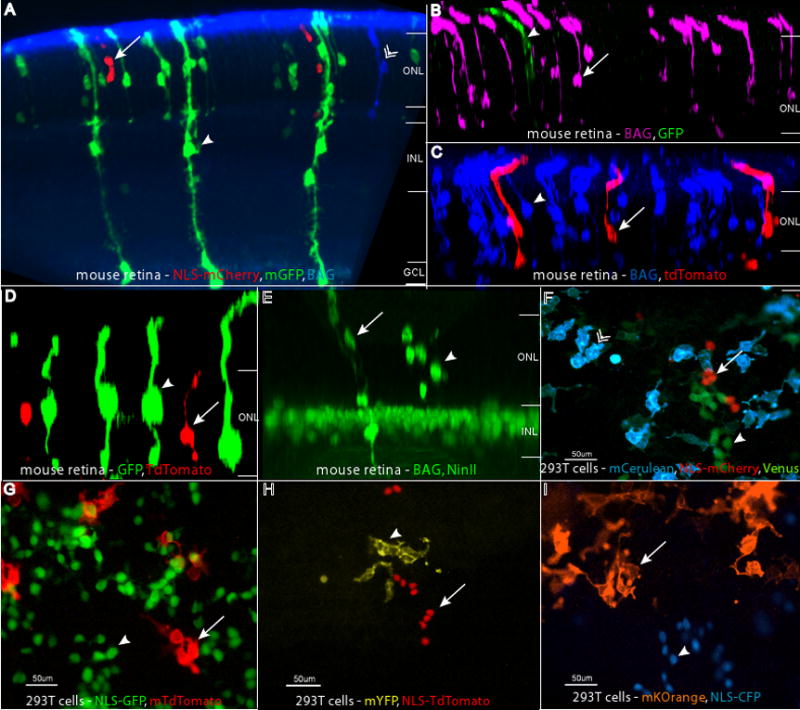
Retroviruses with distinct histochemical and fluorescent reporters. Combinations of BAG, LIA, and QC viruses were injected in vivo into the subretinal space of P0 retinas, and the retinas were examined at P21. The reporter combinations shown are (A) GFP (anti-GFP, in green, arrowhead), mCherry (anti-RFP, in red, arrow), and BAG (anti-β-galactosidase, in blue, chevron), (B), GFP (anti-GFP, in green, arrowhead) and BAG (anti-β-galactosidase, in magenta, arrow) (C), tdTomato (anti-RFP, in red, arrow) and BAG (anti-β-galactosidase, in blue, arrowhead), (D) GFP (anti-GFP, in green, arrowhead) and tdTomato (anti-RFP, in red, arrow), (E) NinII (anti-β-gal, in green, arrowhead) and BAG (anti-β-gal, in green, arrow) with Chx10-NLS-GFP, a gene in the transgenic background, marking bipolar cell nuclei. The retinal layers are labeled in panels A–E as follows: ONL = outer nuclear layer, INL = inner nuclear layer, GC = ganglion cell layer. Panels F–I show images of infected 293T cells. These combinations are: (F) membrane Cerulean (blue, chevron), Venus (green, arrowhead), and NLS-mCherry (red, arrow), (G) NLS-GFP (green, arrowhead) and membrane tdTomato (red, arrow), (H) NLS-tdTomato (red, arrow) and membrane YFP (yellow, arrowhead), and (I) membrane Kusabira Orange (orange, arrow) and NLS-CFP (blue, arrowhead).
The examples in Figure 7 illustrate the advantages of several of these reporter viruses and their combinations. Fluorescent viral reporters provide a fairly easy way to define clonal boundaries. Several viruses with distinct fluorescent properties can be made using the expanding palette of fluorescent reporters, and then one can use a single type of tissue preparation and microscopy to simultaneously detect different clones (Figure 7F–I). This does require a level of expression sufficient for detection of the fluorescent proteins, without immunohistochemical detection. When reporter expression from the viral genome is not sufficiently bright, one can use antibodies to give each protein a different color. For example, in Figure 7A, the fluorescence of already fluorescent GFP and mCherry was enhanced by the use of antibodies, and the non-fluorescent β-galactosidase enzyme was detected by a β-galactosidase antibody, whose signal was converted into Cy5 fluorescence. Depending on which cocktail of viruses is used, the detection of β-galactosidase can be computationally converted into magenta (Figure 7B) or blue (Figure 7C), making quantification easier. The combinations of viruses that will be useful for a particular application will depend upon the requirements for a particular experiment, as discussed further in the Discussion.
Discussion
Clonal analysis of cells with a specific gene expression history
The goal of this study was to evaluate the utility of the cTVA mouse as a means to map lineages from progenitor cells with a specified gene expression history. Integrating the high spatial and temporal resolution of viral infection with gene expression history, the cTVA mouse permits a finer analysis of the relationships between gene expression history and lineage relationships. Viruses can be injected into a particular location at different times throughout development in order to generate a lineage tree from cells with a specific gene expression history. Any existing Cre line can be used. Alternatively, viruses encoding Cre, e.g. Adenovirus with a cell type-specific promoter driving Cre (Maeda et al., 2006), can also be used in conjunction with the cTVA allele. Clones also can be generated by infecting cells with expression of TVA driven directly by a promoter, e.g. not cells with a Cre history, but cells that currently express TVA from a particular promoter. This allows one to build lineage trees through a comparison of clones initiated by progenitors with a history of Cre, as compared to those with current expression from a specific promoter. For this type of analysis, one can again use Adenoviruses, or any other vector, to drive expression of TVA using specific promoters, or use engineered mouse strains (Fisher et al., 1999; Holland and Varmus, 1998; Montaner et al., 2003; Orsulic et al., 2002).
The cTVA mice can be used to generate either broad or narrow patterns of TVA expression, and viruses with different titers can be used to deliver genes to a few or many TVA-expressing cells. The replication-competent avian vectors (RCAS) generated by Hughes and colleagues (Hughes et al., 1987) typically grow to higher titers than replication-incompetent vectors. When one wishes to transduce a gene or an shRNA into cells with TVA expression, rather than perform clonal lineage analysis, high titered preparations of RCAS vectors can be employed to infect more cells. For example, RCAS was used to deliver oncogenes to glial cells in a transgenic mouse which used a glial promoter (GFAP) to drive TVA expression, to create murine models of glioblastoma (Holland and Varmus, 1998; Holland et al., 1998). The flexibility of this system also allows use of other viruses with an EnvA protein, as many small enveloped viruses are promiscuous regarding their envelope proteins. For example, Wickersham et al. showed that rabies virus pseudotyped with a fusion protein of EnvA and the rabies glycoprotein could enter cells through TVA (Wickersham et al., 2007). We have also used lentiviral vectors pseudotyped with EnvA. This broadens the applications that are possible as lentiviruses can infect neurons and other postmitotic cells (Lewis et al., 2001).
Efficiency of EnvA-mediated infection
We observed infection by EnvA pseudotyped viruses in the mouse through TVA, as indicated in previous studies (Fisher et al., 1999; Holland and Varmus, 1998; Seidler et al., 2008). We were able to further investigate the efficiency of this phenomenon quantitatively in vivo and in vitro. By counting clones generated by infection with a cocktail of known titers of EnvA and VSV-G pseudotyped retroviruses, a relative efficiency could be calculated. Additionally, we determined the specificity of EnvA enveloped virus infection for murine cells in vivo and in vitro. If injected alone, the infectivity of EnvA viruses to non-TVA expressing cells was negligible using a number of virions typically used for standard retroviral infections (105 or fewer virions). No infection was observed in all MEF cultures and brains that did not have TVA expression. In the case of the NIH 3T3 cells, there was some background infection when TVA should not have been expressed. This was not due to a problem with the TVA allele being leaky, as the same level of expression was seen in cells that did not have the TVA transgene. The reason(s) for this low level of infectivity in certain tissues and cell lines is unclear, but these observations dictate that one should perform proper controls for each new cell line or tissue tested.
In addition to specificity, it is useful to know the efficiency of viral infection. This was measured by comparing the infection rates of cells transfected with CAG-TVA or cTVA plus a Cre plasmid. The cTVA + Cre combination was less efficient in mediating ASLV-A enveloped viral infection than CAG-TVA, despite equal transfection efficiencies and 100% co-transfection efficiency. This difference is likely due to different TVA expression levels, as TVA is driven by the CAG promoter in the CAG-TVA plasmid, and cTVA is driven by the human UbC promoter. In vivo, we found that the efficacy of TVA-mediated viral entry was roughly 25-fold lower than VSV-G-mediated viral entry in the retina, and in MEF cultures, it was about 550 times lower than VSV-G mediated entry. A similar reduction of infection efficiency of murine cells by EnvA pseudotyped viruses has been previously observed (Lewis et al., 2001). Infection of MEFs was particularly inefficient, for reasons that are unclear, but may be similar to the reasons discussed below.
Reduced efficiency of infection by EnvA pseudotyped viruses in the developing mouse may be due to a number of causes. In the retina, perhaps the TVA receptor is not present on the processes of progenitor cells extending into the subretinal space, where the viruses are injected, or where the processes are exposed to the medium, in the case of retinal explants. The expression of TVA from the UbC promoter might also be poor, though low levels of TVA have been reported to be sufficient for infection (Federspiel et al., 1994). It is also possible that EnvA-mediated entry simply does not give rise to as many infectious integration events in vivo as does VSV-G-mediated entry. For example, VSV-G fusion occurs in the early endosomes (Le Blanc et al., 2005; Sun et al., 2005; Sun et al., 2008), whereas EnvA appears to fuse after entry into the lysosome (Mothes et al., 2000). This could lead to increased genome degradation or alteration of the viral life cycle. It has been previously demonstrated that the infectivity of retroviruses is dependent on the mechanism of entry (Narayan et al., 2003). The further reduced ratio in the MEFs may be due to a limited receptor expression and a low probability of receptor : glycoprotein interaction. At 37°C, MMLV virions quickly decay; the half-life is less than 6 hours (Kwon et al., 2003). When injected subretinally, the virus has much less space to diffuse, and its envelope protein may more frequently contact its cognate receptor than in the MEFs. The reduced efficiency in MEFs may also be a reflection of differences between primary cells and immortalized cells or developing tissues.
A few differences between EnvA and VSV-G should be noted. VSV-G is often used for its efficiency and ability to expand the host range of retroviruses (Burns et al., 1993; Emi et al., 1991; Hopkins, 1993; Yoshida et al., 1997). The VSV-G protein allows VSV to infect a wide range of species and cell types, from invertebrates through vertebrates, and it has been speculated to have a phospholipid for its receptor, though this is not entirely clear (Lyles and Rupprecht, 2007). This is in contrast to EnvA pseudotyped viruses, which can only infect cells expressing the TVA receptor. Viral glycoproteins associate with their cognate receptors (or phospholipids) according to their equilibrium constants (Battini et al., 1996, Yu et al., 1995). Since phospholipids are much more abundant than protein surface receptors, a larger fraction of EnvA virions may remain unassociated with cells. Therefore, both in terms of the number of cells susceptible, as well as cellular and receptor topology, it is expected that VSV-G-mediated viral entry would be more efficient than EnvA-mediated infection. It is also of note that the standard method of viral titering, as determined in cell culture, is not always an accurate measure of the number of infectious retroviral particles. Discrepancies such as target cell type and number, the presence of polycations, temperature, and exposure time may all make titers somewhat inconsistent (Kwon et al., 2003). Mathematical models, as previously demonstrated (Kwon et al., 2002), could be used to more accurately determine the initial concentration of infectious retroviruses, which may yield different ratios in the assays used herein (Chen et al., 1999; Emi et al., 1991; Kwon et al., 2003; Ory et al., 1996; Yang et al., 1995).
A novel mechanism of viral entry
We observed that EnvA viruses could infect non-TVA expressing cells when co-injected with VSV-G enveloped viruses. The infection ratio was roughly 1 : 23 when infection with an EnvA virus was performed on cTVA+/Cre−, cTVA−/Cre+, and cTVA−/Cre− mouse cells relative to the cTVA+/Cre+ condition. These data were not in agreement with the data from infection of the MEFs derived from the cTVA mice, where no infection of EnvA viruses was observed on non-TVA expressing cells. We realized that the difference was likely the inclusion of the VSV-G enveloped control virus with the EnvA virus in the experiments that showed infection of non-TVA expressing cells. We hypothesized that there might be an association between the virions with the different glycoproteins. Indeed, when exclusively EnvA viruses were used to infect a retina that did not express TVA, no infection events were observed. These data support the hypothesis of an association between membranes containing different envelope proteins that permitted entrance of EnvA virions into otherwise non-permissible cells. By this hypothesis, if a VSV-G enveloped virus carried an EnvA virus into the cell, once inside, either viral genome could integrate. If the viral genome pseudotyped with VSV-G integrated, this would be scored as a VSV-G mediated event, and if the EnvA pseudotyped viral genome integrated, as an EnvA-mediated event. It is also possible that both genomes would integrate into the same cell, and both markers would be seen. However, this event is unlikely, as the majority of retroviral particles are non-infectious. As the particle : infectious particle ratio is 143 : 1 (Kwon et al., 2003), the chance of successful co-integration is about 104. This means that only one histochemical marker would likely be detected in these cells. In accord with this calculation, in no instance was a double β-galactosidase/PLAP labeled cell observed.
This series of experiments led to the identification of a novel means of viral entry, whereby EnvA retroviruses entered and infected cells by a non-TVA mediated pathway. From a practical standpoint, this phenomenon is worth being aware of in terms of planning experiments. From a virology standpoint, it is of interest to understand how viral entry is mediated for one virus by another.
Cocktails of viruses with distinctive reporter genes
We have created a number of novel replication-incompetent retroviruses for use in lineage analyses and other types of cell marking applications, such as the labeling of dividing cells in a tissue. Both fluorescent proteins and genes encoding enzymes that can yield colored precipitates from histochemical reactions are of use in these applications. Fluorescent reporters have multiple advantages, including imaging in live tissue. They allow one to screen tissue for infection prior to performing histological analyses. They can be readily combined with detection methods aimed at defining a cell type using antibodies and fluorescent immunohistochemistry. However, as the viral infection leads to only a single copy of the genome, fluorescent proteins are not always expressed at a high enough level to give robust staining of all cells, or all aspects of the defining morphological characteristics of a cell. This can be overcome with immunohistochemical detection of GFP and its derivatives. This however limits the number of such GFP-derivative viruses that can be used in combination, unless one uses combinations that target distinct cell compartments, e.g. membrane-bound, nuclear, and/or cytoplasmic. As we have made various targeted versions of each fluorescent protein, this might suffice for some applications.
The viruses encoding β-galactosidase and PLAP offer the greatest sensitivity for detection of infected cells. In addition, they provide for a very stable label following histochemical detection with Xgal or BCIP/NBT. These reaction products have been stable in stained sections stored at room temperature for at least 7 years in our laboratory. In addition to sensitivity, the histochemical detection methods offer another advantage. Both β-galactosidase and PLAP enzymatic assays can be rapidly and easily carried out, and make the cells easy to detect and score, even in thick tissue sections or whole mount preparations. However, if one wishes to combine immunohistochemistry for a cell type-specific protein in order to better define an infected cell stained with e.g. BCIP/NBT used to detect PLAP, it is difficult to do so. The BCIP/NBT reaction products absorb most wavelengths of light, making fluorescent detection of the cell type-specific protein very difficult. Similarly, the histochemical reaction products can be very dark and it is difficult to see e.g. HRP reaction products on top of the BCIP/NBT or Xgal reaction products. One way to circumvent this problem is to use nuclear β-galactosidase combined with immunohistochemical detection of a membrane or cytoplasmically localized cell type-specific protein. In addition, there are excellent commercial antibodies for both β-galactosidase and PLAP, permitting immunofluorescent labeling of cells infected with viruses encoding these genes, and thus double immunofluorescent detection of the viral label and a cell type-specific protein is possible. These vectors will be freely available from Addgene (http://www.addgene.org) and thus investigators can find optimal combinations of the different vectors for their own applications.
We conclude that the cTVA mice are useful for lineage analysis of cells with a particular gene expression history. The mice permit specific infection of cells with a Cre expression history, and no background was observed in any tissue injected with EnvA virions alone. In addition to lineage analysis, these mice offer a way to introduce genes into cells with a specific gene expression history. For example, viruses carrying shRNAs or gain of function constructs can be delivered to only a subset of cells in a tissue, those with a history of Cre expression.
Supplementary Material
Supplemental Figure 1: Murine NIH 3T3 cells were co-transfected with pCAG GFP:Cre and pCALNL DsRed. After three days, the cells were fixed and DAPI stained. GFP, DsRed, and DAPI were detected by intrinsic fluorescence. The DAPI and GFP-positive cells were counted in each of six 5x magifnication fields and the GFP : DAPI ratio (transfection efficiency) was obtained by averaging these ratios (A). Errors bars indicate ± 1 SD. The Cre efficiency was measured by quantifying the number of DSRed cells relative to GFP:Cre cells (B–E). All 230 GFP:Cre positive cells (B, E) were also DsRed positive (overlay, C, E).
Acknowledgments
We would like to thank S.Dymecki, J. Trimarchi, and N. Billings for technical assistance and helpful discussions. We are grateful to C. Punzo and R. Kanadia for critical review of the manuscript. This work was supported by NIH R01EY008064. K. Beier was supported by the NIH grant F31NS068012. C.L. Cepko is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- Altman J. Proliferation and migration of undifferentiated precursor cells in the rat during postnatal gliogenesis. Exp Neurol. 1966;16:263–278. doi: 10.1016/0014-4886(66)90063-x. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Lo Turco JJ, Rush J, Cepko C. Taurine promotes the differentiation of a vertebrate retinal cell type in vitro. Development. 1993;119:1317–1328. doi: 10.1242/dev.119.4.1317. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P, Young JA, Varmus HE. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- Battini JL, Rodrigues P, Müller R, Danos O, Heard JM. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Ryder EF, Austin CP, Walsh C, Fekete DM. Lineage analysis using retrovirus vectors. Methods in Enzymology. 1993;225:933–960. doi: 10.1016/0076-6879(93)25059-b. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin C, Golden J, Fields-Berry S, Lin J. Lineage analysis using retroviral vectors. Methods. 1998;14:393–406. doi: 10.1006/meth.1998.0594. [DOI] [PubMed] [Google Scholar]
- Cepko C, Pear W. Curr Protoc Mol Biol. Unit9. Chapter 9. 2001. Retrovirus infection of cells in vitro and in vivo; p. 14. [DOI] [PubMed] [Google Scholar]
- Chen CM, Smith DM, Peters MA, Samson ME, Zitz J, Tabin CJ, Cepko CL. Production and design of more effective avian replication-incompetent retroviral vectors. Dev Biol. 1999;214:370–384. doi: 10.1006/dbio.1999.9432. [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Dymecki S, Tomasiewicz H. Using Flp recombinase to characterize expansion of Wnt1- expressing neural progenitors in the mouse. Developmental Biology. 1998;201:57–65. doi: 10.1006/dbio.1998.8971. [DOI] [PubMed] [Google Scholar]
- Emi N, Friedmann T, Yee JK. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Federspiel MJ, Bates P, Young JA, Varmus HE, Hughes SH. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci USA. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields-Berry SC, Halliday AL, Cepko CL. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Orsulic S, Holland E, Hively WP, Li Y, Lewis BC, Williams BO, Varmus HE. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene. 1999;18:5253–5260. doi: 10.1038/sj.onc.1203087. [DOI] [PubMed] [Google Scholar]
- Golden JA, Fields-Berry SC, Cepko CL. Construction and characterization of a highly complex retroviral library for lineage analysis. Proc Natl Acad Sci USA. 1995;92:5704–5708. doi: 10.1073/pnas.92.12.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday AL, Cepko CL. Generation and migration of cells in the developing striatum. Neuron. 1992;9:15–26. doi: 10.1016/0896-6273(92)90216-z. [DOI] [PubMed] [Google Scholar]
- Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci USA. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins N. High titers of retrovirus (vesicular stomatitis virus) pseudotypes, at last. Proc Natl Acad Sci USA. 1993;90:8759–8760. doi: 10.1073/pnas.90.19.8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Kim JC, Dymecki SM. Genetic fate-mapping approaches: new means to explore the embryonic origins of the cochlear nucleus. Methods Mol Biol. 2009;493:65–85. doi: 10.1007/978-1-59745-523-7_5. [DOI] [PubMed] [Google Scholar]
- Kwon YJ, Hung G, Anderson WF, Peng C, Yu H. Determination of infectious retrovirus concentration from colony-forming assay with quantitative analysis. J Virol. 2003;77:5712–5720. doi: 10.1128/JVI.77.10.5712-5720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YJ, Peng C. Engineering analysis of ex vivo retroviral transduction system. Ann Biomed Eng. 2002;30:731–742. doi: 10.1114/1.1484221. [DOI] [PubMed] [Google Scholar]
- Landau NR, Littman DR. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I, Luyet P, Pons V, Ferguson C, Emans N, Petiot A, Mayran N, Demaurex N, Fauré J, Sadoul R, Parton RG, Gruenberg J. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7:653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber SM, Sanes JR. Migratory paths of neurons and glia in the embryonic chick spinal cord. J Neurosci. 1995;15:1236–1248. doi: 10.1523/JNEUROSCI.15-02-01236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levison SW, Goldman JE. Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 1993;10:201–212. doi: 10.1016/0896-6273(93)90311-e. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- Lewis BC, Chinnasamy N, Morgan RA, Varmus HE. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J Virol. 2001;75:9339–9344. doi: 10.1128/JVI.75.19.9339-9344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lyles DS, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Straus SE, Martin MA, Roizman B, editors. Fields virology. 5. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkin; 2007. pp. 1363–1408. [Google Scholar]
- Maeda M, Namikawa K, Kobayashi I, Ohba N, Takahara Y, Kadono C, Tanaka A, Kiyama H. Targeted gene therapy toward astrocytoma using a Cre/loxP-based adenovirus system. Brain Res. 2006;1081:34–43. doi: 10.1016/j.brainres.2006.01.105. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1095. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner S, Sodhi A, Molinolo A, Bugge TH, Sawai ET, He Y, Li Y, Ray PE, Gutkind JS. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi’s sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell. 2003;3:23–36. doi: 10.1016/s1535-6108(02)00237-4. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Narayan S, Barnard RJO, Young JAT. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J Virol. 2003;77:1977–1983. doi: 10.1128/JVI.77.3.1977-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MA, Cepko CL. The dorsal-ventral axis of the neural retina is divided into multiple domains of restricted gene expression which exhibit features of lineage compartments. Dev Biol. 2002;251:59–73. doi: 10.1006/dbio.2002.0791. [DOI] [PubMed] [Google Scholar]
- Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, Cepko CL. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev Dyn. 2008;237:1034–42. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Schorpp M, Jäger R, Schellander K, Schenkel J, Wagner EF, Weiher H, Angel P. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci USA. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleford GM, Varmus HE. Expression of the proto-oncogene int-1 Is restricted to postmeiotic male germ cells and the neural tube of mid-gestational embryos. Cell. 1987;50:89–95. doi: 10.1016/0092-8674(87)90665-9. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Sun X, Belouzard S, Whittaker GR. Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. J Biol Chem. 2008;283:6418–6427. doi: 10.1074/jbc.M708955200. [DOI] [PubMed] [Google Scholar]
- Sun X, Yau VK, Briggs BJ, Whittaker GR. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255:434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- Weiss R, Teich N, Varmus H, Coffin J. RNA tumor viruses. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1985. [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young JAT, Callway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Sullivan HA, Seung HS. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat Protoc. 2010;5:595–606. doi: 10.1038/nprot.2009.248. [DOI] [PubMed] [Google Scholar]
- Yang Y, Vanin EF, Whitt MA, Fornerod M, Zwart R, Schneiderman RD, Grosveld G, Nienhuis AW. Inducible, high-level production of infectious murine leukemia retroviral vector particles pseudotyped with vesicular stomatitis virus G envelope protein. Hum Gene Ther. 1995;6:1203–1213. doi: 10.1089/hum.1995.6.9-1203. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Emi N, Hamada H. VSV-G-pseudotyped retroviral packaging through adenovirus-mediated inducible gene expression. Biochem Biophys Res Commun. 1997;232:379–382. doi: 10.1006/bbrc.1996.5976. [DOI] [PubMed] [Google Scholar]
- Young JA, Bates P, Varmus HE. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985a;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 1985b;353:229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- Yu H, Soong N, Anderson WF. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J Virol. 1995;69:6557–6562. doi: 10.1128/jvi.69.10.6557-6562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Murine NIH 3T3 cells were co-transfected with pCAG GFP:Cre and pCALNL DsRed. After three days, the cells were fixed and DAPI stained. GFP, DsRed, and DAPI were detected by intrinsic fluorescence. The DAPI and GFP-positive cells were counted in each of six 5x magifnication fields and the GFP : DAPI ratio (transfection efficiency) was obtained by averaging these ratios (A). Errors bars indicate ± 1 SD. The Cre efficiency was measured by quantifying the number of DSRed cells relative to GFP:Cre cells (B–E). All 230 GFP:Cre positive cells (B, E) were also DsRed positive (overlay, C, E).


