Abstract
Regulation of protein kinase A (PKA) by binding of cAMP to the regulatory subunit and the resulting release of the active catalytic subunit is a very well established mechanism of kinase activation. We have shown recently that PKA in budding yeast is also subject to an additional level of regulation that that modulates its activity in response to nutrient availability. Nutrient regulation of PKA activity requires a pair of proteins, Gpb1 and Gpb2, that contain several kelch repeats, a sequence motif that predicts that they fold into a β-propeller structure. The regulatory process mediated by Gpb1 and Gpb2 causes an increase in the stability and phosphorylation of the PKA regulatory subunit Bcy1 in response to low extracellular glucose concentrations. Phosphorylation of serine-145 of Bcy1 controls its stability, and other phosphorylation events at the cluster of serines at positions 74–84 correlate with changes in nutrient availability. Here we present data consistent with a model in which the effects of Gpb1 and Gpb2 on Bcy1 are an indirect consequence of their primary effects on the PKA catalytic subunits.
Key words: protein kinase A, kelch repeat proteins, nutritional signaling, Saccharomyces cerevisiae
Introduction
Protein kinase A (PKA) or cAMP-dependent protein kinase, is one of the most well studied kinases in eukaryotic biology. The inactive form of PKA consists of a complex of two catalytic subunits bound to two regulatory subunits.1 The classic mechanism of protein kinase A activation, which is highly conserved in eukaryotes, involves binding of cAMP to the regulatory subunit, resulting in the release of free, active catalytic subunits. Because PKA was first characterized more than 40 years ago,2 it is generally thought that its regulation is now very well understood. However, recent studies in yeast have revealed that there may be something new to learn about this venerable enzyme.
In the yeast Saccharomyces cerevisiae, PKA is required for normal growth, regulation of energy metabolism and control of stress responses.3 PKA is also thought to be involved in the response to nutrients, based on the observation that addition of a fermentable sugar to yeast cells that have been starved for a carbon source results in a spike of cAMP. In yeast, adenylate cyclase is activated by the heterotrimeric G protein β-subunit Gpa2, as well as by Ras proteins (Fig. 1). Gpa2 is coupled to a cell surface receptor, Gpr1, that may directly detect nutrients in the extracellular environment.4–8 The way in which Ras is activated by nutrient signals is not known. Once activated, Gpa2 and Ras stimulate adenylate cyclase to produce cAMP, which activates PKA by the classic mechanism. The yeast PKA regulatory subunit is called Bcy1 and the three isoforms of the PKA catalytic subunit are called Tpk1, Tpk2 and Tpk3. The PKA catalytic subunits phosphorylate transcription factors that regulate growth, energy metabolism and stress responses.
Figure 1.
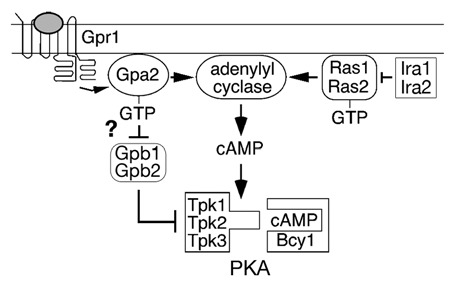
The PKA pathway in Saccharomyces cerevisiae.
A novel regulatory process that affects signaling through the PKA pathway involves a pair of proteins, Gpb1 and Gpb2, that contain several copies of a domain that was originally identified in the Drosophila protein kelch. Sequence analysis of Gpb1 (also called Krh2) and Gpb2 (also called Krh1) has shown that their C-terminal region contains either six9 or seven10,11 kelch repeats. Based on the structure of other kelch repeat proteins, this sequence would predict that they fold into a six- or seven-bladed β-propeller,12 a structure that is known to participate in protein-protein interactions. Gpb1 and Gpb2 also contain a large region of sequence at their N-terminal region that is not homologous to other proteins.
The way in which Gpb1 and Gpb2 regulate signal transduction has been controversial since these proteins were first described. Gpb2 was originally identified based on its ability to interact with the G protein β-subunit Gpa2 in a two-hybrid screen, and Gpb1 was found by its homology to Gpb2.9–11 Proposed mechanisms to explain their effects on signaling include acting as G protein β-subunit mimics10 or G protein effectors,9 functioning to inhibit receptor-G protein coupling,13 to positively regulate Ras GTPase activating proteins,14 and to stimulate the association between PKA regulatory and catalytic subunits.11 One characteristic of Gpb1 and Gpb2 that has been confirmed by several different methods is their ability to bind Gpa2.9–11,15 However, the functional significance of this binding is not well understood.
In the present study, we have focused on the predominant function of Gpb1 and Gpb2, which involves inhibition of PKA activity by a mechanism that does not involve changes in cAMP.11,16,17 Previous work by others has shown that cells lacking Gpb1 and Gpb2 display decreased binding of the PKA regulatory subunit to the catalytic subunits.11 This phenotype could be due to direct effects of Gpb1 and Gpb2 on either the regulatory subunit or the catalytic subunits. Here we investigate the effects of Gbp1 and Gpb2 on the PKA regulatory subunit Bcy1.
Our recent study has shown that Gpb1 and Gpb2 affect both the abundance and phosphorylation state of Bcy1 in response to nutrients.18 In low glucose concentrations, Bcy1 abundance increases due to a process that requires the kelch repeat proteins. The increase in Bcy1 abundance that occurs when nutrients are scarce is due to an increase in protein stability. Stability of Bcy1 is controlled by phosphorylation at serine-145, the site of Bcy1 phosphorylation by PKA catalytic subunits. Phosphorylation of Bcy1 at serine-145 appears to target it for degradation in cells that lack Gpb1 and Gpb2. In addition to the effect of Gpb1 and Gpb2 on Bcy1 stability, the kelch repeat proteins also promote phosphorylation of Bcy1 at an unknown site when cells are grown in low glucose concentrations. The effects of Gpb1 and Gpb2 on Bcy1 stability and phosphorylation are eliminated when the kinase activity of PKA catalytic subunits is inhibited using variants of the Tpk proteins that are sensitive to an ATP analog. Therefore, we undertook to determine whether the effects of Gpb1 and Gpb2 on Bcy1 abundance and phosphorylation are a direct effect on the regulatory subunit or are an indirect effect resulting from their regulation of the catalytic subunits. Regulation of the catalytic subunits could affect Bcy1 abundance and phosphorylation indirectly by several different mechanisms. For example, Gpb1 and Gpb2 could alter the catalytic subunits in a way that results in an increase in the amount of Bcy1 bound to them. Binding of Tpk proteins to Bcy1 could cause Bcy1 to be protected from degradation and to be phosphorylated by another kinase.
Results
To explore further the process by which Gpb1 and Gpb2 inhibit PKA activity, studies were performed to identify the region of Bcy1 that is the target of regulation by Gpb1 and Gpb2. In addition to regulating Bcy1 stability, Gpb1 and Gpb2 also promote Bcy1 phosphorylation at an unknown site when cells are grown in low glucose concentrations.18 Phosphorylation of Bcy1 at this site also occurs when PKA catalytic activity is inhibited in strains containing variants of Tpk1, Tpk2 and Tpk3 that are sensitive to the ATP analog 1NM-PP1.19 Therefore, a study was performed in the Tpk analog-sensitive strain to identify the region in Bcy1 that contains the sites that are phosphorylated when PKA activity is inhibited.
A series of internal deletions were constructed in the BCY1 gene, and these constructs were expressed in the Tpk analog-sensitive strain. Treatment of these cells with 1NM-PP1 revealed that deleting amino acids 78–170 eliminates the slower mobility form of Bcy1 (Fig. 2). Bcy1 lacking amino acids 135–170 retains the ability to be phosphorylated. Therefore, the region containing the phosphorylated amino acids encompasses residues 78–135. It was shown previously that this region of Bcy1 contains a cluster of phosphorylated amino acids that include serines 74, 77, 79, 81, 83 and 84.20 Therefore, additional experiments were carried out to test whether these sites are phosphorylated in a manner that depends on Gpb1 and Gpb2.
Figure 2.
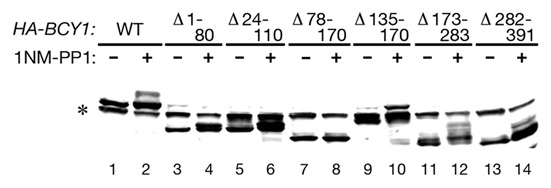
(A) Cell lysates were prepared from Tpk analog-sensitive strain Y3527 (tpk1M164G tpk2M147G tpk3M165G) expressing the indicated HA-BCY1 deletions that were untreated or treated with 1 µM 1NM-PP1 for 90 min. Lysates were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody. (B) Schematic diagram of Bcy1 deletion constructs indicating their ability to undergo phosphorylation when PKA is inhibited. Asterisk indicates non-specific band.
Wild type cells grown in a low concentration of glucose contain a slower mobility form of Bcy1 and gpb1Δ gpb2Δ mutants display a greatly decreased level of this form of Bcy1.18 To determine whether the slower mobility form of Bcy1 is modified by phosphorylation of serines at positions 74–84, alleles of BCY1 were constructed in which either serines 77 and 79 or 83 and 84 were replaced by alanines. The BCY1S77,79A and BCY1S83,84A alleles were expressed in GPB1 GPB2 and gpb1Δ gpb2Δ cells, and Bcy1 was detected in these cells grown in low glucose. Cells lacking Gpb1 and Gpb2 display a greatly decreased level of the slower mobility form of Bcy1 (Fig. 3), confirming our recent results.18 In wild type cells containing the Bcy1S77,79A variant, there is very little of the slower mobility form of Bcy1, and in cells containing the Bcy1S83,84A variant, the slower mobility form of Bcy1 is not detectable. These results demonstrate that phosphorylation of the cluster of serines at positions 74–84 in Bcy1 is responsible for generating the slower mobility form of Bcy1 that is observed in wild type cells grown in low concentrations of glucose.
Figure 3.
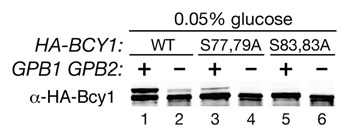
Cell lysates were prepared from the following strains grown in 0.05% glucose to log phase: strain HS287-2C (bcy1Δ) carrying plasmids containing wild type HA-BCY1, HA-BCY1S77,79A or HA-BCY1S83,84A (lanes 1, 3 and 5) and strain HS293-10D (gpb1Δ gpb2Δ bcy1Δ) carrying plasmids containing wild type HA-BCY1, HA-BCY1S77,79A or HA-BCY1S83,84A (lanes 2, 4 and 6). Lysates were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody.
One important question raised by these results is whether alterations in Bcy1 phosphorylation at the serine 74–84 cluster is a primary effect of regulation by Gpb1 and Gpb2 or a consequence of that effect. One way to address this question is to determine whether this serine cluster is present in PKA regulatory subunits from other organisms, as would be expected if phosphorylation at these sites represents a widely conserved regulatory mechanism. Conservation of the regulatory process mediated by Gpb1 and Gpb2 is supported by the observation that the yeast kelch repeat proteins stimulate binding of the mammalian PKA regulatory and catalytic subunits in a two-hybrid assay.11 A survey of the sequences of PKA regulatory subunits from closely related fungi reveals that serines 74, 77, 79, 81, 83 and 84 are not conserved in these species. Therefore, it is not likely that phosphorylation of Bcy1 at the 74–84 cluster is the initial event mediated by Gpb1 and Gpb2.
Another important question raised by these results is the identity of the kinase that phosphorylates the serines at positions 74–84 of Bcy1. It has previously been shown that cells lacking the Yak1 kinase display a very low degree of phosphorylation of the serine 74–84 cluster.20 However, deletion of the YAK1 gene has no effect on the phenotypes conferred by gpb1Δ gpb2Δ mutations.11,18 Therefore, if Yak1 is the kinase that phosphorylates the serines at positions 74–84 of Bcy1, then it is not likely that phosphorylation of Bcy1 at these sites is mechanism by which Gpb1 and Gpb2 control PKA activity.
These results indicate that the nutrient-regulated phosphorylation of Bcy1 that is dependent on Gpb1 and Gpb2 is most likely an indirect consequence of the primary regulatory mechanism mediated by the kelch repeat proteins. If the primary target of Gpb1 and Gpb2 regulation is not the regulatory subunit of PKA, then the most obvious candidate for this target is the catalytic subunit.
Perspective
PKA in yeast is regulated by a pair of kelch repeat proteins that act downstream of adenylate cyclase to directly affect the activity of the kinase.11,16 We have shown recently that cells lacking Gpb1 and Gpb2 display a decrease in the abundance and phosphorylation of the PKA regulatory subunit Bcy1, and that this phenotype increases in severity as the concentration of nutrients is reduced.18 Here we present evidence suggesting that the effect of gpb1Δ gpb2Δ mutations on Bcy1 is most likely an indirect consequence of their effect on the catalytic subunits. Given that PKA is highly conserved in eukaryotes, and that the presence of Gpb1 and Gpb2 can affect the binding of mammalian PKA regulatory and catalytic subunits,11 these results could indicate that the activity PKA is subject to control by a widely conserved mechanism that acts directly on the catalytic subunits.
Consistent with the idea that Gpb1 and Gpb2 directly affect the Tpk proteins, it has been shown that Gpb1 and Gpb2 can form complexes with bacterially synthesized Tpk1, Tpk2 and Tpk3.11 Gpb1 and Gpb2 are predominantly cytoplasmic proteins,13,15 but the Tpk proteins localize alternately to the cytoplasm or the nucleus depending on nutritional conditions.21 These observations raise the intriguing possibility that Gpb1 and Gpb2 could interact with the Tpk proteins when they are cytoplasmic, causing changes in their modification that affect their localization. Moreover, it has been reported that phosphorylation of Tpk1 changes in response to glucose in a manner that is dependent on Gpr1, the receptor that is coupled to the G protein α-subunit Gpa2.22 Thus a speculative pathway can be sketched out in which nutritional changes in the environment cause occupation of a receptor at the cell surface, resulting in activation of a G protein that controls the function of Gpb1 and Gpb2. Interaction of Gpb1 and Gpb2 with activated Gpa2 could cause modification and release of bound PKA catalytic subunits. Released catalytic subunits could then translocate to the nucleus and phosphorylate their substrates. In this model, changes in Bcy1 abundance and phosphorylation would reflect the amount of Bcy1 that is bound to the catalytic subunits. Bcy1 is stable and hyperphosphorylated under conditions of low glucose, and a large proportion of it would be expected to be bound to the catalytic subunits when nutrient availability is low. The amount of Bcy1 bound to the catalytic subunits would be due to the combined effects of the cAMP concentration in the cell and the effect of Gpb1 and Gpb2 on catalytic subunit modification or localization.
In summary, studies by a number of labs have shown that, in addition to its regulation by cAMP, PKA activity is affected by the regulatory process mediated by Gpb1 and Gpb2. Given the broad role of PKA in many physiological processes, it is not surprising to find that its activity is controlled by more than one type of regulation. Future studies will be required to determine how the nutrient signal is transmitted to Gpb1 and Gpb2 and to establish the precise mechanism by which these proteins modulate the activity of PKA regulatory and catalytic subunits.
Acknowledgements
We thank J. Broach, G. Griffioen and S. Palecek and S. Kron for plasmids and strains used in this work. This work was supported by grant GM074242 and by ARRA supplement GM074242-04S1 from the National Institutes of Health.
Abbreviations
- PKA
protein kinase A
- 1NM-PP1
1-(1,1-dimethylethyl)-3-(1naphthalenylmethyl)-1H-pyrazolo[3,4d]pyrimidin-4-amine
References
- 1.Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, et al. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 3.Rubio-Texeira M, Van Zeebroeck G, Voordeckers K, Thevelein JM. Saccharomyces cerevisiae plasma membrane nutrient sensors and their role in PKA signaling. FEMS Yeast Res. 2009;10:134–149. doi: 10.1111/j.1567-1364.2009.00587.x. [DOI] [PubMed] [Google Scholar]
- 4.Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraakman L, Lemaire K, Ma P, Teunissen AWRH, Donaton MCV, Van Dijck P, et al. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 6.Rolland F, de Winde JH, Lemaire K, Boles E, Thevelein JM, Winderickx J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol. 2000;38:348–358. doi: 10.1046/j.1365-2958.2000.02125.x. [DOI] [PubMed] [Google Scholar]
- 7.Welton RM, Hoffman CS. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics. 2000;156:513–521. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Batlle M, Lu A, Green DA, Xue Y, Hirsch JP. Krh1p and Krh2p act downstream of the Gpa2p Gα subunit to negatively regulate haploid invasive growth. J Cell Sci. 2003;116:701–710. doi: 10.1242/jcs.00266. [DOI] [PubMed] [Google Scholar]
- 10.Harashima T, Heitman J. The Gα protein gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol Cell. 2002;10:163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- 11.Peeters T, Louwet W, Gelade R, Nauwelaers D, Thevelein JM, Versele M. Kelch-repeat proteins interacting with the Gα protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc Natl Acad Sci USA. 2006;103:13034–13039. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 13.Harashima T, Heitman J. Gα subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G Protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4557–4571. doi: 10.1091/mbc.E05-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harashima T, Anderson S, Yates JR, III, Heitman J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol Cell. 2006;22:819–830. doi: 10.1016/j.molcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Niranjan T, Guo X, Victor J, Lu A, Hirsch JP. Kelch repeat protein interacts with the yeast Gα subunit Gpa2p at a site that couples receptor binding to guanine nucleotide exchange. J Biol Chem. 2007;282:24231–24238. doi: 10.1074/jbc.M702595200. [DOI] [PubMed] [Google Scholar]
- 16.Lu A, Hirsch JP. Cyclic AMP-independent regulation of protein kinase A substrate phosphorylation by kelch repeat proteins. Eukaryot Cell. 2005;4:1794–1800. doi: 10.1128/EC.4.11.1794-1800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peeters T, Versele M, Thevelein JM. Directly from Gα to protein kinase A: The kelch repeat protein bypass of adenylate cyclase. Trends Biochem Sci. 2007;32:547–554. doi: 10.1016/j.tibs.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Budhwar R, Lu A, Hirsch JP. Nutrient control of yeast PKA activity involves opposing effects on phosphorylation of the Bcy1 regulatory subunit. Mol Biol Cell. 2010;21:3749–3758. doi: 10.1091/mbc.E10-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffioen G, Branduardi P, Ballarini A, Anghileri P, Norbeck J, Baroni MD, et al. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol Cell Biol. 2001;21:511–523. doi: 10.1128/MCB.21.2.511-523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffioen G, Anghileri P, Imre E, Baroni MD, Ruis H. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1449–1456. doi: 10.1074/jbc.275.2.1449. [DOI] [PubMed] [Google Scholar]
- 22.Portela P, Moreno S. Glucose-dependent activation of protein kinase A activity in Saccharomyces cerevisiae and phosphorylation of its TPK1 catalytic subunit. Cell Signal. 2006;18:1072–1086. doi: 10.1016/j.cellsig.2005.09.001. [DOI] [PubMed] [Google Scholar]


