Abstract
The Mps1 family of protein kinases contributes to cell cycle control by regulating multiple microtubule cytoskeleton activities. We have uncovered a new Mps1 substrate that provides a novel link between Mps1 and the actin cytoskeleton. We have identified a conserved human Mps1 (hMps1) interacting protein and have termed Mps1 interacting protein-1 (Mip1). Mip1 defines an uncharacterized family of conserved proteins that contain coiled-coil and calponin homology domains. We demonstrate that Mip1 is a phosphoprotein that interacts with hMps1 in vitro and in vivo and is a hMps1 substrate. Mip1 exhibits dynamic localization during the cell cycle; Mip1 localizes to the actin cytoskeleton during interphase, the spindle in early mitosis and the cleavage furrow during cytokinesis. Mip1 function is required to ensure proper spindle positioning at the onset of anaphase after cells begin furrow ingression. Cells depleted of Mip1 exhibit aberrant mitotic actin filament organization, excessive membrane blebbing, dramatic spindle rocking and chromosome distribution errors during early cytokinesis producing high numbers of binucleate cells. Our data indicate that Mip1 is a newly recognized component of the actin cytoskeleton that interacts with hMps1 and that it is essential to ensure proper segregation of the genome during cell cleavage.
Key words: Mps1 kinase, actin, Mip1, cytokinesis
Introduction
The orderly progression of cells through mitosis requires the activity of several different protein kinases. These enzymes regulate a myriad of cellular events including chromosome behavior, spindle assembly and function, membrane dynamics of the nuclear envelope and the plasma membrane. The mitotic protein kinases include the cyclin-dependent kinases (CDKs), Polo-like kinases (PLKs), Aurora kinases, Bub kinases and Mps1.1 A challenge in understanding any of these protein kinases is to identify their substrates and determine the function of those proteins and how that function is regulated by phosphorylation.
The MPS1 (mono-polar spindle) gene encodes a widely conserved protein kinase whose activity is regulated by autophosphorylation2–4 and protein stability.5,6 MPS1 was identified by a yeast mutation defective in spindle pole body (SPB) assembly and the gene was subsequently shown to be required for SPB duplication.6–9 Several yeast Mps1 SPB substrates have been identified including Spc110, Spc98, Spc42 and Spc29.8,10,11 Initial reports on the localization and role of human Mps1 (hMps1) in centrosome duplication differed,6,12–14 but continued research has shown that some fraction of hMps1 localizes to centrosomes,3 and that it is important for proper centrosome duplication,15–17 at least in part by phosphorylating the centriole protein Centrin 2.18
It is clearly established that Mps1 functions in the spindle assembly checkpoint (SAC).12–14,19–29 Mps1 has also been shown to be important for recruitment of kinetochore checkpoint proteins, regulation of Cdc20 complexes and correction of chromosome kinetochore attachment errors by regulating Aurora B activity through phosphoryaltion of borealin1,30–34 and, in yeast, for efficient microtubule kinetochore attachment by Dam1 phosphorylation.35 Mps1 has also been implicated in a number of other cellular processes including meiosis,36–38 fin and heart development in zebrafish,25,39,40 Smad signaling,41 and DNA damage checkpoint.42,43 Finally, there is some evidence for a role for hMps1 in cytokinesis,6 but to date no molecular link between hMps1 and cytokinesis has been identified.
Here we identify a new hMps1 substrate, Mip1, which localizes to the actin cytoskeleton and the ingression furrow during cytokinesis principally via a C-terminal calponin homology domain. Reduction of Mip1 protein level leads to abnormal actin structures during cell division and defects in spindle positioning during anaphase. This is the first connection between hMps1 and the actin cytoskeleton and suggests hMps1 has additional uncharacterized roles in the cell cycle.
Results
Isolation of a hMps1 binding protein.
We performed a two-hybrid screen using the N-terminal 311 amino acids of hMps1 as bait to identify proteins that interact with hMps1. This segment of the hMps1 protein has been previously shown to target hMps1 to centrosomes and kinetochores.6,13 Isolation and sequencing of three independent positive clones from our two-hybrid assay revealed that they contained the same partial cDNA encoding residues 456–669 of a human ORF termed CytoSpin-A/SPECC1L/KIAA0376. In the absence of data on the function of this protein, we have chosen to name this ORF Mip1 (Mps1 interacting protein-1). To further test whether Mip1 interacts with hMps1 a full-length MIP1 clone fused to six histidine residues was expressed in E. coli and lysates from this strain were incubated with glutathione-sepharose bead-bound hMps1. We found that bead-bound wild-type hMps1 and hMps1-kinase dead mutant (KD) protein, but not GST alone, precipitated Mip1 confirming the interaction in vitro (Fig. 1A). The region of Mip1 isolated in the two-hybrid screen is within a large predicted coiled-coil domain, and we wished to determine whether this is the only segment of Mip1 required for hMps1 binding. To test hMps1 binding by the various domains of Mip1, we divided the MIP1 gene into fragments that would encode the amino-terminal (residues 1–351), central coiled-coil (residues 351–851) or carboxy-terminal (residues 851–1,117) segments. Purification of the Mip1 coiled-coil domain produced in E. coli was hindered by incomplete translation or protease degradation and multiple bands were isolated. Nonetheless, we found only the large coiled-coil domain, which included the original fragment bound by hMps1 in the two-hybrid screen, was specifically bound by hMps1 (data not shown). These in vitro assays strongly support the premise that the interaction between these two proteins is direct and lead us to characterize the Mip1 protein further.
Figure 1.
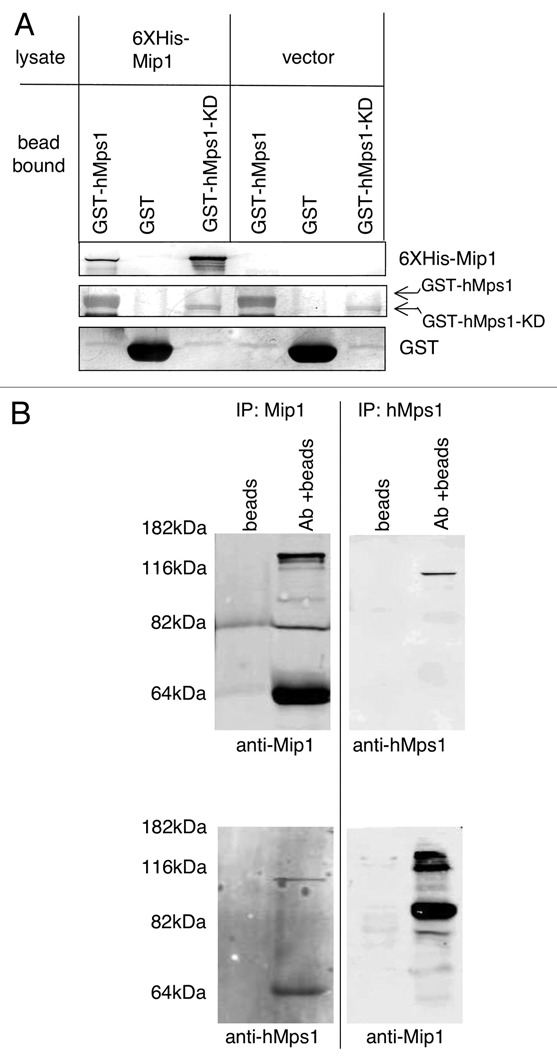
Mip1 interacts with hMps1 in vitro and in cell lysates. (A) Recombinant full length Mip1 binds hMps1 in vitro. Equivalent volumes of glutathione-bead bound GST-hMps1-ha, GST-hMps1-KD-ha or GST alone were added to E. coli lysates from cells expressing 6X-histidine tagged Mip1 or empty vector. The beads were collected, washed and analyzed by SDS-PAGE and immunoblot with anti-6X histidine and anti-GST antibodies. (B) Mip1 and hMps1 co-precipitate from cell lysates. Antibodies towards hMps1 or Mip1 were used to immunoprecipitate proteins from cycling U2OS cells and precipitated material was probed for the presence of Mip1 and hMps1 using either anti-Mip1 or anti-hMps1 antibodies. Multiple bands in Mip1 containing lanes are degradation products.
Mip1 and hMps1 interact in vivo.
To determine whether hMps1 and Mip1 interact in vivo, we generated a polyclonal antibody to the first 351 amino acids of Mip1. This antibody is highly specific for Mip1 and recognizes a band of the predicted molecular weight of Mip1 at 123 kDa in both U2OS and RPE1 cell lines (Sup. Fig. 1A). We used this antibody to precipitate Mip1 from U2OS cells and found that hMps1 coprecipitated (Fig. 1B). Furthermore, Mip1 was co-immunoprecipitated with antibodies specific for hMps1 (Fig. 1B). We obtained similar results from immunoprecipitation experiments with RPE1 cells (data not shown). These results indicate that hMps1 and Mip1 physically interact in human cells.
Mip1 is conserved among metazoans.
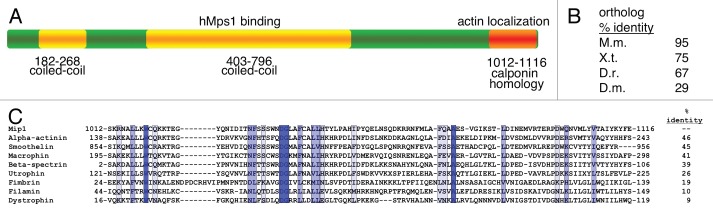
The predicted full-length MIP1 ORF encodes a 1,117 amino acid protein with a molecular mass of 123 kDa. Human Mip1 and its orthologs have been termed Cytospin-A or Spectrin-1-like (SPECC1L) in the NCBI database (Accession number for protein AAX84184). Northern analysis of total RNA from RPE1 and U2OS cells indicated a single transcript approximately 6.5 kb in size (data not shown). This is consistent with NCBI data indicating a MIP1 mRNA transcript of approximately 7 kb (accession number XM_037759). Database searches with the Mip1 amino acid sequence identified orthologs in a number of organisms including mice, Xenopus, zebrafish and Drosophila (Fig. 2). These proteins share similar domain architecture and common structural features including a small coiled-coil domain within the amino terminus, a larger centrally located coiled-coil domain and a carboxy-terminal calponin homology (CH) domain (Fig. 2A). The mouse, Xenopus and zebrafish Mip1 proteins display 95, 75 and 67 percent identity to the human Mip1, respectively (Fig. 2B). Drosophila has two potential Mip1 isoforms and the alignment value shown in Figure 2B is for the shorter 950 amino acid isoform A. Isoform B is closer in length to human Mip1 at 1,094 amino acids but is only 25 percent identical at the amino acid level. In each case, the most highly conserved domain of these proteins is the carboxy terminal segment containing the CH domain. Vertebrate Mip1 proteins exhibit greater than 92 percent amino acid identity in the CH domain, while the CH domains in the Drosophila isoforms are 75 percent identical to the human Mip1 CH domain. Alignment of the human Mip1 CH domain shows moderate homology to CH domains from α-actinin, Smoothelin, Macrophin and Beta-Spectrin (Fig. 2C). A lower degree of homology can be found with CH domains from Utrophin, Fimbrin, Filamin and Dystrophin. The amino end of the Mip1 proteins is less well conserved, but residues 384–456, partially within the larger coiled-coil domain, share substantial homology. This small segment of the human protein is greater than 94 percent identical to the mouse, Xenopus and zebrafish proteins, and 61 percent identical to the Drosophila protein in this region.
Figure 2.
The Mip1 protein is conserved among metazoans. (A) Domain highlights of the Mip1 protein include coiled coil domains, in yellow, recognized by the COILS program (www.ch.embnet.org/software/COILS_form.html) and the calponin homology domain, in orange, identified in the “specific hits” and “superfamilies” report following a blastp search of the Mip1 sequence using the non-redundant protein list at NCBI (blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins&PROGRAM=blastp&BLAST_PROGRAMS=blastp&PAGE_TYPE =BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome, with accession number AAX84184). (B) A list of Mip1 orthologs was identified and compiled using blastp and the non-redundant protein list at NCBI as in (A). Conservation among orthologs is reported as percent identical residues following alignment with the ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/index.html) using accession numbers AAX84188 (Mus musculus), AAX84190 (Xenopus tropicalis), AAX84193 (Danio rerio) and NP_569848 (Drosophila melanogaster, isoform A). (C) Comparison of the Mip1 calponin homology domain to similar domains from alpha-actinin (XP_646979), smoothelin (EAW59925), macrophin (EAX07277), beta-spectrin (1AA2_A), utrophin (1QAG_A), fimbrin (1AOA_A), filamin (EAW72747) and dystrophin (CAI39566) using the ClustalW2 program as in (B). Identical residues-dark blue, conserved residues-blue and semi-conserved residues-light blue.
Mip1 is a hMps1 substrate in vitro and phosphoprotein in vivo.
The Mps1 kinase family has only a handful of known substrates, and we next determined whether Mip1 is a hMps1 substrate in vitro. Full-length Mip1 did not express well in E. coli and only small amounts of the protein were isolated for this assay. Despite this, we were able to generate enough full-length Mip1 to determine that it was a substrate for hMps1 (data not shown). To identify the region of Mip1 phosphorylated by hMps1 we added the three Mip1 segments, defined earlier, to a hMps1 kinase assay. Analysis of these reactions indicated that the coiled-coil domain of Mip1 (residues 351–851) is a hMps1 substrate (Fig. 3A). This is the same Mip1 segment isolated in the two-hybrid screen and found to bind hMps1 directly in our recombinant protein-binding assay. In addition, the amino-terminal residues (aa 1–351) were also weakly phosphorylated by hMps1, while the carboxy-terminal residues (aa 851–1,117) were not strongly phosphorylated by hMps1 (Fig. 3A). We tested whether Mip1 is a phosphoprotein in vivo by treating Mip1 immunoprecipitates with lambda phosphatase. Immunoprecipitated Mip1 migration in SDS-PAGE was accelerated after phosphatase treatment (Fig. 3B). Our results show that hMps1 directly binds and phosphorylates Mip1 and indicate that Mip1 is a phosphoprotein in vivo.
Figure 3.
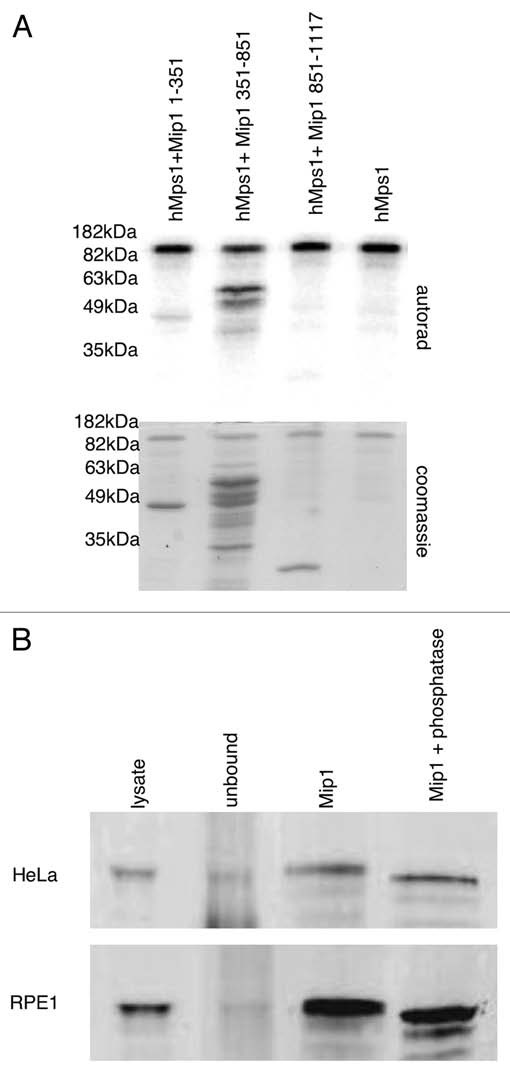
Mip1 is a phosphoprotein. (A) hMps1 phosphorylates Mip1 protein fragments. Approximately 1 µg of each of the indicated Mip1 protein fragments were incubated with 200 ng of hMps1 in the presence of 32P-γATP. The reaction products were analyzed by SDS-PAGE and autoradiography. (B) Immunoprecipitated Mip1 migration is sensitive to the addition of the phosphatase. Mip1 protein was immunoprecipitated from HeLa and RPE 1 cells. Bead bound material was washed with phosphatase buffer and then incubated in the presence or absence of lambda phosphatase for 60 minutes. The reaction products were subjected to SDS-PAGE, transferred to PVDF membrane and immunoblotted with anti-Mip1 antibodies.
Mip1 localizes to distinct sub-cellular structures.
hMps1 is known to localize to both centrosomes and kinetochores and to more clearly characterize the link between Mip1 and hMps1 we determined whether Mip1 localized to either of these structures. Using a green fluorescent protein fusion (GFP-Mip1), we could not detect Mip1 at kinetochores but could detect a low level signal that co-stained with γ-tubulin at centrosomes (data not shown). This centrosome signal was fixation dependent and suggests that Mip1 centrosome localization is transient. However, both endogenous and GFP-Mip1 strongly co-localized with actin filaments in interphase cells (Fig. 4A and B). In addition, treatment of cells with latrunculin A, but not nocodazole, disrupted the Mip1 localization pattern further supporting the conclusion that Mip1 localizes to a part of the actin filament network (data not shown).
Figure 4.
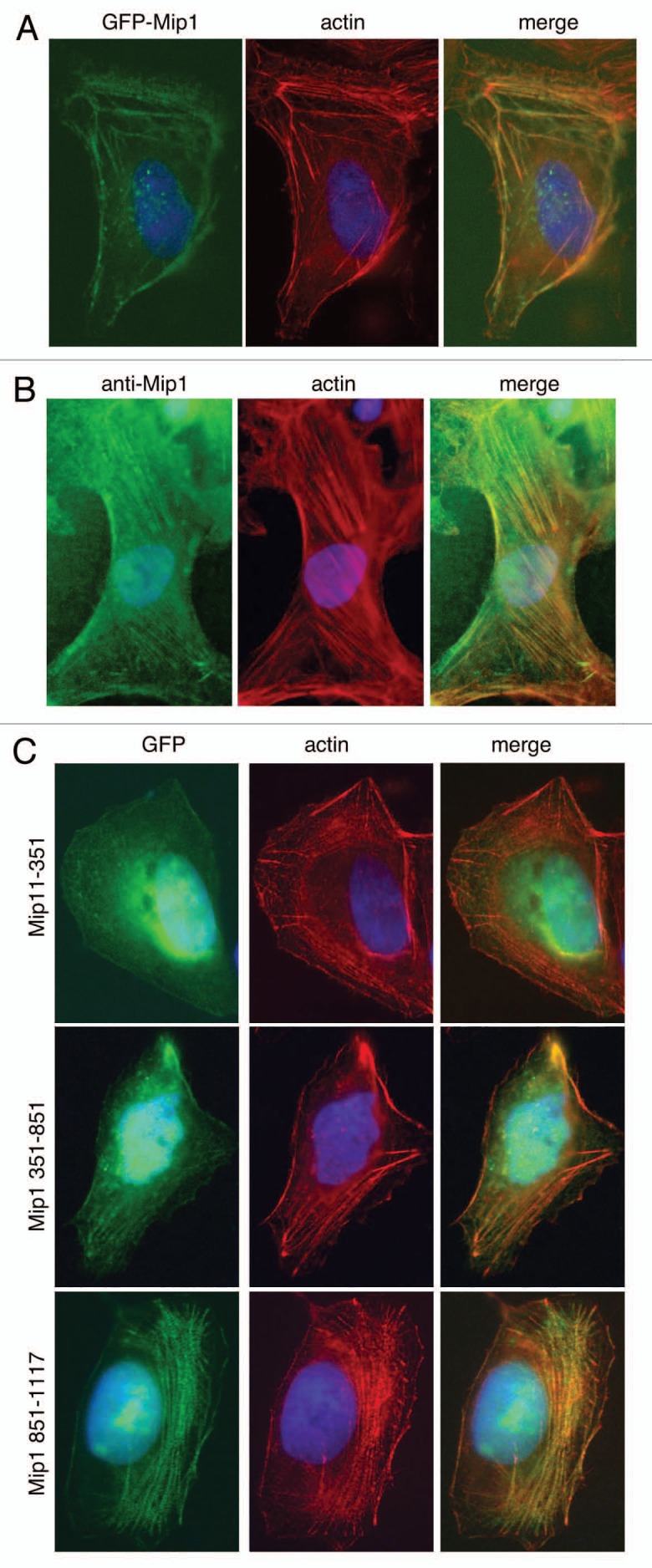
Mip1 colocalizes with the actin network in cells. (A) U2OS cells were transfected with a plasmid expressing a GFP-Mip1 fusion (green). Cells were fixed, counterstained with Alexafluor labeled phalloidin (red) and DAPI (blue) and visualized by microscopy. (B) U2OS cells were fixed and analyzed by immunofluorescence with anti-Mip1 antibodies (green) and counterstained as in (A). (C) U2OS cells transfected with plasmids containing GFP fusions to the indicated Mip1 fragments (green) were fixed and analyzed as in (A).
The CH domain is found in several proteins known to cross-link actin, such as filamin,44 or function as a linker between actin and other cytoskeletal components such as in plectin binding to intermediate filaments45 or MAP2c to microtubules.46 However, the CH domain of calponin is not required for binding to actin.47,48 We expressed the previously described fragments of Mip1 fused to GFP in cells to determine what segment of Mip1 was important for actin localization. This revealed that the amino-terminal segment of Mip1 did not detectably associate with actin, and we detected only a very weak actin association with the middle coiled-coil segment. Conversely, the carboxy terminal CH containing segments did associate strongly with actin filaments (Fig. 4C). Our data indicate that the CH domain of Mip1 is important for the association of Mip1 with actin filaments either through direct binding of Mip1 to actin or via association with other actin binding proteins.
Mip1 was clearly associated with actin filaments during interphase; however, we noticed that Mip1 localization changed dynamically during mitosis (Fig. 5A and B). In early metaphase Mip1 localized within the microtubule network of the spindle reminiscent of NuMa.49,50 As mitosis progressed Mip1 can be seen at the earliest stages of the ingression furrow similar to anillin.49,51,52 Mip1 remains at the furrow through the ingression process and can be seen at the midbody/Flemming body during cytokinesis. This pattern of localization suggests Mip1 may be important for several aspects of mitosis.
Figure 5.
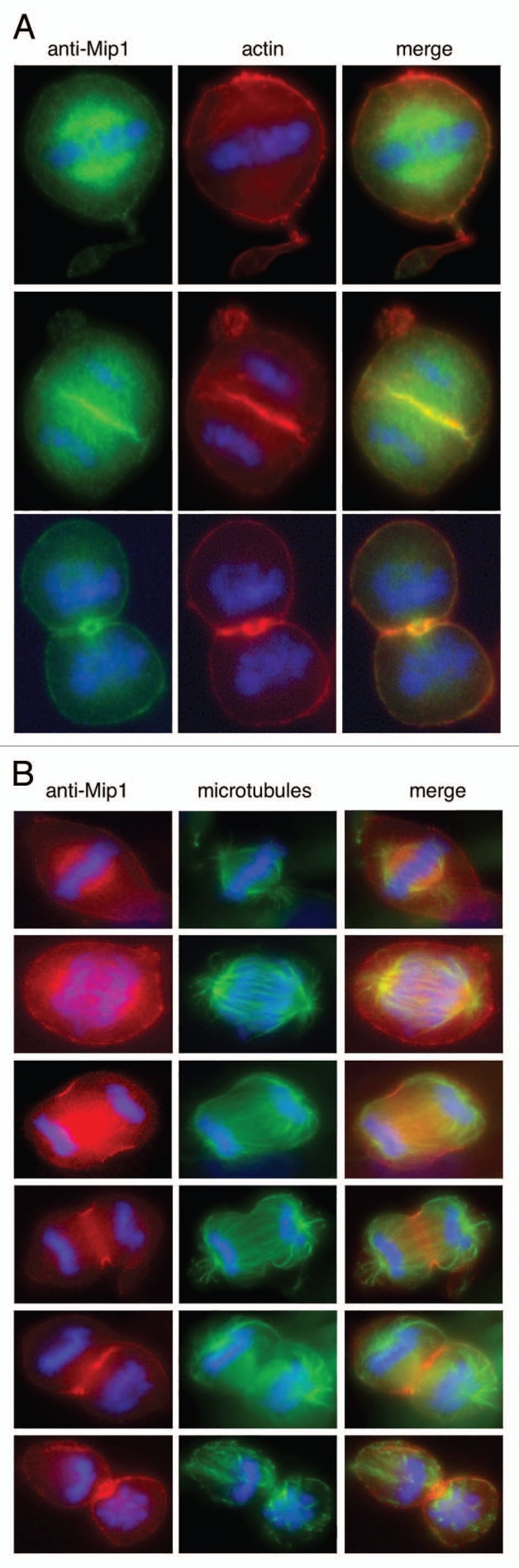
Mip1 localizes to the mitotic spindle during mitosis and the cytokinetic furrow during cytokinesis. (A) U2OS cells were arrested in S-phase for 14 hours with thymidine. Following removal of thymidine containing media and continued incubation for several hours, cells were fixed and analyzed by immunofluorescence with anti-Mip1 (green) and counterstained with alexafluor labeled phalloidin (red) and DAPI (blue) to visualize actin and chromosomes respectively. (B) Cells were treated as in (A), but were analyzed with anti-Mip1 and anti-tubulin antibodies.
Mip1 reduction causes multinucleate cells.
Mip1 localized to the cytokinetic furrow at an early stage and remained there throughout cell division suggesting a role for the protein in cytokinesis. To determine the function of Mip1 we treated cells with Mip1 specific small interfering RNAs (siRNAs) to reduce the level of the protein. Each of two different siRNAs targeting Mip1 was effective in reducing the Mip1 protein level by approximately 80% (Sup. Fig. 1 and data not shown). U2OS cells treated with Mip1 or control siRNAs for 36 hours were fixed and analyzed by immunofluorescence. We detected an increase in binucleate and tetranucleate cells 48-hours after Mip1 siRNA treatment, but not in cells treated with a lamin specific siRNA control. Among the cell populations we analyzed, 24 percent of Mip1 siRNA treated cells were multinucleate whereas in lamin siRNA treated cells only three percent were multinucleate. We found the same results after treating cells with each of the two Mip1 siRNAs. The vast majority of Mip1 siRNA treated multinucleate cells contained either two or four nuclei. Further inspection of Mip1 siRNA treated cells also showed a noticeable increase in mitotic cell blebbing (can be observed in live cell recordings, Sup. Vid. 2). We also noticed a reduction in the number of Mip1 siRNA treated cells remaining on the growth substrate after fixation (data not shown). Importantly, a similar reduction in cell number was not seen with lamin siRNA treated cells. This effect was variable and suggests that knockdown of Mip1 by siRNA was toxic to cells or adversely affected the ability of cells to attach to the growth substrate. Interestingly, Mip1 siRNA treatment did not cause an observable defect in other previously described hMps1 functions such as centrosome duplication or the spindle assembly checkpoint (Sup. Fig. 2).
Mip1 is required for proper actin organization during mitosis.
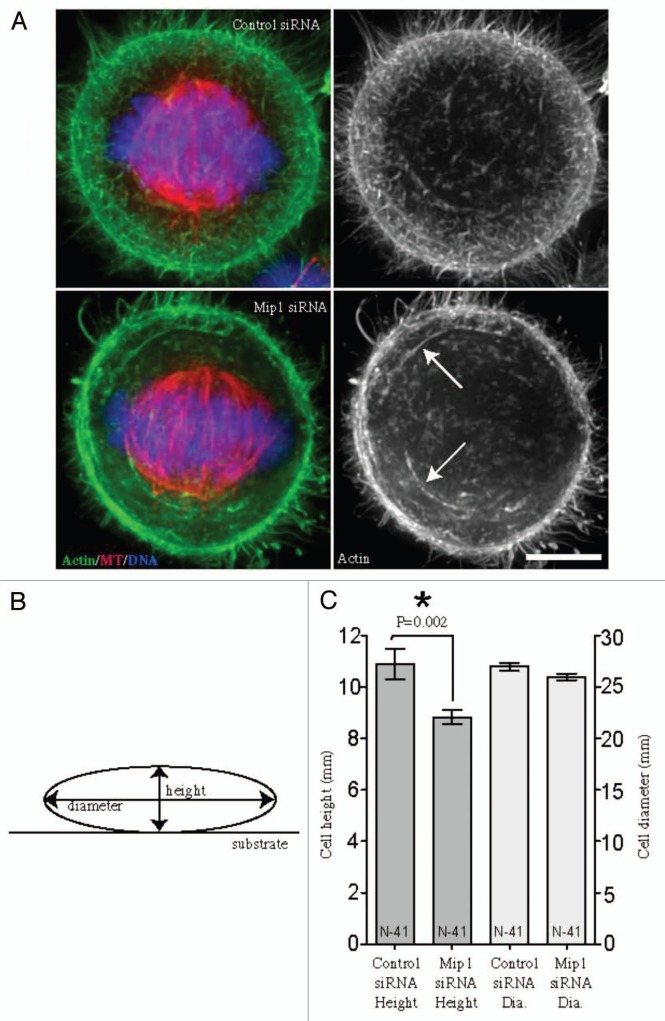
Metaphase cells were evaluated for actin filament organization after Mip1 depletion (Fig. 6). Deep actin filaments and cortical actin enrichment were evident in both control and Mip1-depleted cells. However, the organization of the actin filaments in Mip1-depleted cells consisted of more swirls of thick bundles relative to the cortical meshwork more evident in control siRNA-treated cells (Fig. 6A). Furthermore, the height of the cells measured from the substrate (diagrammed in Fig. 6B) was decreased in Mip1-depleted cells (Fig. 6C), suggesting that actin cortical integrity and cell rounding were compromised.
Figure 6.
Depletion of Mip1 leads to subtle alterations in mitotic cell shape and actin organization. Representative volume views of actin filaments labeled with fluorescein-phalloidin (A) were evaluated in HeLa cells treated with control or Mip1 siRNAs. Metaphase cells depleted of Mip1 exhibited greater numbers of long actin bundles (arrows) rather than the fine actin meshwork more characteristic of control cells. Bar = 5 microns. (B) Cell height and diameter were measured in volume views. (C) Cells depleted of Mip1 exhibited a decreased cell height (left y-axis; dark grey), whereas there was no measurable difference in cell diameter (right y-axis; light grey).
Mip1 functions to position and stabilize the mitotic spindle.
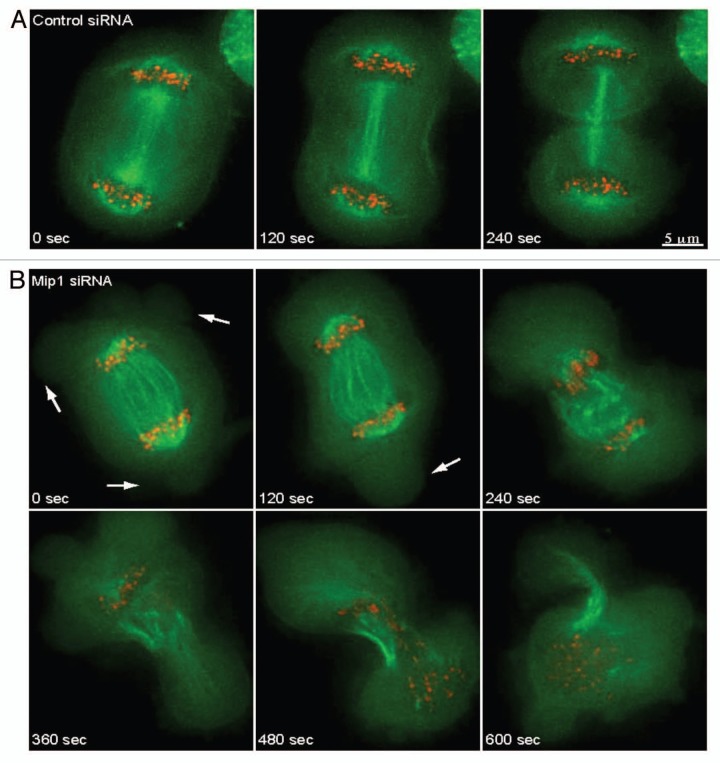
Live cell imaging was performed to characterize the role of Mip1 during cytokinesis and identify the specific defect caused by reducing the level of Mip1 by siRNA. Surprisingly, we found that Mip1 siRNA treated cells display a spindle rocking phenotype at the onset of anaphase (37% of Mip1-depleted, n = 19, compared to 12% of control-depleted anaphase cells, n = 17) (Fig. 7 and Sup. Vids. 1 and 2). Interestingly, this phenotype is similar to what has been reported for cells depleted of anillin or containing hyperstabilized microtubules.53,54 In most of these cells (six of seven cells with spindle rocking) spindles displayed dramatic oscillations that caused a violent disintegration of spindle integrity during furrow ingression that usually resulted in separated chromosomes repositioning themselves into a single daughter cell (five of seven cells). While the mitotic spindle formed correctly and appeared normal through anaphase A in these cells, it did not remain intact during later stages of cytokinesis. Similar to what we notice with fixed cells, Mip1 siRNA substantially increased membrane blebbing at the onset of anaphase. This unexpected violent loss of spindle integrity coupled with the increased membrane blebbing suggests that Mip1 may function both to maintain cell cortex stability and spindle position during cytokinesis.
Figure 7.
Depletion of Mip1 results in spindle rocking and failure to properly distribute chromosomes. HeLa cells expressing GFP-tubulin (green) and mRFP-Cenp-B (red) were analyzed by time-lapse microscopy following treatment with control (A) or Mip1 (B) siRNAs. Cells depleted of Mip1 form large blebs during late anaphase (arrows in B) and fail to distribute chromosomes equally into daughter cells. Scale bar is 5 mm.
Discussion
We have identified a new protein in the actin filament network that is conserved in vertebrates. Our localization studies using GFP fusions and immunofluoresence indicate that the Mip1 protein localizes to several actin-containing structures in the cell. During interphase Mip1 localizes to the filamentous actin network. As cells enter mitosis Mip1 localization changes and Mip1 is present in the metaphase spindle. In anaphase, Mip1 can be seen at the ingression furrow where it remains throughout cytokinesis. In the absence of Mip1, cells displayed several phenotypes including increased membrane blebbing and spindle position instability. These findings coupled with the localization pattern we observed suggest a function for Mip1 during cytokinesis. The disruption of several distinct aspects of cytokinesis suggests Mip1 may interact with central spindle components, the actin-myosin contractile ring, and/or the plasma membrane. Based on the increased membrane blebbing observed in Mip1 siRNA treated cells it seems likely that Mip1 may be important for proper actin filament assembly to stabilize the cell cortex during this period. Although we have no evidence that Mip1 directly binds actin, it is possible that Mip1 cross-links actin and without Mip1 there is insufficient cortex structure to accompany the added stress of membrane insertion.
Reduction of Mip1 leads to phenotypes similar in several ways to those observed after reduction of anillin. Like reduction of anillin, decreased Mip1 levels leads to spindle oscillation, increased membrane blebbing and binucleate cells.53,55 This is consistent with the similar localization patterns of anillin and Mip1 at the ingression furrow.51,52 Unlike anillin, we did not detect furrow regression in cells treated with Mip1 interfering RNAs. Anillin acts as a scaffold for recruitment and function of actin and myosin II during cytokinesis.56 RhoA is a key regulator of ring formation, and anillin also stablizes RhoA furrow localization providing a link between these essential contractile ring components.56,57 An interaction between Mip1 and anillin could explain the complexity of effects we observed with Mip1 reduction in cells.
Alternatively, Mip1 function may be independent of anillin, but require interaction with other cytokinesis components such as actin, myosin or the ezrin, radixin, moesin (ERM) protein family. ERM proteins are required for cortical stability and provide the connection between the actin cortex and plasma membrane.58 For example, loss of moesin function leads to spindle organization and positioning defects, and loss of cortical stability.59 Mip1 reduction leads to some similar effects during mitosis, but Mip1 siRNA treated cells are able to organize proper mitotic spindles that are stable at least until anaphase onset. We compared Mip1 localization to the ERM protein ezrin.60 Ezrin localizes between the membrane and actin cytoskeleton61 and our data indicate Mip1 is internal to the membrane associated ezrin signal in the ingression furrow (data not shown). This observation suggests Mip1 may function at the level of actin cortex rather than as part of the linker between cortex and membrane.
Early studies of the Mps1 kinase focused on its roles in spindle pole body duplication and spindle assembly checkpoint. Since then it has been implicated in several additional cellular activities, and our data open the possibility that this multifunctional kinase may play a role in some aspect of actin cytoskeleton function. We have identified Mip1 as a previously uncharacterized substrate for hMps1 that co-localizes with actin filaments. Our data provide the first potential link between Mps1 and the actin cytoskeleton. The amino-terminus of hMps1 is known to be important for its localization to kinetochores and centrosomes,6,13 and our data indicate the amino-terminus of hMps1 is also important for interaction with the coiled coil domain of Mip1. Coiled coil domains commonly mediate protein-protein interactions and our observations suggest the possibility that Mip1 may function as a scaffold for recruitment of hMps1 to previously unidentified distinct sub-cellular structures.
If Mip1 acts as a scaffold for hMps1 it is not clear where the interaction occurs. We could not reliably detect Mip1 at the centrosome or kinetochores, where hMps1 localizes. Conversely hMps1 is not known to localize to the interphase actin filament network, the ingression furrow or the mitotic spindle. The absence of Mps1 localization at additional sub-cellular structures may reflect the need for better tools to assess hMps1 localization and may explain why we did not detect hMps1 localization at the furrow. Nonetheless, loss of hMps1 function can lead to multinucleate cells and this suggests it may have a role in cytokinesis.6 hMps1 may participate in cytokinesis via the regulation of MIP1, but what role it plays in Mip1 function remains to be discovered.
Materials and Methods
Cell culture, transfection and siRNA.
U2OS, HeLa and RPE1 cells were cultured at 37°C in a humidified chamber in the presence of 5% CO2 in Dulbecco's modified Eagle's Medium (Sigma) containing 10% fetal bovine serum (Hyclone), penicillin and streptomycin (Invitrogen). For overexpression experiments, cells were transfected with the appropriate plasmid, using Effectine (Qiagen) reagent per the manufacturers recommendations. For siRNA, cells were transfected with siRNA using oligofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. Mip1 depletion cells were transfected with 60 µM each of the two following Mip1 specific siRNA equences (separately at 120 µM or in combination at 60 µM) obtained from Sigma: UCA GAU CAG UGA CAG AGC UdTdT paired with AGC UCU GUC ACU GAU CUG AdTdT and CGC AGU CAC CAU AUG GAG CdTdT paired with GCU CCA UAU GGU GAC UGC GdTdT. Equivalent results were obtained with both sets of siRNA. Treatment of cells with either siRNA alone at 120 µM had indistinguishable effects. Control depletions used the negative control siRNA #1 (Ambion).
Cloning, plasmids and primers.
For two hybrid screening, the first 311 residues of human Mps1 were ligated into the pGBKT7 bait plasmid after digestion with BamHI and SalI following PCR with the primers 5′-CGG GGA TCC GTA TGG AAT CCG AGG ATT TAA GTG GC-3′ and 5′-CGG TCG ACG CAC AAC CAA ATC TCG GCA TTC TGA TC-3′. Construction of MIP1 expression plasmids was done by amplification of the human MIP1 ORF from cDNA clones (clone-DKFZp762P042Q, RZPD, Berlin, Germany or clone-hh00388, Kazusa DNA Research Institute, Chiba, Japan). To prepare a 6XHis-Mip1 fusion, the Mip1 ORF was amplified with 5′-GGG GAT CCa TGA AGA AAG CAA GCA GGA GTG-3′ and 5′-GGA ATT CTC AGG TCT CAA AGT ACT TGT AGA TCG-3′ primers, digested with BamHI and EcoRI, and ligated into pRSET-A (Invitrogen) to generate pRSET-MIP1. A MIP1 ORF fusion to green fluorescent protein (GFP) was constructed as follows. First the MIP1 ORF was amplified using the following primers, 5′-GGG GTA CCa TGA AGA AAG CAA GCA GGA GTG-3′ and 5′-ATA AGA ATG CGG CCG CTC AGG TCT CAA AGT ACT TGT AGA TCG-3′, digested with KpnI and NotI, and ligated into the pECE-GFP-hMps1-ha6 backbone replacing the hMPS1 ORF. Following this, a ∼4 kb ClaI-NotI cDNA fragment was used to replace most of the MIP1 coding sequence and add the 3′UTR to generate pECE-GFP-MIP1.
Segments of MIP1 for expression as GST fusions in E. coli were generated using the following primers: Mip1 amino acids 1–351, 5′-GGG GAT CCA TGA AGA AAG CAA GCA GGA GTG-3′ and 5′-GTC GAA TTC TCA CTC ACT GCA CTC ACT GTC-3′, Mip1 amino acids 351–851, 5′-CTC GGA TCC ATG GAG GTC TAC CAG CCC CTC-3′ and 5′-GTG GAA TTC TCA TGC ACT GTC AAA GGA CTT G-3′, and Mip1 amino acids 851–1,117, 5′-CTC GGA TCC ATG GCA TCT CAA GTA CCA AAC CC-3′ and 5′-CGG TCG ACG CAC AAC CAA ATC TCG GCA TTC TGA TC-3′. These fragments were digested with BamHI and EcoRI and ligated into pGEX-6P1 (GE Healthcare) to generate plasmids pGX-MIP1-1-351, pGX-MIP1-351-851 and pGX-MIP1-851-1117. For expression of these same fragments in human cell culture an EcoRI site was inserted just 5′ of the TGA stop codon in pECE-GFP-DKFZ0376 + 3′UTR using the following primers: 5′-CTC GG T ACC ATG GCA TCT CAA GTA CCA AAC CC-3′, 5′-CCT TAT GAT GAA GAA ATC TAA AGC C-3′, 5′-TGC TCA GAA TTC GGT CTC AAA GTA CTT GTA GAT C-3′, 5′-ACC GAA TTC TGA GCA TGC CGG GAG GAG CC-3′ and 5′-TGC TCA GAA TTC GGT CTC AAA GTA CTT GTA GAT C-3′, and a two step PCR amplification reaction. The resulting 1.6 kb fragment was digested with KpnI and ligated into pECE-GFP-DKFZ0376 + 3′UTR digested with KpnI to generate pECE-GFP-MIP1-851-1117. For expression of Mip1 amino acids 1–351 and 351–851, PCR using 5′-GGG GTA CCA TGA AGA AAG CAA GCA GGA GTG-3′ and 5′-GTC GAA TTC TCA CTC ACT GCA CTC ACT GTC-3 (for 1–351) and 5′-CTC GGA TCC ATG GAG GTC TAC CAG CCC CTC-3′ and 5′-GTG GAA TTC TCA TGC ACT GTC AAA GGA CTT G-3′ was performed, fragments were digested with KpnI and EcoRI, and ligated into pECE-GFP-MIP1 digested with EcoRI and partially digested with KpnI to generate pECE-GFP-MIP1-1-351 and pECE-GFP-MIP1-351-851. All plasmid constructs were verified by sequencing, and all plasmids used for cell transfection were isolated using a MAXI DNA isolation kit (Qiagen).
Protein purification.
Purification of glutathione S-transferase fused hMps1 (and kinase dead hMps1-KD) has been described previously in reference 4. GST-Mip1 protein fragments were expressed in BL21(DE3) pLysS (Invitrogen) from the appropriate plasmids. After induction for four hours in 1 mM IPTG, cells were collected by centrifugation and frozen as pellets. Cell pellets were thawed in PBS containing 50 mM NaCl, 5 mM MgCl2, 10% glycerol (vol/vol), 0.2% Triton X-100 (vol/vol), 0.1% 2-mercaptoethanol (vol/vol) and 125 µg/ml egg white lysozyme, and further disrupted with sonication. Lysates were centrifuged for clarification and fusion proteins were isolated through batch purification using glutathione-Sepharose (Amersham Biosciences). After extensive washing, fusion proteins were eluted with 50 mM Tris•HCl (pH 8.0) containing 10% glycerol (vol/vol), 0.1% 2-mercaptoethanol (vol/vol) and 10 mM reduced glutathione. Eluted protein was characterized by SDS-PAGE, protein concentration was determined by BCA assay and peak fractions were pooled and frozen. Where necessary, GST was removed from fusions using the PreScission Protease (GE Healthcare).
Yeast two-hybrid.
hMps1 interacting proteins were isolated by yeast two-hybrid analysis with the Clontech Match-Maker system. Residues 1–311 of hMps1 were ligated into pGBKT7 and transformed into the AH109 strain. This strain was mated to strain Y187 carrying a pretransformed cDNA-prey library from human testis (BD Biosciences Clontech). Diploid strains were selected for growth on appropriate media to identify interacting clones. Interacting clones were isolated, retransformed to confirm interaction and sequenced.
Immunofluorescence.
Cells were fixed in IF fixation buffer (PBS containing 0.2% triton X-100, 0.5 mM MgCl2, 4% formaldehyde (Ted Pella) for 10 minutes at room temperature in the dark. Cells were washed four times in PBS (with 0.5 mM MgCl2) and blocked in PBS containing 0.5% bovine serum albumin, 0.5% NP-40, 1 mM MgCl2 and 1 mM sodium azide. All antibody incubations were performed in blocking buffer followed by four washes with PBS containing 0.5 mM MgCl2. Hoechst or DAPI (Vector labs) dye was used to visualize DNA and visualization of filamentous actin was achieved using either alexaflour 647 labeled phalloidin (Invitrogen) or fluorescein-labeled phalloidin (Molecular Probes). Analysis of centrosome number was performed as described in reference 4, and checkpoint function was assessed by the addition of nocodazole to 200 ng/ml and assessing Mad2 immunofluorescence.
Immunoprecipitation, immunoblotting and coprecipitation.
Immunoprecipitations were performed as in reference 62, with minor changes. Briefly, U2SOS cells were grown in 15 cm dishes and when 80% confluent, were washed, trypsinized, collected by centrifugation and lysed in 1 ml of hypotonic gentle lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100), with mammalian protease inhibitor cocktail (Sigma) for 10 min on ice. Sodium chloride was added to 150 mM, and the extracts were incubated for another 5 min on ice. Cell debris was removed by centrifugation, and the supernatant was gently mixed with the appropriate antibody for at least 4 hours at 4°C. Following this, a 1:1 mix of magnetic protein A and protein G coupled beads (NEB) was added to the lysate and incubated overnight at 4°C. Beads were collected by magnetism, washed eight times with 1 ml of NET-2 (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Triton X-100), and bead bound material was subjected to SDS-PAGE and immunoblotting. Phosphorylation of immunoprecipitated material was evaluated after incubating immunoprecipitates with 1 ul of Lambda phosphatase (New England Biolabs).
For immunoblotting cells were washed, trypsinized, collected by centrifugation and lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.2% SDS, mammalian protease inhibitor cocktail (P8340, Sigma)) on ice for 30 minutes. Lysates were sonicated, protein content was determined using BCA assay, and 4x Laemmli sample buffer was added to 100 ug samples. Protein was electrophoresed on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membrane. After blocking in TBST (0.1% Tween 20) with 6% milk and 1% bovine serum albumin, the blots were probed with the appropriate antibodies.
A polyclonal sera against residues 1–351 of Mip1 generated in E. coli was raised in rabbits by Affinity Bioreagents (Golden, CO). Anti-Mip1 sera were affinity purified using Mip1 residues 1–351 coupled to AminoLink Coupling Gel (Pierce Biotechnology, Rockford, IL) according to manufacturers' instructions. Human Mps1 was probed with rabbit polyclonal IgG sc-540 anti-hMps1 (TTK C-19; Santa Cruz) or anti-Mps1 NT clone 3-472-1 (Upstate Biotechnology Inc.). Blots were washed thoroughly, incubated with the appropriate secondary antibodies, washed again and visualized using the Odyssey imaging system (Li-Cor).
Mip1 and hMps1 protein coprecipitation was characterized using glutathione S-transferase (GST) bead bound wild type or kinase dead hMps1 protein fused to hemaglutinnin (GST-hMps1-ha, described in ref. 4. GSH-bead bound GST-hMps1-ha fusion protein was added to bacterial lysates of cells expressing a 6XHis-Mip1 fusion or empty vector. After incubation at 4°C for 1 hour with gentle mixing, GST-hMps1-ha was collected with glutathione beads (Pharmacia). Bead bound material was analyzed by SDS-PAGE and immunblotted with antibodies to 6X-histidine (Sigma), GST (GE Healthcare) or hMps1 (Santa Cruz).
Kinase assays.
Kinase assays were performed essentially as in reference 4. Briefly, 1 ug of Mip1 protein or protein fragments were incubated with GST-hMps1-HA (0.2 ug) in kinase assay buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 mM dithiothreitol, 1 mg/ml MBP, 0.1 mM ATP) for 30 minutes at 30°C in kinase assay buffer with or without 6x His-MIP1. Reactions were terminated by addition of loading buffer, boiled and analyzed by SDS-PAGE and autoradiography.
Cell imaging.
HeLa cells were cultured on poly-L-lysine-coated 35 mm2 glass coverslip dishes (MatTek Corp.,) and switched to CO2-independent media (Life Technologies) 36 hours after siRNA transfection. Cells were imaged using time-lapse microscopy on a Deltavision RT system (Applied Precision) equipped with a CCD camera, 60x 1.42 NA lens (Olympus) and a 37°C environmental chamber (Applied Precision). Five optical sections with 0.5 µm spacing were acquired at intervals of 20 or 30 seconds. For volume views of fixed cells, 0.3 µm spacing was acquired through the entire volume of the cell on a Deltavision RT system (Applied Precision). Cells were deconvolved and volume views were calculated using SoftWorx (Applied Precision) to measure height, diameter and evaluate actin filament organization. Final image processing and analysis was achieved using ImageJ software (NIH).
Acknowledgements
Thank you to Chad Pearson for helpful suggestions and to Yvette Bren-Mattison, Matt Tarver and Barry Hurlburt for critical reading of this manuscript. This work was supported by a Fellowship Award from the Colorado Tobacco Research Program and Department of Defense Fellowship Award DAMD17-03-1-0404 (to C.P.M.), National Institutes of Health Grants GM51312 (to M.W.) and GM69429 (to L.W.), a Ruth L. Kirschstein National Service Award GM778572 (to J.S.) and Special Fellow award from the Leukemia and Lymphoma Society 3652-11 (to J.S.).
Supplementary Material
References
- 1.Lan W, Cleveland DW. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J Cell Biol. 2010;190:21–24. doi: 10.1083/jcb.201006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Yang Y, Gao Y, Xu Q, Wang F, Zhu S, et al. Structural and Mechanistic Insights into Mps1 Kinase Activation. J Cell Mol Med. 2009;13:1679–1694. doi: 10.1111/j.1582-4934.2008.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyler RK, Chu ML, Johnson H, McKenzie EA, Gaskell SJ, Eyers PA. Phosphoregulation of human Mps1 kinase. Biochem J. 2009;417:173–181. doi: 10.1042/BJ20081310. [DOI] [PubMed] [Google Scholar]
- 4.Mattison CP, Old WM, Steiner E, Huneycutt BJ, Resing KA, Ahn NG, et al. Mps1 activation loop autophosphorylation enhances kinase activity. J Biol Chem. 2007;282:30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- 5.Cui Y, Cheng X, Zhang C, Zhang Y, Li S, Wang C, et al. Degradation of the human mitotic checkpoint kinase Mps1 is cell cycle-regulated by APC-cCdc20 and APC-cCdh1 ubiquitin ligases. J Biol Chem. 2010;285:32988–32998. doi: 10.1074/jbc.M110.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goebl MG, Winey M. The yeast cell cycle. Curr Opin Cell Biol. 1991;3:242–246. doi: 10.1016/0955-0674(91)90147-q. [DOI] [PubMed] [Google Scholar]
- 8.Jaspersen SL, Winey M. The budding yeast spindle pole body: Structure, duplication and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 9.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 10.Holinger EP, Old WM, Giddings TH Jr, Wong C, Yates JR, 3rd, Winey M. Budding yeast centrosome duplication requires stabilization of Spc29 via Mps1-mediated phosphorylation. J Biol Chem. 2009;284:12949–12955. doi: 10.1074/jbc.M900088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki Y, Gombos L, Migueleti SP, Sivashanmugam L, Antony C, Schiebel E. N-terminal regions of Mps1 kinase determine functional bifurcation. J Cell Biol. 2010;189:41–56. doi: 10.1083/jcb.200910027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stucke VM, Baumann C, Nigg EA. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- 15.Kanai M, Ma Z, Izumi H, Kim SH, Mattison CP, Winey M, et al. Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells. 2007;12:797–810. doi: 10.1111/j.1365-2443.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 16.Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasbek C, Yang CH, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environ Mol Mutagen. 2009;50:654–665. doi: 10.1002/em.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang CH, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 Phosphorylation Sites Regulate the Function of Centrin 2 in Centriole Assembly. Mol Biol Cell. 2010;21:4361–4372. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauze' E, Stoelscker B, Luca FC, Weiss E, Schutz A, Winey M. Yeast spindle pole duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poch O, Schwob E, Fraipont F, Camasses A, Bordonne' R, Martin R. RPK1, an essential yeast protein kinase involed in the regulation of the onset of mitosis, shows homology to mammalian dual-specificity kinases. Mol Gen Genet. 1994;243:641–653. doi: 10.1007/BF00279573. [DOI] [PubMed] [Google Scholar]
- 21.Mills GB, Schmandt R, McGill M, Amendola A, Hill M, Jacobs K, et al. Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J Biol Chem. 1992;267:16000–16006. [PubMed] [Google Scholar]
- 22.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 23.Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 24.He X, Jones MH, Winey M, Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 25.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 26.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, Chen Y, Zhao Y, Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc Natl Acad Sci USA. 2007;104:20232–20237. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q, Zhu S, Wang W, Zhang X, Old W, Ahn N, et al. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by mps1 phosphorylation. Mol Biol Cell. 2009;20:10–20. doi: 10.1091/mbc.E08-03-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-CMad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 34.Bourhis E, Lingel A, Phung Q, Fairbrother WJ, Cochran AG. Phosphorylation of a borealin dimerization domain is required for proper chromosome segregation. Biochemistry. 2009;48:6783–6793. doi: 10.1021/bi900530v. [DOI] [PubMed] [Google Scholar]
- 35.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straight PD, Giddings TH, Jr, Winey M. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol Biol Cell. 2000;11:3525–3537. doi: 10.1091/mbc.11.10.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilliland WD, Wayson SM, Hawley RS. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr Biol. 2005;15:672–677. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 38.Poss KD, Nechiporuk A, Stringer KF, Lee C, Keating MT. Germ cell aneuploidy in zebrafish with mutations in the mitotic checkpoint gene mps1. Genes Dev. 2004;18:1527–1532. doi: 10.1101/gad.1182604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 40.Fischer MG, Heeger S, Hacker U, Lehner CF. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr Biol. 2004;14:2019–2024. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Zhu S, Wang W, Clarke DC, Liu X. Activation of Mps1 promotes transforming growth factor-beta-independent Smad signaling. J Biol Chem. 2007;282:18327–18338. doi: 10.1074/jbc.M700636200. [DOI] [PubMed] [Google Scholar]
- 42.Wei JH, Chou YF, Ou YH, Yeh YH, Tyan SW, Sun TP, et al. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J Biol Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- 43.Yeh YH, Huang YF, Lin TY, Shieh SY. The cell cycle checkpoint kinase CHK2 mediates DNA damage-induced stabilization of TTK/hMps1. Oncogene. 2009;28:1366–1378. doi: 10.1038/onc.2008.477. [DOI] [PubMed] [Google Scholar]
- 44.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: Promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Sevcik J, Urbanikova L, Kost'an J, Janda L, Wiche G. Actin-binding domain of mouse plectin. Crystal structure and binding to vimentin. Eur J Biochem. 2004;271:1873–1884. doi: 10.1111/j.1432-1033.2004.04095.x. [DOI] [PubMed] [Google Scholar]
- 46.Roger B, Al-Bassam J, Dehmelt L, Milligan RA, Halpain S. MAP2c, but not tau, binds and bundles F-actin via its microtubule binding domain. Curr Biol. 2004;14:363–371. doi: 10.1016/j.cub.2004.01.058. [DOI] [PubMed] [Google Scholar]
- 47.Gimona M, Mital R. The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J Cell Sci. 1998;111:1813–1821. doi: 10.1242/jcs.111.13.1813. [DOI] [PubMed] [Google Scholar]
- 48.Galkin VE, Orlova A, Fattoum A, Walsh MP, Egelman EH. The CH-domain of calponin does not determine the modes of calponin binding to F-actin. J Mol Biol. 2006;359:478–485. doi: 10.1016/j.jmb.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 49.Sun QY, Schatten H. Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein. Front Biosci. 2006;11:1137–1146. doi: 10.2741/1868. [DOI] [PubMed] [Google Scholar]
- 50.Lydersen BK, Pettijohn DE. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 51.Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 54.Rankin KE, Wordeman L. Long astral microtubules uncouple mitotic spindles from the cytokinetic furrow. J Cell Biol. 2010;190:35–43. doi: 10.1083/jcb.201004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 57.Gregory SL, Ebrahimi S, Milverton J, Jones WM, Bejsovec A, Saint R. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr Biol. 2008;18:25–29. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 58.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: The role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol. 2008;180:739–746. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: Integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 61.Charras GT, Hu CK, Coughlin M, Mitchison TJ. Reassembly of contractile actin cortex in cell blebs. J Cell Biol. 2006;175:477–490. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.