Abstract
Barrett’s esophagus is a premalignant intermediate to esophageal adenocarcinoma, which develops in the context of chronic inflammation and exposure to bile and acid. We asked whether there might be common genomic alterations that could be identified as potential clinical biomarker(s) for Barrett’s esophagus by whole genome profiling. We detected copy number alterations and/or loss of heterozygosity (LOH) at fifty-six fragile sites in 20 patients with premalignant Barrett’s esophagus (BE). Chromosomal fragile sites are particularly sensitive to DNA breaks and have been shown to be frequent sites of rearrangement or loss in many human cancers. 78% of all genomic alterations detected by array-CGH were associated with fragile sites. Copy number losses in early BE were observed at particularly high frequency at FRA3B (81%), FRA9A/C (71.4%), FRA5E (52.4%) and FRA 4D (52.4%), and at lower frequencies in other fragile sites, including FRA1K (42.9%), FRAXC (42.9%), FRA 12B (33.3%) and FRA16D (33.3%). Due to the consistency of the region of copy number loss, we were able to verify these results by quantitative PCR which detected loss of FRA3B and FRA16D, in 83% and 40% of early molecular stage BE patients respectively. LOH in these cases was confirmed via pyrosequencing at FRA3B and FRA16D (75% and 70% respectively). Deletion and genomic instability at FRA3B and other fragile sites could thus be a biomarker of genetic damage in BE patients and a potential biomarker of cancer risk.
INTRODUCTION
Barrett’s esophagus (BE) is a condition in which the normal squamous lining of the esophagus is replaced by a metaplastic columnar (intestinal type) epithelium. BE develops in the context of chronic gastro-esophageal reflux disease (GERD), with repeated cycles of injury and repair in a genotoxic environment of exposure to acid, bile and chronic inflammation(1;2). BE is a pre-malignant condition – it is the only known precursor of esophageal adenocarcinoma (EA), a cancer which is increasing at an exponential rate in the USA. It is estimated that the incidence of GERD within the population is about 10%; Barrett’s esophagus is estimated to develop in 10% of those individuals, and the annual incidence of EA in these patients is estimated to be 0.5–1% per year (3). Barrett’s esophagus is therefore of considerable clinical significance since the five-year survival rate of esophageal adenocarcinoma is only ~10%, unless detected at an early stage, in which case it is curable. It is therefore recommended that BE patients be managed by endoscopic surveillance; however, at present 95% of patients with esophageal adenocarcinoma do not have a prior diagnosis of Barrett’s esophagus (4). It is therefore important to define biomarkers which could be readily applied to patients with GERD to identify those who have BE and are at risk for EA, and would therefore benefit from endoscopic surveillance and/or medical or surgical intervention. Although conventional upper GI endoscopy has become widespread in its applications and availability, it is constrained by the requirement for patient sedation, as endoscopes large enough to allow biopsies are not otherwise tolerated (5). To address this problem, an accurate, sensitive molecular biomarker for the presence of BE would be of great utility.
Widespread genomic instability is believed to facilitate neoplastic progression in BE, as well as many other pre-neoplastic diseases. This process is facilitated the loss and mutation of important cell cycle checkpoint machinery and tumor suppressor loci, such as p16 and p53. In addition, biomarkers of the process of genomic instability itself may be of clinical use. We have documented shortened telomere length and chromosomal instability using fluorescence in-situ hybridization (FISH) in BE (6;7). Although we have previously focused on sites of known tumor suppressors, we and others have shown that chromosomal “fragile sites” and in particular, FRA3B, have an extremely high rate of deletion in BE patients(8;9) and in patients that progress to EA(10). Thus, we hypothesize that fragile sites may serve as a sensitive biomarker of BE and the genotoxic injury that accompanies BE.
Fragile sites are loci that exhibit an increased propensity for sister chromatid exchange, translocation, and breaks under conditions of genotoxic stress (11;12). The susceptibility of these loci to damage is believed to be a consequence of their primary and secondary structure, which affects chromatin organization and can stall DNA replication (13;14). The resulting DNA gaps, breaks, and other chromosomal aberrations at fragile sites impact genomic stability, and often manifest as deletions and translocations. Currently, there are over one hundred documented fragile sites within the human genome, categorized as “common” (present in all individuals) or “rare” (present in less than 5% of the population) (13;14); most are defined cytogenetically and their molecular characterization is not known.
While instability at specific fragile sites has been linked to different cancers (15;16) including breast (17), prostate, and lung (18;19), there is still uncertainty as to whether these fragile site alterations causally contribute to cancer development or are merely “silent markers” of genomic stress. Putative tumor suppressors have been suggested to be located within common fragile sites; the fragile histidine triad gene, FHIT, at FRA3B and the WW-domain containing oxidoreductase, WWOX, at FRA16D have the best documented evidence for a role in cancer progression (20), while most other genes known to be at fragile sites, such as parkin at FRA6E have less clear evidence for roles as tumor suppressors (21). Alternatively, breakage at fragile sites could contribute to repeated cycles of bridge-breakage-fusion, potentially promoting the amplification of oncogenes (22) such as Met within the FRA7G region (23) or the prolactin-inducible protein (PIP) gene (24).
We propose that as a consequence of chronic acid and nitric oxide exposure and stalling the G1/S transition in Barrett’s epithelium combined with impaired DNA repair pathways (25–28), there is an increased instability at fragile sites in this premalignant tissue. In previous studies of BE patients, copy number loss was detected at two fragile sites (FRA3B and FRA13B)(8;9). In this report, we include a larger sampling of new early-stage BE specimens and report genomic instability (copy number loss and/or LOH) at many fragile sites in BE, some of which demonstrate increasing alterations with disease progression.
MATERIALS & METHODS
Patients and Cells
All patients included in this study were participants in the Seattle Barrett’s Esophagus Study Program and were evaluated as previously described (29). The endoscopic biopsies were analyzed for molecular alterations at chromosome arm 9p CDKN2A locus), chromosome arm 17p (TP53 locus), and DNA content tetraploidy and aneuploidy as previously described (29;30). Two sets of patients were examined; genomic DNA was isolated from paired Barrett’s epithelium and gastric samples: 1) 20 patients without high grade dysplasia characterized for chromosome arm 9pLOH and/or 17pLOH in which epithelial cells from selected biopsies were purified by Ki67/DNA content flow sorting. All biopsies were diploid by flow cytometry, measured as previously described (29). The maximum diagnoses for regions within 1 cm of biopsy site were: 6 metaplasia, 9 indefinite, 5 low-grade dysplasia. 17 of the 20 patients were lost-to-follow-up; however, 3 of these patients progressed to low grade dysplasia during surveillance. DNA content tetraploidy and aneuploidy was not detected in 19 of 20 of the baseline endoscopies. 2) 20 patients with early molecular stage BE without chromosome arm 17pLOH or DNA content tetraploidy or aneuploidy (Table 2) were analyzed by PCR and pyrosequencing, in which 1 to 6 biopsies (separated by a minimum of 2 cm longitudinally in the BE segment) were studied from each patient. Patients ranged in age from 36 to 81 years of age (mean, 60.8 years) and BE length ranged from 1 to 12 cm (mean 5.7 cm). Biopsies were analyzed for molecular alterations at chromosome arm 9p (CDKN2A locus), chromosome arm 17p (TP53 locus), and DNA content flow cytometry as previously described (29) and the maximum molecular abnormality for that patient is listed in Table 2. Histologic diagnosis was established by previously published criteria (29). All of the biopsies evaluated were within 1 cm of the maximum diagnosis as shown in Table 2. None of these patients overlapped with those examined previously by array CGH (8).
Table 2. Molecular and histological data for samples analyzed by qPCR and pyrosequencing.
For LOH, multiple sites were tested from patients, where available, relative to paired gastric constitutional. LOH reflects proportion of sites in which LOH or allelic imbalance (see Methods) was detected relative to the total number of sites tested per patient.
| Patient | Diagnosis | 9pLOH | 17pLOH | Aneuploid | Copy # FRA3B | LOH FRA3B | Copy # FRA16D | LOH FRA16D |
|---|---|---|---|---|---|---|---|---|
| 1 | Metaplasia | ND | ND | No | Gain | 0 of 1 | Normal | 0 of 1 |
| 2 | Metaplasia | No | No | No | Loss | 2 of 4 | Normal | 3 of 4 |
| 3 | Metaplasia | No | No | No | Loss | 1 of 1 | Normal | 0 of 1 |
| 4 | Metaplasia | No | No | No | Loss | 1 of 1 | Loss | 1 of 1 |
| 5 | Metaplasia | No | No | No | Loss | 2 of 4 | Loss | 4 of 4 |
| 6 | Metaplasia | No | No | No | Loss | 0 of 1 | ND | 0 of 1 |
| 7 | Metaplasia | No | No | No | Gain | 1 of 1 | Loss | 1 of 1* |
| 8 | Metaplasia | No | No | No | Loss | 1 of 1 | Loss | 1 of 1 |
| 9 | Metaplasia | Yes | No | No | Loss | 6 of 6 | Normal | 4 of 6 |
| 10 | Metaplasia | Yes | No | No | Loss | 0 of 1 | Normal | 1 of 1 |
| 11 | Metaplasia | Yes | No | No | Loss | 3 of 4 | Loss | 3 of 4 |
| 12 | Indefinite | No | No | No | Normal | 2 of 3 | Normal | 1 of 3 |
| 13 | Indefinite | No | No | No | ND | 2 of 2 | ND | 1 of 2 |
| 14 | Indefinite | No | No | No | Loss | 1 of 3 | Normal | 2 of 3 |
| 15 | Indefinite | No | No | No | Loss | 2 of 2 | ND | 1 of 2 |
| 16 | Indefinite | Yes | No | No | ND | 3 of 3 | ND | 3 of 3 |
| 17 | Indefinite | Yes | No | No | Loss | 1 of 1 | Normal | 1 of 1* |
| 18 | Low-Grade | Yes | No | No | Loss | 2 of 2 | ND | 0 of 2 |
| 19 | High-Grade | No | No | No | Loss | 1 of 1 | Loss | 1 of 1 |
| 20 | High-Grade | Yes | No | No | Loss | 3 of 3 | Normal | 2 of 3 |
LOH flank refers to LOH detected in genomic sequence flanking the fragile site, but not within FRA16D itself.
Copy # refers to qPCR data comparing the relative difference in copy number between the BE sample and the paired constitutional samples for 1–2 sites within the respective fragile sites normalized against 2 flanking sites.
Epithelial cells from these biopsies were purified by Ki67/DNA content flow sorting and gastric cells isolated by mincing of whole biopsy material as described previously. DNA was extracted using the Puregene DNA isolation kit (Invitrogen) as described previously (31). Cell lines from BE patients, CPA, CPB, CPC and CPD, and their culture were as previously described (32;33). The Seattle Barrett’s Esophagus Study has been approved by the Human Subjects Divisions of the University of Washington and/or the Fred Hutchinson Cancer Research Center (FHCRC) continuously from 1983 to the present.
Agilent arrays
Genomic DNA
We obtained normal female 46, XX genomic DNA from Promega (Madison WI). The BE cell lines were maintained in modified MCDB 153 as described previously (33). In brief, genomic DNA was prepared from each cell line using the DNeasy Tissue Kit (Qiagen). All DNA samples were gel-verified for quality and assayed using a Nanodrop spectrophotometer to determine concentration and purity.
Sample Labeling
For each CGH hybridization, 1 μg of genomic DNA from the reference (46, XX female) and the corresponding experimental sample was digested with AluI (2.5 units) and RsaI (2.5 units) (Promega). Labeling reactions were performed directly with the restricted DNA and a Bioprime labeling kit (Invitrogen) according to the manufacturer’s directions in a 50 μl volume with a dNTP pool to final concentrations of 120 μM dATP, 120 μM dGTP, 120 μM dTTP, 60 μM dTTP, and 60 μM Cy5-dUTP or Cy3-dUTP (Perkin-Elmer). Cell line samples were labeled with Cy5-dUTP and the reference sample with Cy3-dUTP in each experiment. Labeled targets were subsequently filtered using a Centricon YM-30 filter (Millipore). Experimental and reference targets for each hybridization were pooled, mixed with 50 μg of human Cot-1 DNA (Invitrogen), and 100 μg of yeast tRNA (Invitrogen) to a final volume of 250 μl, then mixed with an equal volume of Agilent 2X in situ Hybridization Buffer.
Oligonucleotide Microarray Processing
Prior to hybridization to Agilent Human Genome 244K CGH arrays (Agilent Technologies), the 500-μl hybridization mixtures were denatured at 100°C for 3minutes and incubated at 37°C for 30 minutes. To remove any precipitate, each mixture was centrifuged at ≥ 14,000Xg for 5 minutes and transferred to a new tube, leaving a small residual volume (≤ 5 μl). The sample was applied to the array using an Agilent microarray hybridization chamber, and hybridization was carried out for 40 hrs at 65°C in a Robbins Scientific rotating oven at 20 rpm. The arrays were then disassembled and washed for 5 minutes at RT in wash 1 (0.5X SSPE/0.005% NLS), followed by 3 minutes at 37°C in wash 2 (0.1X SSPE/0.005% NLS). Slides were dried and scanned at 5 μm resolution using an Agilent scanner. Image analysis was performed using default CGH settings of Feature Extraction Software version 9.1 (Agilent Technologies).
Illumina SNP genotyping
Paired constitutional and endoscopic BE samples were analyzed by the Illumina Infinium assay on high density SNP genotyping Bead Chips per manufacturer’s instructions. In brief, genomic DNA was extracted from Ki67-positive flow sorted epithelium samples as described or unsorted constitutive fundal mucosa from the lower esophageal sphincter (LES) or stomach. These samples underwent whole-genome amplification using standard Infinium protocols. The resultant product was fragmented to ~500 bp by enzymatic digestion, precipitated, resuspended in hybridization buffer, denatured, and hybridized to the BeadChip overnight at 48°C. BeadChips were washed, primer extended, and stained on a Tecan Genesis/EVO robot using a Tecan GenePaint slide processing system. After staining, the BeadChips were washed, immediately coated with a protective reagent, and imaged on Illumina’s BeadArray Reader. The image intensities are extracted, and the resultant data analyzed using Illumina’s BeadStudio 3.0 software.
Two types of SNP genotyping arrays were employed: HumanHap300 BeadChip (317k loci) and a non-commercial multi-sample HumanHap300_Pool 10 BeadChip (33k loci) that contained a subset (single beadpool) of the overall SNPs from the HumanHap300 BeadChip (33k) in a 12-sample format (34). Illumina HumanHap300 arrays were processed and scanned at Children’s Hospital of Philadelphia, and the Illumina Human-1 and Illumina multi-sample 33k BeadChips were processed at Illumina Inc. (San Diego). All sample pairs were analyzed in “paired mode” using BeadStudio 2.0 or 3.0 as described (34).
Copy number estimates and genotype calls for Illumina BeadChips were calculated using the BeadStudio 2.0 output for raw intensity values and subsequent analyses. Log2 intensity ratios for BE samples relative to paired constitutional controls were analyzed per patient for significant regions of copy number loss or gain using the CLAC (Clustering Among Chromosomes) software (35) with a 3 SNP moving window and FDR = 0.01. Copy number differences were manually curated to include only events with an average log2 intensity ratio of > 0.26 or < −0.32, corresponding to a minimum change of 20% between constitutional and test samples. Fragile site names and cytoband locations were downloaded from NCBI MapViewer (Build 36.2; http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9606). Cytoband positions were obtained from the UCSC Genome Annotation Database (hg18; http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/). LOH was determined as a minimum of 20% difference in the relative allele frequency between the paired constitutional and BE samples in regions containing at least one informative SNP. Frequency p-values for the all copy number loss, copy number gain, and LOH were evaluated for each chromosome arm using STAC v1.2 with data outputted in binary format per 1 Mb region and 500 permutations per analysis (36). A p-value cut-off of 0.05 was used for selection of fragile sites with significant regions of loss.
Quantitative PCR
DNA was extracted from gastric or flow purified BE epithelium, and amplified with quantitative PCR using the Biotage Rotorgene RG-3000 using a protocol adapted from Boehm et al., 2004 (37). Initial DNA concentrations were measured using the Nanodrop ND-1000 and adjusted to 4 ng/μl. Cycle threshold (Ct) was determined using a Sybr Green fluorescence threshold of 20%; copy numbers were estimated based on amplification relative to a standard curve (2-fold increments ranging from 0.25 to 4 ng) and prepared with pooled male genomic DNA (Promega #1471). One Ct difference equates to a loss of one gene copy. Individual runs were normalized against multiple genomic DNA control samples to facilitate comparison between reactions with varying efficiencies. Two primers located within region of loss (as determined by SNP genotyping) and one 5′ and one 3′ flanking primer set (on the same chromosome but at least 10 Mb from the fragile site) were used to compare copy number. Primer sequences are provided in Supplementary Table 1. Copy number estimates were evaluated per chromosome, with flanking regions normalized to 2 copies. The ratio between copy number for BE relative to gastric within the fragile site was then used to identify copy number gains (ratio ≥ 1.2) or copy number loss (ratio ≤ 0.8). All samples were run in triplicate.
Pyrosequencing
LOH at fragile sites was independently confirmed on 20 Barrett’s esophagus patients by SNP pyrosequencing. Patient data is summarized in Table 3; DNA from multiple sites at 2 cm intervals within a single BE segment was evaluated when material was available. For each patient, genomic DNA was extracted from flow sorted endoscopic biopsies and unsorted gastric biopsies. Pyrosequencing (forward, reverse, and sequencing) primers were designed using the PSQ Assay Design 1.0 software to select primers with similar melting temperatures, with products ranging in size from 80 to 120 bp, and overall scores above 85. BLAT search (http://genome.ucsc.edu/cgi-bin/hgBlat) was used to confirm unique sequence for each PCR product. Primers selected to interrogate fragile site FRA3B (see Supplementary Table 1) had a mean HapMap minor allele frequency = 0.338 and FRA16D (see Supplementary Table 1) had a mean HapMap minor allele frequency = 0.299. PCR reactions were performed using standard PCR protocol (1X PCR buffer, 1.5 mM MgCl2, 0.2 ng DNA, 5 mM dNTP, 5 μM primers, 1.25 units AmpliTaq) and amplified using this program: 95°C, 4 min, then 45 cycles at 95°C, 15 sec; 57°C, 30 sec; 72°C, 15 sec. Pyrosequencing reactions were purified over vacuum prep and mixed with sequencing primer as per manufacturer’s instructions. All reactions were performed on the Biotage PSQ HS 96. Allele frequencies were outputted as relative percentages using Biotage PSQ software and genotypes were assigned based on the relative allele frequency between constitutional and experimental samples. Informative calls were evaluated as relative ratios of 50:50 ± 10%. Allelic imbalance (AI) was assigned to SNP’s which were informative in the constitutional and showed relative allele frequencies of greater than 66:33 to 89:11 in the BE sample. Loss of heterozygosity (LOH) was determined as ratios 90:10 or greater in the BE sample with an informative ratio in the constitutional sample.
Clustering Analysis
Average copy number estimates for each fragile site region in Figure 1A were analyzed in Cluster version 2 using complete linkage and hierarchical clustering with uncentered correlation. Heat maps were visualized using TreeView version 1.60.
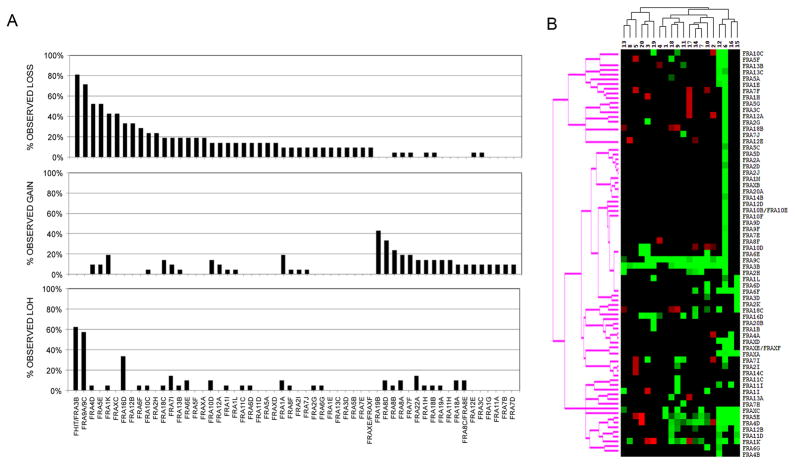
Figure 1. Copy loss, copy gain, and LOH at fragile sites in BE samples.
(A) Shown is frequency of copy number loss (top panel), copy number gain (middle panel), or LOH (bottom panel) within the respective cytoband regions for fragile sites with alterations in two or more patients. Copy number changes were determined using the CGH-Miner software and LOH using the relative allele frequency between the BE sample and the paired constitutional sample. (B) Clustering analysis for fragile site copy number gain and loss by patient. Black indicates normal copy number; red, copy gain; green, copy loss for individual fragile sites.
RESULTS
Instability at multiple fragile sites detected in endoscopic BE samples
To examine whether copy number losses could be detected in fragile sites from early stage BE clinical samples, genome-wide analyses were performed using DNA isolated from 20 patients without high-grade dysplasia selected from the Seattle Barrett’s Esophagus Surveillance Program. DNA extracted from diploid BE epithelium was Ki67/DNA content flow sorted and analyzed for genome-wide abnormalities using multi-sample Illumina 33K arrays relative to paired gastric constitutional DNA.
We first evaluated whether copy number alterations or LOH would be associated with alterations at predicted fragile sites defined solely by cytoband location since most fragile sites are thus far uncharacterized. Copy number loss, copy number gain, and/or LOH were indeed detected in two or more patients at genomic regions associated with 56 different fragile sites (Figure 1). Overall, copy number loss was detected at an average of 9.8 fragile sites per patient within this set and 2 of the 20 patients showed copy loss at over 20 different chromosomal fragile sites. As shown in Figure 1A, top panel, the most frequent and significant losses were observed at FRA3B (81%, FRA9A/9C (71.4%), FRA4D (52.4%), FRA5E (52.4%), FRA1K (42.9%), FRAXC (42.9%), FRA16D (33.3%), and FRA12B (33.3%). Note: While there was no coverage for p16/CDKN2A on the 33K arrays, we believe that the deletion observed in the region of FRA9A/9C was likely at p16/CDKN2A consistent with earlier reports that this locus is frequently mutated or deleted in BE. The regions of deletion seen in fragile sites FRA3B and FRA16D were surprisingly small and consistent; the consensus regions of copy change being 400 kb and 230 kb, respectively. The compact nature of these changes is further illustrated in Figures 2 and 3.
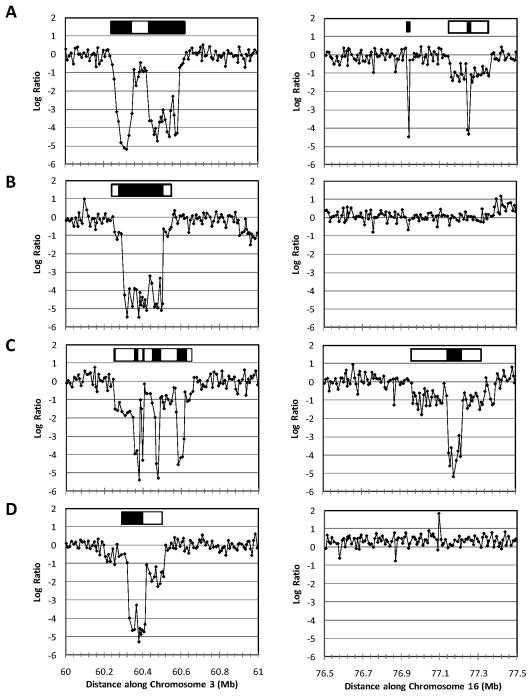
Figure 2. Copy number loss at common fragile sites detected in BE cell lines.
Copy number plots showing FRA3B (left) and FRA16D (right) for individual BE cell lines (A, CPA; B, CPB; C, CPC; D, CPD) analyzed on Human Genome (244K) arrays. A log ratio of -1 represents one-copy number loss and a log2 ratio of approximately −4 corresponds to homozygous two copy number loss in at least 95% of cells. Open and closed boxes above each plot indicate approximate extents of one- and two-copy number loss, respectively.
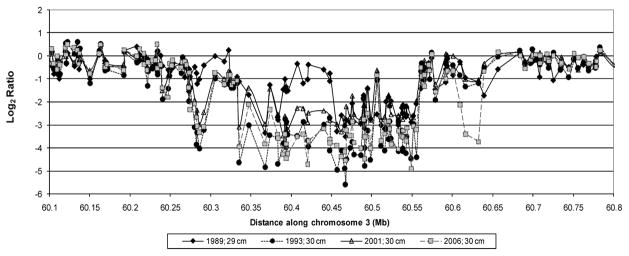
Figure 3. HumanHap300 (317k) array analysis of BE biopsies at the same level (±1cm) from one patient over 4 endoscopies from 1989 to 2006.
Closed diamonds, 1989; closed circle, 1993; open triangle, 2001; open square, 2006. In 1989 the BE consensus FRA3B region showed a region of 1-copy number loss (b) flanked by 2-copy number loss (a), with an adjacent region of 1-copy number loss (c). In 1993 (a and b) merged into a region of uniform 2-copy number loss (f), flanked by new regions of 2-copy (d) and 1-copy (e) loss. In 2006 the region of 1-copy number loss at (c) lost its second copy (g).
On average, copy gain was present at an average of 4.5 fragile sites per patient (Figure 1A, middle panel). Individual sites of gain were observed at low frequency except at FRA19B (42.9%), FRA8D (33.3%), and FRA8B (23.8%). LOH was observed at an average of 2.6 sites per patient (Figure 1A, bottom panel), and was detected at higher than expected frequency in three fragile sites, FRA3B (61.9%; p-value = 0.002), FRA9A/9C/p16/CDKN2A (57.1%; p-value = 0.002), and FRA16D (33.3%; p-value =0.002). However, the total number of LOH calls made with the 33K arrays may be underestimated, as the lower number of informative SNPs on these arrays reduces the ability to detect small regions of LOH in comparison to determination of copy change, which is based on all 33K probes.
Figure 1B illustrates a clustering analysis of patients showing copy number gain or loss at individual fragile sites; the highest correlation was detected for losses at FRA3B and FRA9A/9C/p16/CDKN2A. We observed that a subset of patients (patient # 6, 12, 15, and 16) had copy number loss detected at several fragile sites including FRAXC, FRA5E, FRA4D, FRA12B, FRA11D, and FRA1K although interestingly, the most frequent copy number losses at FRA3B/FHIT and FRA9A/9C/p16/CDKN2A were not detected in patients 15 & 16. The most striking copy number losses were observed for patient #6. Future studies which correlate clusters of fragile site instability with patient outcome should be revealing; however, that information was unavailable for the majority of this patient set.
Since most fragile sites are uncharacterized, we asked whether conserved copy number and genotypic alterations could be detected in more defined regions within the cytobands; therefore, we used the frequency and footprint analysis of the Significance Testing for Aberrant Copy number (STAC) algorithm as described by Diskin et al., 2006 (36). We postulated that if the fragile site instability were a consequence of the shared environment of inflammation and oxidative stress created in Barrett’s esophagus and enriched in specific DNA sequences, then conserved regions of copy number change should be detectable amongst our patient set. Results of the STAC analysis are shown in Table 1, which displays the subset of the fragile sites at which copy number losses, copy number gains or LOH were statistically significant by this method. Seventeen regions of copy loss, gain or LOH were identified within putative fragile sites. Moreover, there were an additional 10 regions of copy number loss, gain or LOH located within 10 Mb of cytoband regions previously identified as fragile sites (Table 1, middle). These 27 regions in or near fragile sites represented 78.1% of all genomic alterations identified by STAC analysis as significantly altered in these 20 paired patient samples. Significant copy gain events were detected at FRA 8D, FRA11H, FRA19B, and FRA22A. 29.5% of all SNPs detected as having LOH on the Illumina 33K arrays were located within just 3 fragile site regions, FRA3B, FRA9A/9C/p16/CDKN2A, and FRA16D.
Table 1. Regions of copy change and LOH.
Shown are significant regions of copy number loss or LOH. Regions within cloned fragile sites (top set), associated with fragile sites (2nd from top), near fragile sites (within 10 Mb, 3rd from top), or at non-fragile sites (bottom set) are shown. Cytogenetic band locations and the chromosome positions (start and end) are noted; the latter indicates the largest extent of the chromosomal region identified by STAC as being statistically altered. The frequency of each alteration is shown. Frequency and footprint p-values were generated in STAC.
| Alteration(s) | Cytoband(s) | Start (Mb) | End (Mb) | Fragile Site | Type | Distance to Fragile Site | Frequency | Frequency p value | Footprint p value |
|---|---|---|---|---|---|---|---|---|---|
|
CLONED FRAGILE SITES
| |||||||||
| copy loss | 3p14.2 | 59 | 62 | FRA3B | common | 80.00% | 0.001 | 0.004 | |
| copy loss | 18q22.1 | 63 | 66 | FRA18C | rare | 35.00% | 0.001 | 0.004 | |
| copy loss | 16q23.3-24.1 | 76 | 78 | FRA16D | common | 35.00% | 0.001 | 0.004 | |
| copy loss | 7q31.2 | 117 | 118 | FRA7G | common | 20.00% | 0.001 | 0.002 | |
| LOH | 3p14.2 | 59 | 61 | FRA3B | common | 60.00% | 0.001 | 0.002 | |
| LOH | 16q23.3-24.1 | 77 | 79 | FRA16D | common | 35.00% | 0.001 | 0.002 | |
|
| |||||||||
|
ASSOCIATED WITH FRAGILE SITES
| |||||||||
| copy loss | 9p21 | 21 | 23 | FRA9A/9C | rare | 75.00% | 0.001 | 0.008 | |
| copy loss | 5p14 | 23 | 30 | FRA5E | common | 55.00% | 0.001 | 0.004 | |
| copy loss | 4p15 | 33 | 36 | FRA4D | common | 45.00% | 0.001 | 0.004 | |
| copy loss | 5p13 | 29 | 30 | FRA5A | common | 45.00% | 0.0010 | 0.0040 | |
| copy loss | Xq22.1 | 98 | 99 | FRAXC | common | 35.00% | 0.013 | 0.01 | |
| copy loss | 1q31 | 185 | 187 | FRA1K | common | 30.00% | 0.001 | 0.006 | |
| copy loss | 12q21.3 | 81 | 85 | FRA12B | common | 30.00% | 0.013 | 0.002 | |
| copy loss | 10q21 | 67 | 69 | FRA10C | common | 15.00% | 0.071 | 0.006 | |
| copy gain | 8q24.3 | 144 | 146 | FRA8D | common | 25.00% | 0.385 | 0.006 | |
| copy gain | 19p13 | 0 | 5 | FRA19B | rare | 35.00% | 0.011 | 0.01 | |
| LOH | 9p21 | 21 | 23 | FRA9A/9C | rare | 55.00% | 0.001 | 0.012 | |
|
| |||||||||
|
NEAR FRAGILE SITES
| |||||||||
| copy loss | 5q14.3 | 83 | 90 | (FRA5B/5D) | common | 1.9 Mb | 50.00% | 0.001 | 0.004 |
| copy loss | 3q25-26.1 | 163 | 170 | (FRA3D) | common | 1.8 Mb | 45.00% | 0.001 | 0.004 |
| copy loss | 13q31.1 | 81 | 86 | (FRA13B) | common | 9.9 Mb | 40.00% | 0.001 | 0.004 |
| copy loss | 11p13 | 38 | 39 | (FRA11E) | common | 1.6 Mb | 35.00% | 0.033 | 0.004 |
| copy loss | 7q21.11 | 84 | 86 | (FRA7E) | common | 4.9 Mb | 35.00% | 0.001 | 0.002 |
| copy loss | 6q21 | 101 | 104 | (FRA6F) | common | 0.8 Mb | 30.00% | 0.045 | 0.006 |
| copy loss | 7p22 | 9 | 11 | (FRA7B) | common | 1.8 Mb | 20.00% | 0.017 | 0.012 |
| copy loss | 11q22.1 | 97 | 98 | (FRA11F) | common | 8.1 Mb | 20.00% | 0.005 | 0.004 |
| copy loss | 11q14.2 | 89 | 90 | (FRA11F) | common | 1.1 Mb | 20.00% | 0.005 | 0.004 |
| copy gain | 10q26.3 | 134 | 135 | (FRA10F) | common | 6 Mb | 15.00% | 0.023 | 0.03 |
|
| |||||||||
|
NON-FRAGILE SITES
| |||||||||
| copy loss | Xp21.1 | 31 | 33 | 45.00% | 0.001 | 0.006 | |||
| copy loss | 3p12.1 | 83 | 87 | 40.00% | 0.001 | 0.004 | |||
| copy loss | Xq25 | 125 | 126 | 40.00% | 0.003 | 0.01 | |||
| copy loss | 6q22.31 | 124 | 125 | 35.00% | 0.003 | 0.004 | |||
| copy loss | 21q21.1 | 20 | 22 | 25.00% | 0.01 | 0.04 | |||
| copy gain | 9q34.2 | 135 | 138 | 20.00% | 0.001 | 0.004 | |||
| copy gain | 20q13.33 | 60 | 61 | 20.00% | 0.011 | 0.002 | |||
Fragile site copy loss and LOH in early molecular stages of BE confirmed by q-PCR and pyrosequencing
To confirm the results observed by SNP genotyping, copy number and LOH were evaluated at two fragile sites using q-PCR and pyrosequencing in 20 BE patients selected as having the earliest molecular stage of disease: without LOH on chromosome arm 17p and without DNA content tetraploidy and aneuploidy (30). Patient and biopsy information is provided in Table 2. For each case, DNA isolated from a BE biopsy that was Ki67/DNA content flow sorted was compared to a reference gastric DNA sample taken from the same individual. Fragile sites FRA3B and FRA16D were selected for this study, since, as noted above, the SNP genotyping analysis indicated that these were commonly deleted sites with small and consistent consensus regions of copy change; this allowed molecular analysis to be performed with the minimum number of PCR targets for qPCR and pyrosequencing (see Methods). As shown in Table 2, copy number loss was detected at FRA3B and FRA16D in 83.3% (15 of 18) and 40% (6 of 15) of cases, respectively. Copy number gain was detected at FRA3B in 11.1% (2 of 18) of patients tested. Of note, copy changes were just as frequent in cases without dysplasia as in cases with a maximum diagnosis of indefinite for dysplasia (FRA3B, p>0.29; FRA16D, p>0.44). Pyrosequencing of SNPs spaced within FRA3B or FRA16D, compared to SNPs taken from flanking regions was performed in one to six biopsies per patient, based on availability. When multiple BE samples were analyzed, samples were taken at 2-cm intervals within the BE segment. LOH or allelic imbalance (AI) was detected in at least one SNP and at least one BE biopsy within FRA3B in 75.0% (15 of 20) of patients (Table 2). At the FRA16D site, we detected LOH or AI in 70.0% of patients (14 of 20). These results confirm the high frequency of copy loss and LOH observed by whole genome analysis.
Loss at fragile sites detected in BE cell lines
To further validate these results, we tested for the extent of copy number losses at FRA3B and FRA16D in populations of BE cells grown in culture, cell lines CPA, CPB, CPC, and CPD (32;33) on higher density Agilent 244K SNP arrays. Figure 2 shows copy number plots at two common fragile sites, FRA3B and FRA16D. Well defined interstitial one- and two-copy deletions were present at FRA3B in all four cell lines tested, and moreover, in each case, at least 2–4 discreet one- or or two-copy number loss events were visible. At FRA16D, two copy losses were flanked by regions of one copy number loss in CPA and CPC whereas normal copy numbers were observed at the FRA16D locus in cell lines, CPB and CPD. A small region of copy number loss was also detected in two of four cell lines at FRAXC (data not shown).
Evolution of FRA3B instability over time
Having shown that copy loss at fragile sites was present at very high frequency in earlier molecular stages of this disease, we next asked whether a temporal and spatial analysis would reveal evolving instability within fragile sites in these patients. Endoscopic biopsies were examined from a single low risk patient (without dysplasia, p53LOH, DNA content tetraploidy or aneuploidy) followed during surveillance (29) over a 16 year time span. Biopsies from three different levels: 26cm, 29cm, 32cm (± 1 cm) within a single BE segment were analyzed using Illumina HumanHap300 BeadChips. Figure 3 illustrates the regions of greatest instability at FRA3B observed longitudinally at the 29 cm level. More than 8 distinct copy loss events were seen within FRA3B/FHIT, seven of which were observed over a 4 year time span. Corresponding LOH was detected in regions of 1-copy (but not 2-copy) loss (data not shown). Similar regions of 1-copy loss were detected at the other levels in the regions of 60.3 to 60.35 Mb and 60.55 to 60.6 Mb on chromosome 3. 2-copy loss was detected in a 240 kb region from 60.33Mb to 60.57 Mb on this chromosome at all three levels at 1993. The copy loss event at 60.6 to 60.63 Mb appeared with less frequency—at 29/30 cm, 2-copy loss was observed but at 26 and 32 cm, 1-copy loss was observed. This patient also had copy loss at other fragile sites (e.g., FRA16D and FRA7G), some of which showed additional lesions over time (data not shown). Thus, in this patient, FRA3B clearly showed the greatest rate of change and evolution over time.
DISCUSSION
Using SNP genotyping analysis to profile for LOH and copy number, with confirmation by quantitative PCR and pyrosequencing, we have demonstrated that copy number alteration and/or LOH at chromosomal fragile sites are frequent and early events in BE neoplasia. Copy number loss and LOH was validated at FRA3B and FRA16D using PCR and pyrosequencing. Copy number loss at FRA3B is a particularly common early event, being observed in 80% of 20 early-stage endoscopic biopsies. The fragile site deletions that are reported were not observed in gastric samples from these patients, and thus, these abnormalities are not constitutional polymorphisms and are specific to the premalignant columnar BE tissue.
While instability at fragile sites has been reported in a number of different cancers, this study demonstrates high frequency of copy number loss and LOH within multiple defined regions associated with chromosomal fragile sites in a premalignant tissue. Fragile sites may be particularly sensitive to damage in Barrett’s epithelium due to chronic acid exposure, which promotes an anti-proliferative effect on Barrett’s epithelium and increases stalling at the late replicating foci (38). While trinucleotide repeat expansion has been reported as the mechanism for rare fragile site instability, recent data suggests that the distinctions between mechanisms of instability at common vs. rare fragile sites are not so stringent, as aphidocolin induced instability was observed at both common and rare fragile sites (39). We suggest that fragile site instability in BE may be similar in this regard.
It appears that regions of copy loss and LOH in BE can be narrow and well-conserved, in at least in a subset of fragile sites, and this is most evident in BE at FRA3B. While published studies of various cancers have reported deletions at FRA3B, these deletions range in size from 300 kb to over 2 Mb (40–42) and deletions consistently constrained to this specific sub-region of FRA3B (60.2–60.6 MB, corresponding to FHIT exons 4–5) have not previously been reported in the literature. Both the high frequency and the uniformity of the alterations in BE may reflect a common etiology of genotoxic stress; in BE this is likely oxidative damage, both as a direct effect of bile acids (15;43;44) and secondarily from oxygen and nitrogen free radicals produced by inflammation. The primary, secondary and tertiary structure of fragile sites is thought to promote stalling of DNA replication forks, which can induce recombination and strand breakage at fragile sites (45). If instability at fragile sites does indeed reflect the history of genotoxic stress in these patients, then it may serve as a biomarker of an individual’s history of such damage (45). Importantly, breaks at fragile sites are induced by replication stresses in vitro (39;46) suggesting that fragile sites may be the first regions of the genome to be mutated during chronic carcinogenic exposure in vivo (13). Our data supports the idea of a specific DNA damage profile, similar to asbestos-related lung cancer, where a significant association between copy changes at 11 fragile sites (including FRA19B, FRA22A, and FRA11H) and asbestos-associated alterations by SNP genotyping has been reported (19).
Thus, measurement of deletion and LOH at fragile sites merits evaluation as a biomarker of cancer risk in BE patients. This biomarker could also play a role as an intermediate indicator of the success of chemopreventative strategies in BE. Such applications could include the use of non-steroidal anti-inflammatory drugs (NSAIDs) which reduce inflammation and have been shown to be protective for development of EA (47), as well as long-term acid suppression, a common therapy for BE, which has been shown to decrease cellular proliferation via downregulation of Mcm2 expression (48), and could potentially stabilize fragile sites by alleviating replicative stress.
There are several challenges to the development of robust assays that are sufficiently sensitive to detect rare sequences in a background of normal sequence: technical challenges in detecting rare sequences against a normal background and the challenge of defining a novel sequence in the target cells of interest. While some genetic alterations have been reported to be present in premalignant BE tissue (2;49–51), none have thus far been present at a sufficiently high frequency to enable their use in screening of at-risk populations. We have described a genetic lesion that is present in ~90% of early stage BE. Therefore, this is a unique case in which we know in advance with ~90% sensitivity the identity of a DNA sequence abnormality in the target cells, with the opportunity to increase this to ~100% by analysis of several additional fragile sites. With the exception of viral sequences (such as HPV in cervical cells), a genetic marker of preneoplastic cells has heretofore not been available for a premalignant tissue. Future expansion of these studies to include patients with the spectrum of disease progression and known outcomes would shed more light onto the prognostic value of this type of genomic instability in predicting development of dysplasia and cancer in BE.
Supplementary Material
Acknowledgments
We would like to thank Jasmine Gallaher for technical assistance. Illumina, BeadArray, and Infinium are registered trademarks or trademarks of Illumina, Inc. EVO and GenePaint are registered trademarks or trademarks of Tecan, AG. We would like to thank Biotage for use of the pyrosequencer. This work was supported by National Institutes of Health (P01 CA91955 to P. S. R and C. C. M; 2 R44 CA103406-02 to K. L. G.; T32 AG00057 to L. A. L.). C. C. M. and R.K. were also supported in part by a PhRMA Foundation Informatics Research Starter Grant, the Pew Charitable Trust, and the Commonwealth Universal Research Enhancement Program from the Pennsylvania Department of Health.
References
- 1.Jenkins GJ, Doak SH, Parry JM, et al. Genetic pathways involved in the progression of Barrett’s metaplasia to adenocarcinoma. Br J Surg. 2002:824–37. doi: 10.1046/j.1365-2168.2002.02107.x. [DOI] [PubMed] [Google Scholar]
- 2.Sihvo EI, Salminen JT, Rantanen TK, et al. Oxidative stress has a role in malignant transformation in Barrett’s oesophagus. Int J Cancer. 2002:551–5. doi: 10.1002/ijc.10755. [DOI] [PubMed] [Google Scholar]
- 3.Wild CP, Hardie LJ. Reflux, Barrett’s oesophagus and adenocarcinoma: burning questions. Nat Rev Cancer. 2003:676–84. doi: 10.1038/nrc1166. [DOI] [PubMed] [Google Scholar]
- 4.Morales TG, Sampliner RE. Barrett’s esophagus: update on screening, surveillance, and treatment. Arch Intern Med. 1999:1411–6. doi: 10.1001/archinte.159.13.1411. [DOI] [PubMed] [Google Scholar]
- 5.Nietert PJ, Silverstein MD, Mokhashi MS, et al. Cost-effectiveness of screening a population with chronic gastroesophageal reflux. Gastrointestinal Endoscopy. 2003:311–8. doi: 10.1067/mge.2003.101. [DOI] [PubMed] [Google Scholar]
- 6.Finley JC, Reid BJ, Odze RD, et al. Chromosomal instability in Barrett’s esophagus is related to telomere shortening. Cancer Epidemiol Biomarkers Prev. 2006:1451–7. doi: 10.1158/1055-9965.EPI-05-0837. [DOI] [PubMed] [Google Scholar]
- 7.O’Sullivan JN, Finley JC, Risques RA, et al. Telomere length assessment in tissue sections by quantitative FISH: image analysis algorithms. Cytometry. 2004:120–31. doi: 10.1002/cyto.a.20006. [DOI] [PubMed] [Google Scholar]
- 8.Lai LA, Paulson TG, Li X, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007:532–42. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 9.Michael D, Beer DG, Wilke CW, et al. Frequent deletions of FHIT and FRA3B in Barrett’s metaplasia and esophageal adenocarcinomas. Oncogene. 1997:1653–9. doi: 10.1038/sj.onc.1201330. [DOI] [PubMed] [Google Scholar]
- 10.Fang JM, Arlt MF, Burgess AC, et al. Translocation breakpoints in FHIT and FRA3B in both homologs of chromosome 3 in an esophageal adenocarcinoma. Genes Chromosomes Cancer. 2001:292–8. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1095>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984:1199–204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland GR, Richards RI. The molecular basis of fragile sites in human chromosomes. Curr Opin Genet Dev. 1995:323–7. doi: 10.1016/0959-437x(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang YH. Chromatin structure of human chromosomal fragile sites. Cancer Lett. 2006:70–8. doi: 10.1016/j.canlet.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 15.O’Keefe LV, Richards RI. Common chromosomal fragile sites and cancer: Focus on FRA16D. Cancer Letters. 2006:37–47. doi: 10.1016/j.canlet.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 16.Arlt MF, Miller DE, Beer DG, et al. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes & Cancer. 2002:82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadian M, Wistuba II, Fong KM, et al. Analysis of the FHIT gene and FRA3B region in sporadic breast cancer, preneoplastic lesions, and familial breast cancer probands. Cancer Research. 1997:3664–8. [PubMed] [Google Scholar]
- 18.Fong KM, Biesterveld EJ, Virmani A, et al. FHIT and FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing aberrations. Cancer Research. 1997:2256–67. [PubMed] [Google Scholar]
- 19.Nymark P, Wikman H, Ruosaari S, et al. Identification of specific gene copy number changes in asbestos-related lung cancer. Cancer Research. 2006:5737–43. doi: 10.1158/0008-5472.CAN-06-0199. [DOI] [PubMed] [Google Scholar]
- 20.Kuroki T, Tajima Y, Furui J, et al. Common fragile genes and digestive tract cancers. Surg Today. 2006:1–5. doi: 10.1007/s00595-005-3094-4. [DOI] [PubMed] [Google Scholar]
- 21.Smith DI, Zhu Y, McAvoy S, et al. Common fragile sites, extremely large genes, neural development and cancer. Cancer Letters. 2006:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Hellman A, Zlotorynski E, Scherer SW, et al. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 23.Miller CT, Lin L, Casper AM, et al. Genomic amplification of MET with boundaries within fragile site FRA7G and upregulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006:409–18. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- 24.Ciullo M, Debily MA, Rozier L, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet. 2002:2887–94. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- 25.Feagins LA, Zhang HY, Hormi-Carver K, et al. Acid has antiproliferative effects in nonneoplastic Barrett’s epithelial cells. Am J Gastroenterol. 2007:10–20. doi: 10.1111/j.1572-0241.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HY, Zhang X, Hormi-Carver K, et al. In non-neoplastic Barrett’s epithelial cells, acid exerts early antiproliferative effects through activation of the Chk2 pathway. Cancer Research. 2007:8580–7. doi: 10.1158/0008-5472.CAN-07-2023. [DOI] [PubMed] [Google Scholar]
- 27.Hong MK, Laskin WB, Herman BE, et al. Expansion of the Ki-67 Proliferative Compartment Correlates with Degree of Dysplasia in Barretts-Esophagus. Cancer. 1995:423–9. doi: 10.1002/1097-0142(19950115)75:2<423::aid-cncr2820750202>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Reid BJ, Sanchez CA, Blount PL, et al. Barretts-Esophagus - Cell-Cycle Abnormalities in Advancing Stages of Neoplastic Progression. Gastroenterology. 1993:119–29. doi: 10.1016/0016-5085(93)90017-7. [DOI] [PubMed] [Google Scholar]
- 29.Reid BJ, Blount PL, Rubin CE, et al. Flow-cytometric and histological progression to malignancy in Barrett’s esophagus: prospective endoscopic surveillance of a cohort. Gastroenterology. 1992:1212–9. [PubMed] [Google Scholar]
- 30.Galipeau PC, Li XH, Blount PL, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. Plos Medicine. 2007:342–54. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulson TG, Galipeau PC, Reid BJ. Loss of heterozygosity analysis using whole genome amplification, cell sorting, and fluorescence-based PCR. Genome Res. 1999:482–91. [PMC free article] [PubMed] [Google Scholar]
- 32.Palanca-Wessels MC, Barrett MT, Galipeau PC, et al. Genetic analysis of long-term Barrett’s esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology. 1998:295–304. doi: 10.1016/s0016-5085(98)70480-9. [DOI] [PubMed] [Google Scholar]
- 33.Palanca-Wessels MC, Klingelhutz A, Reid BJ, et al. Extended lifespan of Barrett’s esophagus epithelium transduced with the human telomerase catalytic subunit: a useful in vitro model. Carcinogenesis. 2003:1183–90. doi: 10.1093/carcin/bgg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Kim Y, Pollack J, et al. A method for calling gains and losses in array CGH data. Biostatistics. 2005:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]
- 36.Diskin SJ, Eck T, Greshock J, et al. STAC: A method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006:1149–58. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boehm D, Herold S, Kuechler A, et al. Rapid detection of subtelomeric deletion/duplication by novel real-time quantitative PCR using SYBR-green dye. Hum Mutat. 2004:368–78. doi: 10.1002/humu.20011. [DOI] [PubMed] [Google Scholar]
- 38.Feagins LA, Zhang HY, Quinones M, et al. Acid exerts an anti-proliferative effect in non-neoplastic Barrett’s epithelial cells by delaying cell cycle progression. Gastroenterology. 2006:A77. [Google Scholar]
- 39.Mrasek K, Schoder C, Teichmann AC, et al. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. International Journal of Oncology. 2010:929–40. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- 40.Lisitsyn NA, Lisitsina NM, Dalbagni G, et al. Comparative genomic analysis of tumors: detection of DNA losses and amplification. Proc Natl Acad Sci U S A. 1995:151–5. doi: 10.1073/pnas.92.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kameoka Y, Tagawa H, Tsuzuki S, et al. Contig array CGH at 3p14.2 points to the FRA3B/FHIT common fragile region as the target gene in diffuse large B-cell lymphoma. Oncogene. 2004:9148–54. doi: 10.1038/sj.onc.1208136. [DOI] [PubMed] [Google Scholar]
- 42.Huebner K, Garrison PN, Barnes LD, et al. The role of the FHIT/FRA3B locus in cancer. Annual Review of Genetics. 1998:7–31. doi: 10.1146/annurev.genet.32.1.7. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins GJ, D’Souza FR, Suzen HS, et al. Deoxycholic acid (DCA) at neutral and acid pH, is genotoxic to oesophageal cells through the induction of ROS: the potential role of antioxidants in Barrett’s oesophagus. Carcinogenesis. 2006 doi: 10.1093/carcin/bgl147. [DOI] [PubMed] [Google Scholar]
- 44.Payne CM, Weber C, Crowley-Skillicorn C, et al. Deoxycholate induces mitochondrial oxidative stress and activates NF-{kappa}B through multiple mechanisms in HCT-116 colon epithelial cells. Carcinogenesis. 2006 doi: 10.1093/carcin/bgl139. [DOI] [PubMed] [Google Scholar]
- 45.Glover TW, Arlt MF, Casper AM, et al. Mechanisms of common fragile site instability. Hum Mol Genet. 2005:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 46.Durkin SG, Ragland RL, Arlt MF, et al. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proceedings of the National Academy of Sciences of the United States of America. 2008:246–51. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan TL, Dong LM, Blount PL, et al. Non-steroidal anti-inflammatory drugs and risk of neoplastic progression in Barrett’s oesophagus: a prospective study. Lancet Oncol. 2005:945–52. doi: 10.1016/S1470-2045(05)70431-9. [DOI] [PubMed] [Google Scholar]
- 48.Lao-Sirieix P, Roy A, Worrall C, et al. Effect of add suppression on molecular predictors for esophageal cancer. Cancer Epidemiology Biomarkers & Prevention. 2006:288–93. doi: 10.1158/1055-9965.EPI-05-0528. [DOI] [PubMed] [Google Scholar]
- 49.Galipeau PC, Prevo LJ, Sanchez CA, et al. Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett’s) tissue. J Natl Cancer Inst. 1999:2087–95. doi: 10.1093/jnci/91.24.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez MV, Artimez ML, Rodrigo L, et al. Mutation analysis of the p53, APC, and p16 genes in the Barrett’s oesophagus, dysplasia, and adenocarcinoma. J Clin Pathol. 1997:212–7. doi: 10.1136/jcp.50.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koppert LB, Wijnhoven BP, van Dekken H, et al. The molecular biology of esophageal adenocarcinoma. J Surg Oncol. 2005:169–90. doi: 10.1002/jso.20359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.