Direct reprogramming of somatic cells into a pluripotent state has attracted enormous interest in biomedical research. So-called induced pluripotent stem (iPS) cells were recently generated by ectopic expression of four transcription factors (TFs; Oct4, Sox2, Klf4 and Myc) in both mouse and human somatic cells.1,2 These iPS cells closely resemble embryonic stem (ES) cells in terms of global gene expression, epigenetic profile, long-term self-renewal capacity and developmental potential. Consequently, the iPS technique promises unprecedented opportunities for studying the fundamental biology underlying epigenetic reprogramming and for the advancement of disease modeling, drug discovery and personalized cell therapy.3
However, the conventional iPS cell technique has several critical drawbacks that hinder many practical applications of the technology. First, permanent genome modification of reprogrammed somatic cells by the exogenous genes themselves (i.e., any of the four reprogramming factors) or even fragments from delivering vectors could be very harmful (potentially oncogenic, for example). To date, progress has been made in overcoming these safety concerns by using non-integrating gene delivery approaches or reprogramming proteins.4 Second, the reprogramming process induced by the four factors generally entails a step-wise progression of non-specific/stochastic events that give rise to pluripotent cells as one of many possible cell fate destinations,5 resulting in low efficiency and slow kinetics. This intrinsically random nature of the reprogramming process may also present ‘hidden’ risks for iPS cells, such as residual epigenetic memory, inappropriately induced epigenetic changes and accumulation and selection of other subtle genetic and epigenetic abnormalities during the reprogramming process.
An alternative approach to address such complications is to use small molecules that can enhance reprogramming efficiency and kinetics and/or functionally replace exogenous reprogramming transcription factors.6,7 For example, small molecules directly modulating epigenetic enzymes or mechanisms, including inhibitors of histone deacetylases, histone methyltransferases, histone demethylases and DNA methyltransferases, were found to promote reprogramming. In addition, chemical screens and follow-up mechanistic studies have identified critical signaling pathways involved in the process of generating iPS cells, including Ca2+-dependent mechanisms, Wnt/β-catenin pathway activation and TGFβ pathway inhibition.
A major advance has been the recent discovery of small molecule conditions that can functionally replace three of the four master reprogramming factors (i.e., Sox2, Klf4 and Myc), thereby enabled the generation of iPS cells using Oct4 alone, and from multiple abundant mouse and human primary cell types.8–11 These studies not only defined significantly improved conditions for generating iPS cells, but also provided new insights into the molecular mechanisms governing the reprogramming process. For instance, the mechanism of action for PS48, a small molecule activator of PDK1 that was recently identified as an enhancer of reprogramming, appears to be the induction of glycolytic gene expression and subsequent facilitation of a metabolic transition from mitochondrial oxidation (mainly used by adult somatic cells) to glycolysis.11 This study demonstrates that modulation at the metabolic level represents a fundamental mechanism in reprogramming.
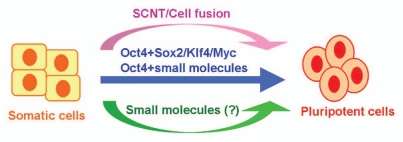
Clearly, a completely small-molecule-based reprogramming condition would be much more practical for iPS cell generation. Importantly, small molecule-based reprogramming would represent a fundamentally different process and method in comparison to somatic cell nuclear transfer (SCNT) and defined transcription-factor-based reprogramming (using genes/ proteins/mRNAs) (Fig. 1), both of which essentially rely on the direct actions of powerful master transcription factors derived from various natural sources (e.g., from oocytes, ectopically expressed in cells or introduced in other forms). In contrast, synthetic small molecules would act to indirectly modulate transcription and epigenetic modification to induce reprogramming, as these compounds do not have specific DNA-recognition domains or transcription regulatory domains. Furthermore, small molecules provide opportunities to fundamentally change the TF-based reprogramming process from non-specific toward a truly directed process.
Figure 1.
Approaches for cellular reprogramming. Somatic cells can be reprogrammed to pluripotent cells through somatic cell nuclear transfer (SCNT), cell fusion or ectopic expression of defined transcription factors (e.g., Oct4, Sox2, Klf4 and Myc). The recent breakthrough in the generation of iPS cells using Oct4 and chemical compounds represents a major step toward a completely small molecule-based reprogramming condition.
Acknowledgements
We thank Dr. Jem Efe and other members of Ding lab for helpful discussions. SD is supported by funding from Fate Therapeutics, California Institute for Regenerative Medicine, NICHD, NHLBI and NIMH/NIH, Prostate Cancer Foundation, Esther B. O'Keeffe Foundation and The Scripps Research Institute.
Comment on: Zhu S, et al. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015.
References
- 1.Takahashi K, et al. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Stadtfeld M, et al. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna JH, et al. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, et al. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng B, et al. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Li W, et al. Trends Pharmacol Sci. 2010;31:36–45. doi: 10.1016/j.tips.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, et al. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. Cell Res. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan X, et al. Stem Cells. 2011 [Google Scholar]
- 11.Zhu S, et al. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]