Abstract
The ability to “knockout” specific genes in mice via embryonic stem (ES) cell-based gene-targeting technology has significantly enriched our understanding of gene function in normal and disease phenotypes. Improvements on this original strategy have been developed to enable the manipulation of genomes in a more sophisticated fashion with unprecedented precision. The rat is the model of choice in many areas of scientific investigation despite the lack of rat genetic toolboxes. Most recent advances of zinc finger nucleases (ZFNs) and rat ES cells are diminishing the gap between rat and mouse with respect to reverse genetic approaches. Importantly, the establishment of rat ES cell-based gene targeting technology, in combination with the unique advantages of using rats, provides new, exciting opportunities to create animal models that mimic human diseases more faithfully. We hereby report our recent results concerning finer genetic modifications in the rat, and propose their potential applications in addressing biological questions.
Key words: genetic manipulation, gene targeting, conditional knockout, transgenic animal, rat model, p53 knockout rat, embryonic stem cells
Introduction
The laboratory rat (Rattus norvegicus) was the first animal species brought into scientific studies and is also one of the most widely used animal models in biomedical research, as reflected by the number of scientific publications on it.1,2 Since the 19th century, scientists have used rats in many experimental studies that contributed greatly to our basic knowledge of physiology, pathology and pharmacology.2,3 Given its physiological similarities to humans, the rat is currently the primary animal model in many preclinical tests, especially those related to cardiovascular disease,4 diabetes,5 breast cancer,6 chronic inflammatory diseases7 and age-related diseases.8 Furthermore, most behavioral studies have been performed on rats because, compared with mice, their behavior is more social, intelligent, complex and skilled.9 Scientists have managed to teach rats complex behavioral paradigms and tasks. As a result, rats are now also entering the field of cognitive neuroscience, where the use of monkeys is predominant.10,11
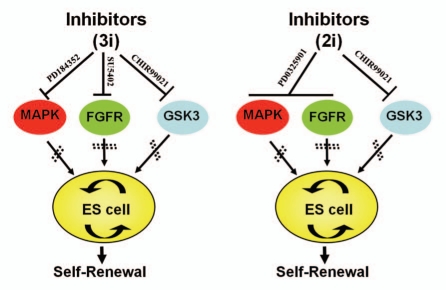
Over the past two decades, however, the development of rat models has lagged far behind that of mouse models, mainly due to significant advances in mouse genetic toolboxes. In the 1980s, embryonic stem (ES) cells were successfully derived from mice, and later technologies were developed enabling the performance of gene-targeting in ES cells.12–14 Ever since then, this ES cell-based gene-targeting strategy has been extensively used to create loss-of-function mutations and gene replacement on predetermined gene loci in mice. Thousands of mouse gene knockout models have been generated and they have become powerful tools for investigating gene function and relevant phenotypes. This technology was previously limited to mice only because of the absence of germline-competent rat ES cell lines. In 2008, we developed a chemically-defined basal culture system that contains serum-free N2B27 medium and small molecule inhibitors (3i: CHIR99021, PD184352 and SU5402 or 2i: CHIR99021 and PD0325901), resulting in successful derivation and maintenance of germline-competent rat ES cells (Fig. 1).15–17 These rat ES cells can be genetically modified and robustly propagated in culture, while retaining the ability to contribute to germline-competent chimeras, as recently demonstrated by the generation of p53 gene knockout rats by gene-targeting via homologous recombination.18 The development of ES cell-based gene-targeting technologies in the rat has opened the door for a bright future of rat genetic manipulations.
Figure 1.
The ground state of rat ES cell self-renewal under N2B27 medium supplemented with small inhibitors. Left: PD184352, SU5402, CHIR99021 encompass the 3i condition in which PD184352 inhibits mitogen-activated protein kinase (MAPK), whereas SU5402 and CHIR99021 specifically inhibits fibroblast growth factor receptor (FGFR) tyrosine kinase and glycogen synthase kinase 3 (GSK3), respectively. Right: 2i condition in which PD184352 and SU5402 can be replaced by a more potent inhibitor, PD0325901.
Although gene knockout models are extremely important for investigating gene dysfunction and recapitulating human inherited disease, creation and utilization of finer genetic modifications, including subtle mutations, knock-in, inducible and conditional knockout models-is increasingly preferred in current scientific research. In this article, we report the latest results of our efforts towards finer genetic modifications in the rat, and explore the unique advantages, as well as the potential applications, of these rat models in biomedical research.
Exploration on p53 Knockout Rats
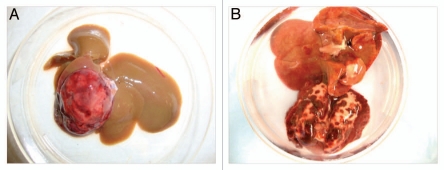
p53 is one of the most famous tumor suppressor genes identified, and its mutations have been implicated in as many as half of all human cancers.19 p53 knockout mice are susceptible to spontaneous tumors without induction, and therefore widely used in basic cancer research for screening carcinogenic compounds, as well as chemotherapeutics.20 Our tracking record on p53 knockout rats has also confirmed that they are highly predisposed to malignant tumors. In our preliminary study, all 14 p53 homozygote rats died or had to be sacrificed within 120 days. Lymphoma is the most frequent tumor type in p53 knockout mice (71%, 44/56).21 Surprisingly, we did not observe any cases of lymphoma in either heterozygous or homozygous rats. The predominant tumor type for p53 knockout rats is hepatic hemangiosarcoma (71%, 10/14), and fewer cases of lung hemangiosarcoma (Fig. 2). In six months, 10 of the 39 p53 heterozygote rats (25.6%) developed tumors.
Figure 2.
Representative tumors developed in p53 knockout rats. (A) Hepatic hemangiosarcoma. (B) Lung hemangiosarcoma.
Although these knockout mouse and rat models can exhibit some characteristics of human p53 mutation, they do not mimic it completely. For example, they develop a different range of tumors than humans do. Tumor spectrum analysis of p53 mutant mice suggested that sarcoma is the most common tumor type in heterozygotes, whereas lymphoma is most common in homozygotes.21 In comparison, humans tend to develop epithelial cell-derived cancers such as breast and ovarian cancers.22,23 This phenotypic difference limits the utility of conventional knockout rodents as models of human cancer. Furthermore, germline knockout of some tumor suppressor genes, such as BRCA1 and BRCA2, is embryonically lethal. In the case of p53, which is dispensable in embryonic development, knockout rodents are highly tumor-prone and usually die within a few months after birth, preventing a faithful recapitulation of sporadic tumor onset and progression (unpublished data).21 Most human cancers originate from an accumulated series of somatic cell mutations, as well as changes in the microenvironment and signaling pathways. This progressive transformation process deviates from cells of knockout animals with initial mutation through germline transmission, which alters the microenvironment permanently and may activate compensatory mechanisms to offset the “knockout” effect during embryogenesis.24 This also accounts for the undesired background of sarcoma (and lymphoma) in p53 knockout mice (rats) which complicates the investigation of cancers in other tissues because many mouse models are built on p53 deficient background. Therefore, the conditional knockout strategy, which induces somatic mutations in a spatially and temporally-specific fashion, has superseded the conventional knockout strategy and become an optimal choice for sophisticated tumor models. For example, a mouse breast cancer model was a conditional knockout of BRCA1 or BRCA2 in mammary gland epithelium in p53 knockout mice.25,26 Since rat mammary tumors resemble many characteristics of human mammary tumors, a rat breast cancer model based on conditional knockout strategy would be an invaluable tool to identify drug targets in combination with the vast amount of research data on pharmacology and toxicology in the species.6
Conditional Knockout
Despite their great advantages, conventional gene-targeting technologies exposed their drawbacks in the extreme complexity of the mammalian genetic system. For example, embryonic lethality is one of the most common impediments to analyzing gene function in later development. And functional redundancy or developmental compensatory mechanisms may also hinder the analysis. On the other hand, in knockout animals where the genetic information was altered irreversibly through germline, the study of genes that function uniquely in a specific tissue or organ may be complicated by the effects of gene disruptions in all the cells of the animal. To circumvent these issues, conditional knockout animals have been developedbycombiningEScell-basedgenetargeting with site-specific recombinases. This technology has revolutionized the area of reverse genetics by allowing a high flexibility of genetic modification that can be switched “on” or “off” in a certain type of cells and/or at a particular developmental period.
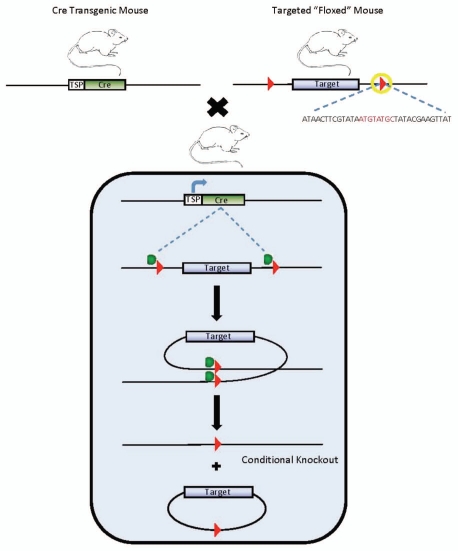
Cre-loxP site-specific recombination is the most frequently used system to generate conditional knockout mice. Alternatively, Flp-FRT works in a similar manner as described elsewhere in references 27 and 28. Cre recombinase from bacteriophage P1 directs a conservative recombination between two loxP recognition sites, a 34 bp consensus sequence consisting of an 8 bp nonpalindromic core flanked by two 13 bp inverted repeats.29 Therefore, various types of sophisticated genetic modifications, such as gene deletion, insertion, chromosome engineering and subtle point mutations, can all be achieved by simply manipulating the orientation and location of the loxP sites. Conditional knockout mice are generated by intercrossing two transgenic mouse lines, one bearing Cre recombinase transgene under the control of a promoter directing tissue and/or time-specific expression, and the other with homozygosity of “floxed” (flanked by loxP) alleles derived from gene-targeted ES cells (Fig. 3).30–32 The double transgenic progeny will have the conditional allele modified only in cells/tissues or developmental stages that express Cre, precluding embryonic lethality and allowing additional accuracy to address biomedical questions. One of the advantages of this system is that only recombinase and loxP-harboring sequences are required for DNA rearrangement. Additionally, it is believed that Cre recombinase has minimal unwanted effects as the mouse (and rat) genome does not contain endogenous loxP sites, providing an ideal background for site-specific recombination.
Figure 3.
Conditional knockout strategy via Cre-mediated DNA recombination. Cre-transgenic mouse is mated with mouse carrying floxed alleles. In the offspring mice, Cre transgene will be activated in targeted tissue. Cre catalyzes the recombination between two in cis loxP sites (red arrowheads), and excises the target gene from chromosome. loxP sequence(indicated by yellow circle) consists of an 8 bp nonpalindromic core flanked by two 13 bp inverted repeats. TSP: tissue-specific promoter.
Unfortunately, the application of Cre-loxP technology was previously restricted to mice. Various reports have demonstrated the feasibility of the Cre-loxP system in the Cre-activated reporter line by crossing to Cre-expressing transgenic rats33 or via somatic delivery of Cre-expressing viruses.34 Cre transgenic rats can be generated using methods such as pronuclear injection. However, the production of “floxed” alleles requires gene-targeting in germline-competent rat ES cells, which had been unavailable for a long time. We have recently derived authentic rat ES cells and generated the first knockout rats via homologous recombination in ES cells.16–18 Our work has finally overcome the last and most critical obstacle to extending this well-developed Cre-loxP technology to rats, and to accelerating the development of rat disease models as well as the pursuit of rat functional genomics.
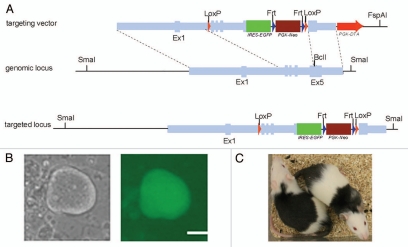
We chose the POU family transcriptional factor Oct4, a well-studied pluripotent gene expressed in early embryos, germline cells and undifferentiated ES cells as the target gene for our experiments. In vivo expression of Oct4 is essential for the initial development of pluripotential capacity in the inner cell mass (ICM) and formation of pluripotent stem cells.35 Oct4 is also one of the four transcription factors used to create induced pluripotent stem (iPS) cells, together with Sox2, Klf4 and c-Myc, indicating its capacity to reprogram differentiated cells into an ES cell-like state.36 Nevertheless, this makes the investigation of potential roles of Oct4 in somatic stem cells impossible because of embryonic lethality. Here we present some data on our efforts to generate tissue-specific Oct4 knockout rats. To construct the targeting vector, a 9 kb fragment containing the rat Oct4 gene was amplified from Dark Agouti (DA) rat genomic DNA and subcloned into a pL611 cloning vector (Fig. 4A). The ires-eGFP-Frt-PGK-neo-Frt-LoxP cassette was introduced into the immediate downstream of the stop codon of Oct4 gene. In this way, the selection cassette can be excised to create a “clean” modification. Another loxP site was also introduced ahead of exon 2, which works together with the remaining loxP site to induce the removal of exon 2–5 of the Oct4 gene under conditional Cre expression in vivo. Additionally, our strategy allows for the eGFP gene to be placed under Oct4 endogenous promoter control, serving as a reporter for targeting events in ES cells (Fig. 4B) and also knockout animals. eGFP removal also accompanies Cre-mediated Oct4 “knockout,” which has been confirmed by in vitro Cre transfection into targeted ES cells (data not shown). The targeting vector was linearized, transfected into DA rat ES cells via electroporation and selected with G418. Resistant ES cell colonies were picked and expanded individually, followed by PCR screening and sequencing analysis. The quality of correctly targeted ES cells was optimized by subcloning. Karyotypically normal ES cells were microinjected into 25 blastocysts collected from E4.5 Fischer 344 rats and subsequently transferred to pseudo-pregnant Sprague-Dawley rats. Four chimeras were produced and their germline transmission capacity is yet to be determined (Fig. 4C).
Figure 4.
Oct4 conditional knockout in the rat. (A) Schematic diagram showing the strategy of Oct4 conditional knockout. The five exons of rat Oct4 gene with flanking genomic sequence are shown. The targeting vector contains of 7.8 kb Oct4 gene. The ires-eGFP-Frt-PGK-neo-Frt-loxP cassette is inserted the downstream of the Oct4 gene locus at BclI restriction site. Another loxP site is also introduced between exon 1 and 2. (B) Oct4-GFP-positive DA rat ES colonies after electroporation and drug selection. Bar, 50 µm. (C) Chimeras produced by blastocyst injection of Oct4-GFP targeted rat ES cells.
Knock-In Reporter System in the Rat
We aim to generate two Rosa26 knock-in rat strains that may facilitate the efforts within the scientific community to establish conditional knockout rats and create disease models. The Rosa26 locus was originally cloned and characterized in gene-trap studies in mouse ES cells.37 It was believed to encode nuclear RNA of unknown function. Gene-targeted mice carrying homozygous Rosa26 locus disruption are viable and phenotypically normal.38 The Rosa26 locus is amenable to genetic modification as it can be targeted with high efficiency and is expressed universally in mice. Various kinds of Rosa26 knock-in mouse strains have been produced and become useful tools for biomedical research.39–41 Therefore, such strains in the rat will be of great help for the research community.
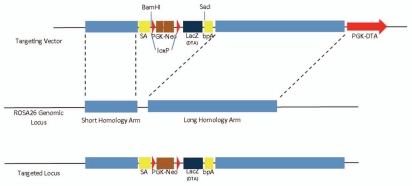
Targeting of the rat Rosa26 locus can be achieved using the pBigT plasmid, which consists of a splice acceptor (SA) sequence, a loxP-flanked PGK-neo selectable marker and a polyadenylation sequence for transcription stop (bpA) (Fig. 5). We constructed two vectors in which lacZ and diphtheria toxin fragment A (DT-A) genes were inserted after loxP flanked PGK-neo sequence, respectively. The Rosa26-lacZ system serves as a rat Cre expression reporter strain when it is crossed with a tissue-specific Cre-expressing transgenic rat strain.39 Currently very few Cre transgenic rats are available in the research community, and researchers may have to establish such strains on their own. Consequently, it is necessary to verify Cre activity and specificity, and ensure that Cre was not prematurely activated during embryonic development before generating conditional knockout rats. On the other hand, the Rosa26-DTA knock-in system will help generate rat disease models via Cre-mediated cell depletion strategy, as previously described in reference 42. Functional cell loss is frequently observed in human diseases, such as midbrain dopaminergic cell loss in Parkinson's disease43 and beta cell loss in diabetes.44 This inducible DTA allele can be used to induce cell type-specific depletion and therefore contribute to the modeling of human diseases and to the study of regeneration.
Figure 5.
Schematic diagram showing the design of Rosa26 knock-in targeting vector. SA-PGK-neo-LacZ(DTA)-bpA is inserted into rosa26 genomic locus. New restriction sites of BamHI and SacI are introduced after targeting event and can be used for Southern blot screening.
Zinc Finger Nuclease: An Alternative Approach?
While the ES cell-based gene-targeting technique has shed light on the promise of sophisticated genetic alterations in rats, an alternative strategy was already developed for generating knockout rats in a targeted manner.45 Zinc Finger Nucleases (ZFN) are engineered proteins that link an assembly of zinc-finger type protein modules, which are pre-designed to sequence-specific DNA binding ability, to a restriction endonuclease domain of FokI. In practice, usually three to six ZFN modules are joined together, with each of them capable of binding to triplets of DNA. Because the FokI endonuclease must dimerize to cleave double strand DNA, two ZFN coding sequences are required; this brings the specificity up to 36 bases, which is believed to be sufficient enough to achieve single site specificity over the whole genome. After the specificity of ZFNs in cell culture system has been confirmed, ZFN coding mRNA is injected into a one-cell stage embryo. The protein will be expressed and bind to the locus it is designed to target, causing a DNA double strand break (DSB) via the cleavage activity of FokI. In eukaryotic cells, DSBs will be repaired by either error-free homologous recombination when provided with a homologous template, or error-prone non-homologous end joining (NHEJ) that often produces small deletions or insertions.46,47 Therefore, ZFN modules can be designed to target coding exons of the gene of interest, and take advantage of NHEJ to disrupt the gene usually by introducing out-of-frame mutations.
Since the first report of knockout rats via ZFN technology in 2009, dozens of rat genes have been “knocked out” in this way (www.sageresearchmodels.com). However, the mutations induced by error-prone NHEJ are unpredictable and limited to causing “gene disruption.” On the other hand, modifications have been done to allow subtle mutations and gene knock-in to be performed by invoking homologous recombination. In this case, a simultaneous delivery of donor DNA template is required. After DSB induction, the innate homology directed repair (HDR) pathway will be artificially activated under homology-containing donor DNA provision, giving rise to the precise integration (or replacement for subtle gene modification) of exogenous sequence at the ZFN cleavage site. Several studies have been performed to induce gene targeting via ZFN-mediated homologous recombination in cultured cells,48 ES cells or iPS cells,49,50 Drosophila,51 plants,52 zebrafish53 and mouse zygotes.54 Importantly, a recent report tested targeted integration in rat embryos with ZFN technology.55 The authors were able to knock-in an eight base pair NotI restriction site (6.7–12.5% targeting efficiency), or a 1.5 kb human phosphoglycerate kinase (PGK) promoter-driven GFP cassette (2.4–8.3% targeting efficiency) into the target site.
As compared with the ES cell-based targeting strategy, ZFN technology in rats has unique advantages, including higher targeting efficiency and a shortened time course to obtain homozygotes by bypassing the chimera stage. However, some concerns remain to be addressed regarding sophisticated genetic manipulations using ZFN technology. First, ZFN-mediated gene-targeting is performed in rat embryos, necessitating laborious screening for positive mutations in adult animals. Second, a selection mechanism is not included in such a strategy; thus, rare targeting events cannot be enriched. Third, it has been shown to be difficult to knock-in large DNA fragments (>1 kb) into target sites. Finally, ZFN has not been successful for the generation of conditional knockout rats. The insertion of two loxP sites may have to be handled sequentially and further optimization of this targeting strategy is expected.
Biomedical Relevance of Rat Models
Although the lack of efficient tools to manipulate the rat genome has made the mouse the leading rodent for genetic research, recent advances in transgenic technology have opened new paths for rats to return back to center stage. Starting with the Rat Genome Project launched in 1995 by the National Institutes of Health (NIH), rat genomic resources have been expanding unprecedentedly, including the landmark sequence data of the Brown Norway (BN) rat genome,56 with more and more other strains being added.57 The rapid development of reference databases has greatly facilitated establishment of rat models. Genetically modified rats are precious platforms for the study of human physiology and disease. For example, as compared with transgenic mice, transgenic rat models of Huntington disease not only present a more typical adult patient pathology, but are also more suitable for in vivo metabolic and structural imaging.58 Also, a triple transgenic rat model of Alzheimer disease can successfully mimic the human disease phenotype of amyloid deposition.59 With the availability of genome sequences of commonly used strains, genetic modifications in rats will rise up to a new level with exquisite specificity and fidelity.
Loss-of-function models are critical for investigating gene function and regulation in vivo. Over the years, many strategies for manipulating rat genes were adapted from the mouse genetic toolbox, including conventional transgenesis by pronuclear injection,60 RNA interference,61 N-ethyl-N-nitrosourea (ENU) mutagenesis62,63 and transposon mutagenesis.64–66 Most importantly, ZFN technology and ES cell-based gene-targeting enable the rat to enter a true “knockout” era. As research advances, an increasing number of scientists are hoping to study gene function in a conditional manner, either in a defined temporal window, or in a specific type of cell or tissue. For example, since the rat is the species of choice in pre-clinical studies of diabetes, a pancreas-specific conditional gene knockout rat model would be important for drug discovery and pharmacological experiments. For neuro-scientists, the rat is now playing a new role in optogenetics—an emerging field that is revolutionizing this area of research by combining genetic approaches with optics to (in)activate a particular class of neurons using laser light with a degree of specificity that conventional electrophysiology does not allow.67 In 2009, the first rat optogenetic study reported manipulation of different circuit elements in Parkinson disease rat models for the purpose of identifying relevant target cell types for Deep Brain Stimulation (DBS).68
A lack of adequate animal models has been regarded by the pharmaceutical industry as one of the major hurdles to drug discovery, especially drugs for the central nervous system (CNS). More sophisticated genetic modification will allow scientists to establish reliable rat models of psychiatric and neurological disorders and generate “humanized” rats after cell type-specific depletion. Progress in such fields will greatly promote elucidation of patho-physiologic mechanisms and drug discovery.
Acknowledgements
This work was funded by a NIH/NCRR grant (R01RR025881). We thank J.Z. Stoller for Rosa26 knock-in vector construction; USC Stem Cell Core for technical assistance; N. Wu and Y. Yan for blastocyst injection; G. Chester for ordering rats; and R. Montano and colleagues for rat husbandry. Guanyi Huang is a research fellow funded by the California Institute for Regenerative Medicine (CIRM) Stem Cell Training Program (#TG2-01161).
References
- 1.Dwinell MR. Online tools for understanding rat physiology. Briefings in bioinformatics. 2010;11:431–439. doi: 10.1093/bib/bbp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krinke G. The laboratory rat. San Diego, CA: Academic Press; 2000. [Google Scholar]
- 3.Baker HJ, Lindsey JR, Weisbroth SH. The Laboratory rat. New York: Academic Press; 1979. [Google Scholar]
- 4.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- 5.Lee JH, Yang SH, Oh JM, Lee MG. Pharmacokinetics of drugs in rats with diabetes mellitus induced by alloxan or streptozocin: comparison with those in patients with type I diabetes mellitus. J Pharm Pharmacol. 2010;62:1–23. doi: 10.1211/jpp.62.01.0001. [DOI] [PubMed] [Google Scholar]
- 6.Smits BM, Cotroneo MS, Haag JD, Gould MN. Genetically engineered rat models for breast cancer. Breast Dis. 2007;28:53–61. doi: 10.3233/bd-2007-28106. [DOI] [PubMed] [Google Scholar]
- 7.Holmdahl R, Lorentzen JC, Lu S, Olofsson P, Wester L, Holmberg J, et al. Arthritis induced in rats with nonimmunogenic adjuvants as models for rheumatoid arthritis. Immunol Rev. 2001;184:184–202. doi: 10.1034/j.1600-065x.2001.1840117.x. [DOI] [PubMed] [Google Scholar]
- 8.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 9.Jacob HJ. Functional genomics and rat models. Genome Res. 1999;9:1013–1016. doi: 10.1101/gr.9.11.1013. [DOI] [PubMed] [Google Scholar]
- 10.Uchida N, Mainen ZF. Speed and accuracy of olfactory discrimination in the rat. Nat Neurosci. 2003;6:1224–1229. doi: 10.1038/nn1142. [DOI] [PubMed] [Google Scholar]
- 11.Kepecs A, Uchida N, Zariwala HA, Mainen ZF. Neural correlates, computation and behavioural impact of decision confidence. Nature. 2008;455:227–231. doi: 10.1038/nature07200. [DOI] [PubMed] [Google Scholar]
- 12.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 14.Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 20.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 21.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 22.Osborne RJ, Merlo GR, Mitsudomi T, Venesio T, Liscia DS, Cappa AP, et al. Mutations in the p53 gene in primary human breast cancers. Cancer Res. 1991;51:6194–6198. [PubMed] [Google Scholar]
- 23.Naito M, Satake M, Sakai E, Hirano Y, Tsuchida N, Kanzaki H, et al. Detection of p53 gene mutations in human ovarian and endometrial cancers by polymerase chain reaction-single strand conformation polymorphism analysis. Jpn J Cancer Res. 1992;83:1030–1036. doi: 10.1111/j.1349-7006.1992.tb02717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 26.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 27.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 28.O'Gorman S, Fox DT, Wahl GM. Recombinasemediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 29.Sauer B, Henderson N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989;17:147–161. doi: 10.1093/nar/17.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brakebusch C, Grose R, Quondamatteo F, Ramirez A, Jorcano JL, Pirro A, et al. Skin and hair follicle integrity is crucially dependent on beta1 integrin expression on keratinocytes. EMBO J. 2000;19:3990–4003. doi: 10.1093/emboj/19.15.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 32.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 33.Ueda S, Fukamachi K, Matsuoka Y, Takasuka N, Takeshita F, Naito A, et al. Ductal origin of pancreatic adenocarcinomas induced by conditional activation of a human Ha-ras oncogene in rat pancreas. Carcinogenesis. 2006;27:2497–2510. doi: 10.1093/carcin/bgl090. [DOI] [PubMed] [Google Scholar]
- 34.Sato Y, Endo H, Ajiki T, Hakamata Y, Okada T, Murakami T, et al. Establishment of Cre/LoxP recombination system in transgenic rats. Biochem Biophys Res Commun. 2004;319:1197–1202. doi: 10.1016/j.bbrc.2004.04.204. [DOI] [PubMed] [Google Scholar]
- 35.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 38.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 40.Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci USA. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 42.Brockschnieder D, Lappe-Siefke C, Goebbels S, Boesl MR, Nave KA, Riethmacher D. Cell depletion due to diphtheria toxin fragment A after Cre-mediated recombination. Mol Cell Biol. 2004;24:7636–7642. doi: 10.1128/MCB.24.17.7636-7642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozano AM, Lang AE, Hutchison WD, Dostrovsky JO. New developments in understanding the etiology of Parkinson's disease and in its treatment. Curr Opin Neurobiol. 1998;8:783–790. doi: 10.1016/s0959-4388(98)80122-0. [DOI] [PubMed] [Google Scholar]
- 44.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, et al. Knockout rats via embryo micro-injection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 48.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 49.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- 53.Amacher SL. Emerging gene knockout technology in zebrafish: zinc-finger nucleases. Brief Funct Genomic Proteomic. 2008;7:460–464. doi: 10.1093/bfgp/eln043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 56.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 57.Abbott A. Return of the rat. Nature. 2009;460:788. doi: 10.1038/460788a. [DOI] [PubMed] [Google Scholar]
- 58.von Horsten S, Schmitt I, Nguyen HP, Holzmann C, Schmidt T, Walther T, et al. Transgenic rat model of Huntington's disease. Hum Mol Genet. 2003;12:617–624. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- 59.Leon WC, Canneva F, Partridge V, Allard S, Ferretti MT, DeWilde A, et al. A novel transgenic rat model with a full Alzheimer's-like amyloid pathology displays pre-plaque intracellular amyloid-beta-associated cognitive impairment. J Alzheimers Dis. 2010;20:113–126. doi: 10.3233/JAD-2010-1349. [DOI] [PubMed] [Google Scholar]
- 60.Cozzi J, Anegon I, Braun V, Gross AC, Merrouche C, Cherifi Y. Pronuclear DNA injection for the production of transgenic rats. Methods Mol Biol. 2009;561:73–88. doi: 10.1007/978-1-60327-019-9_5. [DOI] [PubMed] [Google Scholar]
- 61.Dann CT, Alvarado AL, Hammer RE, Garbers DL. Heritable and stable gene knockdown in rats. Proc Natl Acad Sci USA. 2006;103:11246–11251. doi: 10.1073/pnas.0604657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Boxtel R, Gould MN, Cuppen E, Smits BM. ENU mutagenesis to generate genetically modified rat models. Methods Mol Biol. 2010;597:151–167. doi: 10.1007/978-1-60327-389-3_11. [DOI] [PubMed] [Google Scholar]
- 63.Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, Wang YR, et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21:645–651. doi: 10.1038/nbt830. [DOI] [PubMed] [Google Scholar]
- 64.Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, et al. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 65.Kitada K, Keng VW, Takeda J, Horie K. Generating mutant rats using the Sleeping Beauty transposon system. Methods. 2009;49:236–242. doi: 10.1016/j.ymeth.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 66.Izsvak Z, Frohlich J, Grabundzija I, Shirley JR, Powell HM, Chapman KM, et al. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods. 2010;7:443–445. doi: 10.1038/nmeth.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 68.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]