Abstract
Genome instability continuously presents perils of cancer, genetic disease and death of a cell or an organism. At the same time, it provides for genome plasticity that is essential for development and evolution. We address here the genome instability confined to a small fraction of DNA adjacent to free DNA ends at uncapped telomeres and double-strand breaks. We found that budding yeast cells can tolerate nearly 20 kilobase regions of subtelomeric single-strand DNA that contain multiple UV-damaged nucleotides. During restoration to the double-strand state, multiple mutations are generated by error-prone translesion synthesis. Genome-wide sequencing demonstrated that multiple regions of damage-induced localized hypermutability can be tolerated, which leads to the simultaneous appearance of multiple mutation clusters in the genomes of UV-irradiated cells. High multiplicity and density of mutations suggest that this novel form of genome instability may play significant roles in generating new alleles for evolutionary selection as well as in the incidence of cancer and genetic disease.
Key words: hypermutability, evolution, DNA damage and repair, single-strand DNA, double-strand breaks
Multiple Mutations and Adaptive Changes
Even a single mutation in DNA can alter biological functions to the detriment or benefit of a cell or organism. Thus an important balance must be maintained between limiting mutation frequency, thereby reducing the risk of harmful changes and allowing a level of mutagenesis that can provide sufficient opportunity for rare adaptive changes that fuel evolution. In general, the rates of spontaneous mutations on a genome scale are limited by the systems of replication fidelity and repair.1,2 Studies using mutation reporters, mutation accumulation on the evolutionary and population scales as well as intergeneration sequence comparisons indicate that the mutation rate per genome duplication in various species is low. At most there is only a single new mutation generated per several tens to hundreds of cell divisions.3–7 This low mutation rate makes accumulation of multiple mutations over just a few generations rather unlikely; however, multiple mutations are implicated in several diseases and in evolution. Even less likely would be the incidence of simultaneous changes in a single gene. Importantly, multiple mutations in a gene are expected to have the strongest biological effects via reduction in gene function, increase in gene function or even creation of a novel function.
Stronger potential for biological effect of multiple mutations is evident for the case of gene inactivation, since the majority of base pair substitutions (bps) and even some small insertions and deletions (indels) would leave the gene functioning at a biologically sufficient level. This is because a second mutation in the gene increases not only the chance that one of the mutations is in itself deleterious, but also creates the potential for the two mutations to work in concert to reduce the gene's function. Multiple mutations also are more likely to generate changes that increase fitness. Studies, aimed to generate enzymes with enhanced activity or even with a new type of activity, have established that these effects are achieved mostly by multiple mutations.8,9 Importantly, multiple mutations can show sign epistasis—a condition where individual changes within a beneficial multiply mutated allele are neutral or even deleterious when analyzed separately.10,11 On an evolutionary scale, these observations translate into a requirement for multiple mutations to avoid fitness valleys, where steps with reduced fitness in the succession of mutation events that would eventually result in alleles with high fitness and to follow fitness ridges, where the successive mutations occur through steps that do not lead to fitness reduction.12,13 As established by comparisons across a wide range of taxons, sign epistatsis as well as fitness ridges and valleys remain important features of current protein evolution.14 However, fitness valleys are not an impediment to adaptive evolution if advantageous alleles with multiple mutations can occur by simultaneous or closely timed mutation events. Multiple mutations that appear to be simultaneous or coordinated in time (chronocoordinate) have been detected in normal mouse and human tissues15,16 and in tumors.17–19 While the fraction of mutations that appear to be chronocoordinate is small, they may play bigger roles in some types of cancers, especially those associated with a high density of DNA damage (see below).
Localized Hypermutability (LHM)
Since beneficial mutations represent a tiny subset of all possible changes, very few multiple mutations are expected to produce high fitness. In order to obtain a set of simultaneous mutations in specific nucleotides within a single ORF that would enhance gene function, individual changes would need to occur with a very high probability. For example a rate at 10−9 per nt per cell generation calculated for budding yeast6,7 isolation of one cell with simultaneous mutation of just three specific bases becomes practically impossible (10−27), because it would require unrealistic amounts of biological material (for example 1027 yeast cells would weigh 100 trillion kilograms). Several orders of magnitude greater mutation rates are required to make simultaneous multiple mutations plausible. For example, the rate of 10−4 per nt would allow simultaneous mutation of three specified nucleotides to be found among 1012 cells, which corresponds to just 100 grams of yeast (or around 1 kg of human cells). However, such high mutation probabilities are impossible on a genome-wide scale even for a single cell generation. A minimal estimate for 40,000 one-kilobase ORFs (in a diploid human genome) yields around 4,000 mutations, which would create an intolerable mutation load by coincidence of allelic recessive lethals and/or inactivation of haplo-insufficient genes.
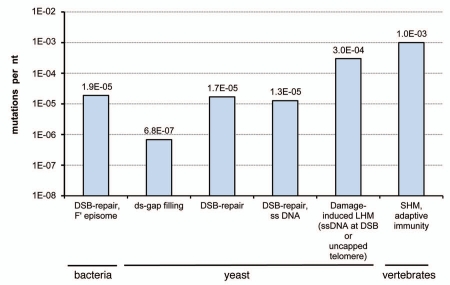
Genome-wide mutation overload can be avoided if high mutation densities are generated only in small regions of a genome—a phenomenon we define as localized hypermutability (LHM). Among the possible sources of region-specific LHM are at-risk motifs capable of forming DNA structures that are poor substrates for DNA repair and mutation avoidance systems20,21 as well as specific chromatin organization.22 Over the past years it has became clear that DNA double-strand breaks (DSBs) and DSB-repair can be important sources of LHM that is not restricted to a certain region in a genome but can occur whereever breaks happened to locate. Studies of adaptive mutagenesis in E. coli by Rosenberg et al.23,24 first indicated that DSB-repair could be mutagenic. Soon after that Strathern and colleagues established in the model yeast system using a defined site-specific DSB repaired by homologous recombination that the repair in fact is associated with up to several hundred fold increase in mutation frequency in the area around a DSB.25,26 Later, the hypermutability of DNA adjacent to a DSB was confirmed in E. coli, establishing the generality of the phenomenon across microbial taxons.27,28 Recently, we confirmed one of the sources of LHM suggested by the Strathern group, which is hypermutability of long single-strand DNA (ssDNA) formed by strand-biased 5′3→′ DNA degradation (resection).29 A second proposed source, error-prone DNA synthesis creating two new strands in the course of repairing a double-strand gap, was demonstrated recently by Haber and colleagues.30 Recently, another form of DSB-repair—break-induced replication, was also demonstrated to be mutagenic.31 In all of these yeast and E. coli systems, rates of mutation per nt in the absence of exogenous DNA damage (“spontaneous”) was close to 10-5 per nt, comparable to the value initially obtained by the Strathern group32 (see also Fig. 1). Not surprisingly this level of hypermutability produced only single mutations in reporter ORFs.
Figure 1.
Mutation frequencies associated with various kinds of localized hypermutability. Mutation frequencies were derived from the following publications: DSB-repair, F′ episome-E. coli mutations selected in F′ codA,B genes with a mutation target size of 848 bp was used for calculations;84 Double-stranded (ds) gap repair—mutations in the yeast URA3 gene occurring during a double-stranded (ds) gap repiair,30 a minimal estimate of mutation target of 125 bp as determined by;85 DSB-repair—frequency of spontaneous mutations in the vicinity of a DsB repaired by homologous recombination.32 A site-specific DsB was induced next to the chromosomal CAN1 gene and repaired by homologous recombination with a truncated copy of CAN1 in the same chromosome. Unlike a double-strand gap in reference 30, DSB ends homologous to adjacent segments of DNA sequence were used for repair. this system did not allow distinction between hypermutability in transiently formed ssDNA (as in ref. 29), or hypermutability during repair of a ds-gap (as in ref. 30). DSB-repair in ssDNA—in this system the CAN1 reporter gene was placed in the vicinity of a DSB that was repaired by a short oligonucleotide, so the CAN1 sequence did not participate in recombination and the most likely hypermutable intermediate was transient ss DNA (ref. 29 and Fig. 2A). The probability of CAN1 mutations in both DSB-repair systems was calculated based on the frequency data corrected for the minimal estimate of mutation target (236 bp as determined by ref. 85). Damage-induced LHM (ssDNA at DSB or uncapped telomere). Average frequencies of mutations induced by Uv-C (45 J/m2) and MMS (30 min in 11.8 mM (0.1%) MMS) in yeast ssDNA around DsB or next to uncapped telomere were taken from references 29 and 45. SHM, adaptive immunity. Approximate frequency of mutations associated with somatic hypermutation in the Ig-genes was taken from references 34 and 35.
Damage-Induced LHM
The rates of spontaneous LHM associated with DSB repair were close to the estimated value of in vivo error rates in yeast cells carrying a double defect in DNA polymerase proofreading and post-replicative mismatch repair (MMR) (1.5 × 10−6 per nt in the URA3 gene).33 Importantly, the combination of proofreading and MMR defects did not produce multiple URA3 mutations even though cells were grown for several generations. There are no indications that significantly higher in vivo error rates could be achieved during synthesis on long undamaged templates that would be capable of producing simultaneous multiple mutations within a single ORF. However, very high mutation density can be achieved if LHM is associated with DNA damage. A dramatic example of programmed, damage-induced increase of mutation frequency by about a million-fold as compared to genome-wide rate is well established for a small region of the Ig-locus in genomes of immune B cells (reviewed in refs. 34 and 35 and Fig. 1). This somatic hypermutation (SHM) is confined to a small region within the Ig locus. SHM is driven by activation induced deaminase (AID), a specialized enzyme which converts some cytosines in the SHM region into uracils. Since this region is involved in determining the affinity to an antigen, SHM results in a very fast accumulation of multiple mutant alleles providing sufficient material for selecting cells producing antibodies with several orders of magnitude greater affinity to the antigen. Because of specially organized cell division control, cells expressing high affinity antibodies also have a proliferative advantage over cells producing low affinity antibodies. Thus SHM also increases the fitness of a cell through multiple beneficial mutations in a single allele. Since the variety and frequency of mutations is so high, SHM also increases the frequency of gene inactivation. Such inactivating mutations likely occur in Ig mutated cells but are eliminated by the selection for high affinity alleles. Albeit at much lower efficiency, AID expressed in immune cells is mutagenic for several other genomic regions, which makes these regions prone to undesired changes.34,36,37 However, since SHM is mostly confined to a small region within the Ig-locus, it can produce multiple mutant alleles with high fitness without excessive generation of lethal or low fitness alleles in other genes.
Can Damage-Induced LHM Operate in a Non-Programmed Mode?
The existence of multiple powerful repair systems enables living cells to repair vast numbers of DNA lesions within a single cell cycle. The number of lesions due to endogenous damage in normal human cells is estimated to be in the tens of thousands per day38 and the tolerable number of lesions that can be caused by exogenous sources can be orders of magnitude greater, reaching a density of one lesion per several thousand nt.39–42 Unrepaired lesions often lead to mutations, if copied by error-prone translesion synthesis (TLS) DNA polymerases.43,44 A cell with a high density of DNA damage would inevitably die if it lacks DNA repair across the genome. However, lack of repair of lesions in a small region of the genome may be tolerated as in the case of SHM. In this situation, error-prone TLS during copying of region with multiple lesions can produce a stretch of multiple mutations. One potential source of LHM due to inhibited DNA repair is damaged ssDNA. Since most DNA repair systems operate exclusively on double-strand DNA (dsDNA), damage in ssDNA often would be left unrepaired and lead to mutation. Coincidence in both the formation of large stretches of ssDNA and induction of DNA-damage could, in principle, be a source of LHM, if the damaged ssDNA is capable of recovery to dsDNA state. However, if long ssDNA with multiple lesions is lost due to degradation or cell death triggered by checkpoint activation, the opportunity for multiple mutations will be lost.
We sought to determine if long stretches of ssDNA formed around a DSB or at uncapped telomeres can recover to generate cells with multiple mutations.29 For this purpose we developed special genetic systems in a model eukaryote, the yeast S. cerevisiae where stretches of long ssDNA can be formed around inducible site-specific DSB or uncapped telomeres (Fig. 2). In these model systems, the frequencies of damage-induced LHM were comparable to those observed in programmed SHM within the Ig-locus (Fig. 1). Damage-induced LHM caused by two different kinds of damaging agents, ultraviolet light (UV) and methylmethane sulfonate (MMS) relied completely on the error-prone TLS polymerase Polζ. Strand-biased mutation spectrum of UV-induced mutations indicated that mutations are caused by TLS in the damaged ssDNA.29 In subsequent work we found that MMS-induced mutations are caused by ssDNA-specific damage (predominantly N-3-methyl cytosine), indicating that the damage was inflicted after the DNA had become single-stranded.45 Importantly, we observed a large number of strand-biased multiple mutations (up to 6 widely-spaced changes in a 4 kb ORF). These findings provided the first demonstration of damage-induced LHM via a mechanism that tolerates multiple, simultaneous lesions in ssDNA. Thus, yeast cells are able to generate simultaneous multiple mutations in a single ORF by the mechanism of damage-induced LHM in transient ssDNA. The experiments described below highlight additional biologically important features of this phenomenon.
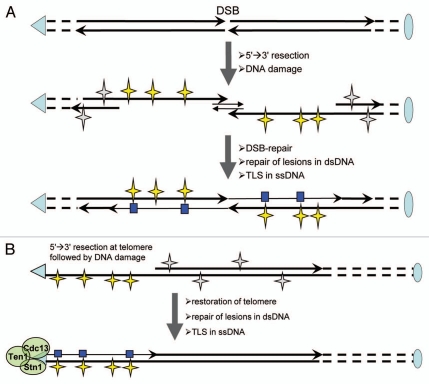
Figure 2.
Damage-induced localized hypermutability associated with transient regions of single-strand DNA generated at double-strand breaks and uncapped telomeres. The experimental approach and conclusions summarized in this figure were described in references 29 and 45. (A) Double-strand break (DSB). Long ssDNA can be generated around a DsB by 5′→3′ resection if DSB repair is delayed. While damage in dsDNA (gray stars) can be repaired by major repair pathways, such as base excision repair, nucleotide excision repair, post-replication repair, damage in ssDNA (yellow stars) often remains unrepaired. Repair of an inducible site-specific DSB was triggered by adding oligonucleotides complementary to the ends of the break. Trans-lesion DNA synthesis (TLS) is required to create a complementary strand on the damaged DNA template. The TLS events can generate wild-type sequence; however, if TLS is error-prone mutations (blue boxes) can be created at many DNA damage sites. (B) Uncapped telomere. Long ssDNA can be generated by 5′→3′ resection at telomeres that transiently lost their capping protein complex. This uncapping was achieved by shifting a cdc13-1 mutant to a non-permissive temperature (37°C). Restoration of the telomere cap and dsDNA was allowed by shifting back to permissive temperature (23°C) after applying DNA damage (Uv or MMS). Multiple mutations were generated by error-prone TLS as in (A). Mutations were initially detected29 in a reporter LYS2 gene (not shown), placed at 2.2 kb from left telomere of the chromosome V, based on lysine auxotrophy. Mutations in the other genomic areas of these lysine auxotrophs were identified in the current study by extended sequencing of the chromosome V subtelomeric region and later by whole genome sequencing (see Results).
Results
We performed additional sequencing in the mutant strains, isolated in our previous study from conditions with the high level of UV-induced LHM within the reporter LYS2 gene placed into subtelomeric region of chromosome V (ref. 29 and Fig. 2B). Formation of subtelomeric ssDNA in these experiments was triggered by shifting cdc13-1 mutant yeast cells to non-permissive temperature 37°C, which lead to telomere uncapping followed by 5′→3′ resection. While the mutants were isolated based on lysine auxotrophy indicative of inactivation of the LYS2 function, we proposed that they carry additional mutations in the left subtelomeric region of chromosome V as well as in other subtelomeric regions. Additional targeted and genome-wide sequencing provided important information about the size and distribution of LHM regions as well as about density of mutations.
Density of mutagenic damage in ssDNA.
In the systems that we have developed, only a fraction of cells that have ssDNA in the region of the mutation reporter at the time of acute DNA damage have the potential for hypermutation. Therefore, the density of mutations in the hypermutable fraction was estimated based on the distribution of single and multiple mutant alleles (refs. 29 and 45 and Sup. Table 1). Damage-induced LHM completely depended on error-prone TLS by Polζ (Rev3, Rev7) and on Rev1. Similar to other kinds of error-prone TLS, damage-induced LHM also depended on PCNA-K164 ubiquitylation. The DNA polymerase Polη (RAD30) provides an error-free TLS pathway for the major UV-lesion cyclobutane dimers, that can compete with error-prone TLS by Polζ and Rev.43 Therefore damage-induced LHM could be further enhanced in the absence of Polη. However, in our initial study we did not detect a statistically significant change in the overall frequency of mutations in the reporters of ssDNA-associated mutagenesis when Polη was deleted.29 We note that the frequency of mutant alleles of a reporter gene in a population that contains cells with LHM spanning a reporter depends upon the mutation density within the LHM region, on the LHM fraction in the population as well as several other parameters. Thus direct measurement of mutation density by sequencing reporter ORFs provides more accurate estimate of LHM (discussed in refs. 29 and 45). Therefore, we have sequenced LYS2 ORFs from 22 UV-induced lys2 mutants in the rad30Δ derivative of the cdc13-1 strain with a subtelomeric LYS2 reporter on the left arm of chromosome V (Table 1 and Sup. Table 1). We chose the subtelomeric LYS2 reporter because of the ORF is twice as large as the DSB-associated CAN1-mutation reporter. The densities of mutations within the lys2 mutant ORFs were very similar between Rad+ and rad30Δ backgrounds. One explanation of this similarity is that the Polζ/Rev1-dependent error-prone TLS dominates in damaged ssDNA of wild type cells, while Polη error-free TLS operates only as a supplemental mechanism. The prevailing role of Polζ/Rev1 TLS may be associated with special conditions with checkpoint activation, which is characteristic for cells experiencing DSB or uncapped telomere. Both damage checkpoint and cell cycle controls have been implicated into regulation of TLS by specialized DNA polymerases.46 Importantly, a density of approximately one mutagenic UV-lesion per 3 kb in the LHM segment is close to the expected density of pyrimidine dimers in DNA of our treated yeast cells based on prior estimates.39,41 We conclude that the main source of UV-induced LHM is lack of repair rather than a higher density of UV-damage in ssDNA and that most lesions give rise to mutations via TLS.
Table 1.
Table 1. Multiple mutations and lys2 mutation spectra resulting from UV irradiation of cdc13-1 yeast strains.
| Genotype1 | Number of mutations in a mutant | Total # of mutants | D2 (mut/kb) | Indel.3 | Compl.4 | Sub., Py5 | Sub., Pu6 | Total # of mutations | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | T | C | A | G | ||||||
| WT | 7 | 16 | 5 | 4 | 1 | 1 | 34 | 0.31 | 19 | 5 | 34 | 17 | 6 | 0 | 81 |
| rad30Δ | 6 | 9 | 5 | 1 | 0 | 1 | 22 | 0.28 | 6 | 6 | 20 | 14 | 1 | 2 | 49 |
The data for the Rad+ (WT) strain are from reference 29, the data for rad30Δ strain are given in Supplemental Table 1.
D (mut/kb), densities of mutations in the 4396 nt LYS2 reporter were calculated as described in reference 45 using the formula: D=(n2+ 2n3+ 3n4 + 4n5 + 5n6)/(M − 4.396), where nx is the observed number of mutants with x mutations and M is the total number of mutants. Calculations were made on the assumptions that (1) the vast majority of mutants originate from the hypermutable fraction; and (2) within this fraction individual mutations occur with equal probability in every cell.
Indel: Small insertions and deletions.
Comp: Complex mutations.
Sub Py and Sub Pu: Number of simple base substitutions in pyrimidine (Py: T or C) or purine (Pu: A or G) nucleotides in the non-resected strand.
Regions of damage-induced LHM can be large.
The mutation reporters used in our previous studies did not allow detection of damage-induced LHM regions greater than 4 kb. This would be sufficient for detecting simultaneous multiple mutations within nearly any yeast ORF because the vast majority of genes in this organism lack introns. However, most genes in higher eukaryotes are much longer due to the presence of introns. For example, the sizes of human genes range from several hundred nucleotides to more than a megabase, with a median around 25–30 kb.47,48 Thus LHM would need to span tens of kilobases to generate multiple mutations in the ORF of an average mammalian gene.
In order to determine the extent of the UV-induced LHM area we employed capillary ABI (Applied Biosystems, Foster City, CA), technology to sequence 30 kb regions adjacent to the left telomere of chromosome V, where subtelomeric LHM was observed in the LYS2 reporter. We examined twelve lys2 mutants of the Rad+ strain obtained in a previous study in reference 29, after UV-irradiation of cdc13-1 cells arrested at non-permissive temperature (37°C). This condition inhibits telomere capping and results in formation of long ssDNA tails by way of 5′→3′ resection.49–51 For the no LHM control, the same region was sequenced from nine lys2 mutants isolated after UV-irradiation of the culture kept at permissive 23°C temperature, a condition in which ssDNA is not formed. Sequenced regions of each of the control strains contained only the single mutation in the LYS2 which gave rise to the Lys- phenotype selected in the experiment. In contrast, lys2 mutants isolated from UV-induced LHM conditions contained up to 11 mutations with tracts of multiple mutations spanning over 17 kb from the telomere (Fig. 3 and Sup. Tables 2 and 3). The mutation density in the LHM regions was constant over telomere-proximal 12 kb and declined beyond that. Similar to our previous observation29 and in agreement with UV damage specificity,52 most mutations were base substitutions with a strong bias toward changes of pyrimidines in the strand that would be retained after 5′→3′ resection from the uncapped telomere. Thus, our results with extended sequencing are in agreement with LHM originating from damaged ssDNA formed by resection from the uncapped left telomere of chromosome V. Since UV damage is comparable for ssDNA and dsDNA53,54 we cannot distinguish between damage to ssDNA formed by resection versus damage to dsDNA right before resection. It is worth note that in the follow-up study of DSB associated LHM caused by methylmethane sulfonate (MMS), a mutagenic agent with ssDNA-specific spectrum, we found that MMS-induced LHM is associated with lesions in ssDNA rather than with damage occurring in dsDNA immediately before resection.45 Furthermore, the strand bias observed in our previous work29 as well as in the current study makes it unlikely that LHM could be due to long-term inhibition of repair in subtelomeric dsDNA persisting through the next round of DNA replication. Such a pathway would result in lys2 mutations originating from both DNA strands and thus a mutation spectrum lacking strand bias. Altogether, long-range sequencing demonstrated that the eukaryotic yeast cell is capable of restoring at least 15 kb of single-strand DNA containing over 10 mutagenic lesions to functional dsDNA with multiple mutations. This indicates that the area of damage-induced LHM can encompass an average size human gene and produce sets of mutations scattered over the entire ORF.
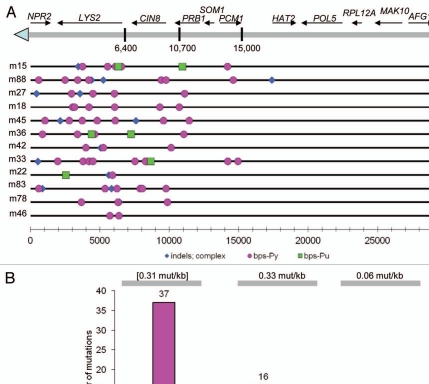
Figure 3.
UV-induced mutations in a subtelomeric area. (A) Presented are mutations in a 30 kb terminal region of the truncated left telomere region of chromosome 5 (details of the construction are described in ref. 29) in twelve lys2 mutants induced by UV-light in G2 arrested at 37°C cdc13-1 cells. Not shown are nine lys2 mutants induced by UV-light in control G1 stationary cells that were kept at permissive temperature (23°C). Each of nine mutants from control set contained only one lys2 mutation across the sequenced 30 kb region (Sup. Table 1). Each horizontal line under the map of the region corresponds to the sequence of an individual mutant. Symbols correspond to a nucleotide change in the single strand that would remain after resection of the complementary strand. See Supplemental Tables 2 and 3 for complete information about all identified mutations. (B) The average mutation densities (numbers above top grey lines) and total numbers of different mutation types (graph) in the 12 sequenced lys2 mutants originating from G2-arrested cdc13-1 cells treated by Uv-light. All mutants were picked by inactivation of LYS2 function; therefore calculation of mutation density in the LYS2 segment (number in brackets) was based on the formula and assumptions described in footnotes for Table 1. The average mutation densities in the other two regions, where all mutations were unselected, were calculated as the total number of mutations divided by the total number of nucleotides sequenced in these regions.
Genome-wide landscape of damage-induced mutations in cells with LHM.
In our previous study, UV-induced LHM was observed only at a subtelomeric LYS2 reporter but not in other LYS genes scattered across the yeast genome.29 This indicated that the mutation load due to UV-induced mutagenesis in the rest of the genome is low. In order to verify this, we explored the genome-wide landscape of UV-induced mutations in several of the yeast clones that were isolated from the subtelomeric LHM experiments and used for long-range sequencing described in the previous section.
Recent advances in high-throughput whole genome sequencing have made it possible to use the entire genome as a reporter in studies of spontaneous and induced mutagenesis in a number of species.55–61 A reference sequence is created from the DNA of the cells that are closely related to the clones or tissues in which mutagenesis is explored. With the current level of technology, mutations can be identified (called) with confidence only in unique or moderately repeated parts of the genome. In the case of yeast this could be as much as 80–90% of the genome because rDNA repeats (about 10% of the genome) are excluded from the analysis and the rest of the reference sequence contains gaps at moderately repeated sequences. The sequence reads from individual clones are then aligned against the reference sequence and mutations are called using special software packages. As a last step in the identification of damage-induced mutations, all changes that are found in more than one clone are removed from the list. These identical mutations could result either from errors in the reference sequence or could arise during propagation of the population that was a source of the reference sequence.
We used the Illumina GAIIx (San Diego, CA) sequencing platform and CLC Genomics Workbench (GWB) 4.0 (CLC Bio, Katrinebjerg, Denmark) software to build the reference sequence of the strain DAG760 and to call mutations from Illumina reads of five clones isolated from the condition with UV-induced subtelomeric LHM (cdc13-1 cells, G2-arrested at non-permissive 37°C) as well as from three control clones obtained after UV-mutagenesis of cdc13-1 cells that stopped at G1 after growth at permissive temperature 23°C (see Materials and Methods and Sup. Materials). The clones were separated from the reference population of cells by 25–30 cell generations. Based on the recent measurements of genome-wide spontaneous mutations in non-mutagenized yeast, it is expected that each clone would contain at most 1–2 new mutations.57,59 We confirmed the expected low level of genome-wide incidence of detectable mutations in non-mutagenized yeast cultures using our sequencing tools and software. In agreement with published data there was only one new base substitution mutation detected in the genomes of six non-mutagenized clones separated by ∼25 cell generations from the reference population. This contrasts with 16–38 new mutations found in each of the sequenced clones isolated after UV-mutagenesis (Fig. 4A and Sup. Table 4). The accuracy of the reference sequence and mutation calling was confirmed by comparing changes identified within Illumina/CLC GWB mutation reports (Sup. Table 4) with mutations in the left 30 kb subtelomeric region of chromosome V that were identified by conventional Sanger (ABI) sequencing (see above; total of 240 kb sequenced for eight isolates; Sup. Tables 2 and 3). All 47 mutations in that region called in high-throughput Illumina sequencing were also identified by Sanger capillary (ABI) sequencing. Importantly, there were only three mutations identi- fied by capillary sequencing that were not called by Illumina (Sup. Table 3). These mutations were actually present in the majority of reads, but were not called by the CLC GWB software due to the stringency of our mutation calling parameters. These three mutations represented minor categories within UV-induced spectra: two mutations were complex combination of base substitutions and indels and one was a simple indel. Thus, there was only a minimal discrepancy between the total of 240 kb sequence obtained by capillary ABI versus the corresponding Illumina generated sequence information.
Figure 4.
Mutations identified by whole-genome sequencing in the individual genomes of UV-irradiated yeast cells. (A) Distribution of UV-induced mutations between subtelomeric and internal regions of yeast chromosomes. Subtelomeric regions were defined as 25 kb from either end of the reference chromosome sequence (see text). Shown are the total numbers of simple base substitutions, indels and complex changes for each type of regions. All mutations are listed in Supplemental Table 4. (B) Numbers of mutations in subtelomeric regions of each chromosome. (ss) indicates mutants obtained from cdc13-1 cells arrested in G2 at 37°C, a condition triggering formation of ssDNA at uncapped telomeres; (ds) indicates mutants obtained from control G1 stationary cdc13-1 cells at 23°C; under these conditions formation of subtelomeric ssDNA is not expected.
The total numbers of UV-induced mutations in each genome was dramatically less than the number of damaged nucleotides expected for doses of UV used in our experiments (∼6,000 lesions per genome based on refs. 39 and 41). This indicates an overall high repair capacity across the genome as compared with the lack of repair resulting in LHM, associated with ssDNA formed at uncapped telomeres. In our experiments, cdc13-1 cells were held at non-permissive temperature for 6 h before UV-irradiation. In prior studies, ssDNA was detected 10–30 kb from telomeres in cdc13-1 cells arrested in G2 by shifting to non-permissive 37°C temperature for 6 h.49 Questions about continuity and size distribution for ssDNA regions created by 5′→3′ resection in this system have yet to be addressed. The resection rate in yeast was measured carefully only with site-specific DSBs, where the 5′→3′ DNA degradation proceeded at a rate of approximately 4 kb per hour.51 While conditions may differ for the resection at uncapped telomeres, this value leads to an estimate of around 25 kb of ssDNA formed by 5′→3′ resection in the cdc13-1 G2-arrested cells and provides an opportunity to address mutations in the subtelomeric vs. internal regions of the genome. In the absence of subtelomeric ssDNA formation, 76 out 84 (90%) UV-induced mutations were located in internal regions of chromosome (93% of the sequenced genome), while only 8 (10%) mutations mapped to subtelomeric regions (comprising the remaining 7% of the sequenced genome) (Fig. 4A). In contrast, the fraction of subtelomeric mutations was 46–69% of all changes induced by UV in cells in which there was an opportunity for generation of subtelomeric ssDNA. Many of the mutations in subtelomeric regions were due to changes in the vicinity of the left telomere of chromosome V, where selection was applied to inactivation of the LYS2 reporter (Fig. 4B). There were several clusters of 2–4 unselected mutations (a total of 16 mutations in clusters) in other subtelomeric regions of cdc13-1 G2-arrested cells, while there were no clusters in subtelomeric regions of the control G1 cells (p < 0.02 by two-tailed Fisher's exact test). This is consistent with the observation of multiple resected telomeres in populations of cdc13-1 G2-arrested cells.62 We conclude that uncapping and resection occurs at multiple telomeres in a single cell. Moreover, multiple areas of damage induced-LHM, associated with regions of transient ssDNA, can occur within the same cell.
To address the genome-wide incidence of unselected mutations, the subtelomeric region adjacent to left telomere of chromosome V was excluded from further calculations, because it contained mutant lys2 alleles selected within our experimental design. Densities of UV-induced internal mutations in control as well as in G2-arrested cdc13-1 cells were in agreement with mutation frequencies of the LYS2 subtelomeric reporter in the absence of ssDNA formation.29 The densities of subtelomeric mutations in three control isolates did not show a statistically significant difference from that of internal UV-induced mutations. In contrast, the density of subtelomeric mutations in cdc13-1 G2 arrested cells was approximately 16-fold greater than the mutation density at the internal regions of the same genomes, suggesting that several regions of LHM could be tolerated in the same cell (Fig. 5A). The density of mutations in the left subtelomeric region of chromosome V, where initial lys2 mutations were selected, was 7-fold greater than the density of unselected UV-induced mutations in other subtelomeric regions (compare Fig. 3B with 5A). The unselected subtelomeric mutation clusters were also shorter than those in the left subtelomeric region of chromosome V, to which mutation selection was applied. This could be explained by incomplete detection of mutations in moderately repeated segments that are often present in subtelomeric regions63 and/or by shorter stretches of ssDNA in the majority of subtelomeres. In order to verify that increased mutation density in subtelomeric regions was due to mutations induced by UV in ssDNA, we summarized data about bases mutated in the reference genome (Fig. 5B). In agreement with the well established mutagen specificity of UV, the majority of mutations associated with model LHM reporters in the vicinity of uncapped telomere or DSB were identified as changes of pyrimidines in the ssDNA formed by resection (reviewed in ref. 29 and Fig. 3). Thus, if increased density of unselected subtelomeric mutations is due to ssDNA generated by 5′→3′ resection in telomeres, the same kind of bias is expected. For the format of genome-wide analysis, it is important to note that the sequence and mutations throughout the entire chromosome are reported in the 5′ to 3′ direction of the top strand. Therefore, it would contain the actual sequence of ssDNA generated by resection from right telomeres and the sequence complementary to ssDNA retained after resection from left telomeres. As expected, there were more base substitutions in pyrimidines of the top strand reported in right subtelomeric regions of cdc13-1 G2 arrested cells, while for the left telomeres there were more substitutions reported in purines of the top strand (Fig. 5B; p < 0.02 by two-tailed Fisher's exact test). In summary, whole genome sequencing confirmed that density of UV-induced mutations in G2-arrested cdc13-1 cells is high in subtelomeric regions, while remaining at a baseline level throughout the rest of the genome. Importantly, based on incidence of unselected mutation clusters yeast cells are capable of tolerating multiple areas of UV-damaged ssDNA.
Figure 5.
Density of unselected mutations and purine/pyrimidine bias in subtelomeric and internal regions of yeast chromosomes revealed by whole genome sequencing. The letter “m” precedes the number of specific mutant strains. In order to obtain parameters for unselected mutations, all changes in the 25 kb left subtelomeric region of chromosome V containing the LYS2 mutation reporter were excluded from calculations. (A) Density of mutations calculated for the total of base substitutions, indels and complex changes. (B) Purine/pyrimidine bias of simple base substitutions. Mutations were categorized based on nucleotide changes in the 5′→3′ (top) strand throughout the chromosome reference sequence.
Discussion
Our results establish that the combination the three factors contribute to damage-induced LHM generating widely spread clusters of multiple mutations without excessive mutation load in the rest of the genome: (1) high capacity of DNA damage repair, (2) toleration of large regions of damaged ssDNA and (3) highly efficient error-prone translesion synthesis (TLS) during restoration of damaged ssDNA to dsDNA. Presented below are several important questions and implications related to the phenomenon of damageinduced LHM.
Mechanisms of damage-induced LHM.
In our experiments LHM was observed after restoration of damaged ssDNA formed at unprotected DNA ends such as DSBs and uncapped telomeres (refs. 29, 45 and this study). While the length of ssDNA can be extensive at unprotected DNA ends in prokaryotic and eukaryotic microbes, it appears to be much shorter in mammalian cells.51,64–66 The prevailing view is that the resection machinery is conserved across eukaryotes but the end-resection capacity is limited under normal conditions in mammalian cells. However, it is worth noting that unlike microbial systems, studies of resection in mammalian cells generally rely on microscopic detection of ssDNA-interacting proteins or antibodies rather than high-resolution monitoring of ssDNA formation. Also, there may be fewer resection tracts in mammalian cells due to DSB repair by non-homologous end-joining (NHEJ) or microhomology-mediated end-joining (MMEJ) in these cells. These pathways can eliminate substrates for end-resection since they act efficiently on blunt or minimally degraded DNA ends. Even if normal resection tracts in mammalian cells are shorter than in yeast, the presence of proteins associated with resection suggest that long resection tracts may be possible especially in conditions limiting factors that might inhibit resection (reviewed in ref. 67 and references therein).
Another potential source of long ssDNA is uncoupling between leading and lagging strands of the replication fork; these can occur spontaneously and/or in response to blockage of DNA polymerase by DNA damage.68,69 Thus it is important to identify genotypes and conditions where frequency, size and persistence of ssDNA regions generated by replication fork uncoupling would be extended over the norm. In addition, long stretches of ssDNA whose origin is unknown have been identified in cultured cancer cells.70 The origin and mechanisms producing this form of ssDNA are unknown. However, if this ssDNA can be restored to dsDNA, this could be an additional source of damage-induced LHM.
In principle, damage-induced LHM need not to be associated with ssDNA. It could originate from any cause that would inhibit DNA repair from the time of damage through the next DNA replication. While several factors such as chromatin state, nucleosome position or transcription status might affect the efficiency of DNA repair and/or mutation frequency (see Introduction), there has been no evidence of strong, multi-fold mutator effects caused by these factors leading to clusters of simultaneously occurring multiple mutations. Importantly, closely-spaced multiple mutation clusters of LHM were detected in our experiments only under situations in which ssDNA was generated. Clusters were not observed in whole genomes of G1 stationary cells or in internal chromosomal regions of G2-arrested, UV-irradiated cdc13-1 cells (Sup. Table 4) where long stretches of ssDNA are unlikely to occur. We note that the size of the dataset we have presented is not sufficient to exclude the infrequent incidence of clusters not associated with uncapped telomeres. In general, we anticipate that our understanding of pathways and molecular mechanisms of damage-induced LHM will be greatly expanded as more genomewide mutagenesis data becomes available.
Can damage induced LHM occur in natural conditions?
Examples of widely spread clusters of multiple mutations (mutation showers) have been detected among the mutation spectra in mice;15,16 however, the mechanisms generating these clusters were not addressed. The hypermutability and mutation clusters in our experiments (refs. 29 and 45 and this study) were caused by damaging arti- ficially formed ssDNA around an inducible site specific DSB or in the vicinity of uncapped telomeres in G2-arrested cdc13-1 mutant yeast. Similarly, ssDNA can be formed by resection at unprotected ends of spontaneous or damage-induced DSBs. Importantly, a vast number of DNA damaging agents can induce both DSBs and mutagenic base or nucleotide damage.1 For example, we demonstrated that base alkylation by methyl methanesulfonate (MMS) results in DSBs via faulty repair of closely-opposed lesions71,72 as well as in a high frequency of base substitutions near ssDNA near artificially induced site-specific DSB.45 However, multiple mutations were found only rarely among spontaneous or damage-induced forward mutations with regular mutation reporters. Mutation reporters designed to detect low frequencies of clustered multiple mutations are under development in our lab.
Does damage-induced LHM contribute to natural genetic variation and disease?
Mutations are the primary source of sequence variation in evolution. Localized increases in the number of mutations accumulated during human evolution from a common ancestor with chimpanzees have been associated with meiotic DNA breaks.73–77 These studies have also identified a number of human accelerated regions (HARs) in which over the past ten million years of primate evolution many more mutations have accumulated than over preceding hundred million years of mammalian evolution. An association was detected between hotspots of meiotic recombination in human males and HARs. Another distinct feature of HARs is a mutation bias of A-T or T-A pairs changing into G-C or C-G pairs. One explanation is that there is biased gene conversion in which G-T and C-A mismatches are more frequently corrected toward G-C and C-G as compared with correction towards A-T and T-A. This would lead to increased fixation of G-C and C-G mutant base pairs. However, HARs could also reflect increased mutability around meiotic DSBs which can be further enhanced by endogenous damage to ssDNA formed around breaks. Recently, based on analysis of vast amounts of human sequencing data, it was concluded that the increased rates of base substitutions over evolutionary, population and even single tumor or cell line timescales are associated with rearrangement breakpoints, and could thereby be associated with hypermutability of break-associated ssDNA78 and references therein. Increased frequency of mutations around rearrangement breakpoints was also reported for prostate cancer genomes.79 In another study, increased mutation rates in the human evolution line were associated with late replicating regions of the genome, which could also be associated with a higher frequency of breakage during mitotic divisions in the germline and/or with increase in ssDNA formation.80 Bringing all these findings together, including our observation that spontaneous and damage induced mutation frequencies are dramatically increased in ssDNA as compared to dsDNA, we propose that error-prone translesion synthesis during restoration of damaged ssDNA may be a significant source of mutations in nature. Future studies integrating model system experiments with genotoxic factors and whole genome mutation analyses will shed light on the role of damage-induced LHM in evolution, the biology of species, as well as human health and disease.
Materials and Methods
Yeast strains and methods.
Yeast strains construction as well as genetic and molecular biology methods were as described in references 29 and 45. The genotype of the strain DAG760 was as follows—MATα ade5-1 his7-2 leu2-3,112 trp1-289 ura3Δ lys2Δ (in chromosome II); wild type LYS2 was inserted between NPR2 and CIN8 close to the de novo left telomere of the chromosome V.
Re-sequencing the 30 kb region adjacent to the left telomere of chromosome V.
The schematics of the region structure are shown on Figure 3A. Details of construction are described in reference 29. Primer pairs (Sup. Table 5) to generate overlapping amplicons for re-sequencing yeast DNA regions were designed using a Perl script, which first called on the RepeatMasker81 program to generate repeat masked sequences, which were then used for primer design via Primer3.82 Target primer Tm was 61°C. Target amplicon size was 500 bp with 150 bp overlap. Primers were obtained from IDT (Coralville, IA). Liquid handling for the resequencing protocol was automated on the BioMek FX robot (Beckman Coulter) using a magnetic bead based purification system (Agencourt Bioscience). PCR and cycle sequencing reactions at 1/64 Big Dye reaction scale (cycle sequence version v1.1, Applied Biosystems) were performed on the MJ Tetrad 225 (BioRad). Bead cleaned cycle sequencing reactions were run on 48 capillary ABI 3730 sequencers (Applied Biosystems). Sequence data files were uploaded into the PolyPhred program83 for quality analysis and polymorphism detection.
Building reference sequence and mutation calling based on illumina reads.
Libraries were prepared from genomic DNA of the yeast strain DAG760 for sequencing on the Illumina GAIIx (San Diego, CA). One library contained fragments around 160 nt and was run on three GAIIx lanes with paired end 35 nt reads. Another library was created with the fragments around 4,500 nt and was run on four GAIIx lanes with mate paired end 51 nt and 76 nt reads. All data were pooled together for building a reference sequence using CLC Genomics Workbench 4.0.2 2 (CLC Bio, Katrinebjerg, Denmark). The data were first aligned against the yeast reference sequence of S288c strain. For this first alignment we allowed for random distribution of not uniquely aligned reads. A consensus sequence build based on this alignment contained all small and large repetitive elements from the genome and all variation form the reference like SNPs and small indels. After extraction of consensus sequence we were able to use it for next steps of the DAG760 reference construction. Second alignments of all sequencing reads to the extracted consensus sequence (pre-reference) were performed in order to detect errors in previous alignment. In this second alignment all reads which matched in more than one site in the pre-reference genome were ignored. 38 SNPs (base substitutions) not detected in first alignment were identified and manually corrected. This resulted in the master DAG760 reference sequence. All sequenced mutant strains used for experiment were aligned to master reference sequence of the DAG760 genome. Parameters for SNP or indel discovery were restrictive for the quality of both substituted bases and surrounding bases but allow for minimum 80% of the variation frequency. We identified SNPs (located usually in sites with repeatedly low coverage), which were common for most of the strains. These common SNPs were excluded from next steps of analysis.
Acknowledgements
We are thankful to Drs. Jan Drake, Jana Stone and Kin Chan for critical reading of the manuscript. The work was supported by funds the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Project ES065073, P.I.-M.A.R.) and by grants from the National Institute of Environmental Health Sciences (P30ES010126), NIH (RC1 ESO18091) and the University Cancer Research Fund (UCRF) to P.A.M.
Supplementary Material
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. Washington: ASM Press; 2006. [Google Scholar]
- 2.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondrashov FA, Kondrashov AS. Measurements of spontaneous rates of mutations in the recent past and the near future. Philos Trans R Soc Lond B Biol Sci. 2010;365:1169–1176. doi: 10.1098/rstb.2009.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake JW. Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 2009;5:1000520. doi: 10.1371/journal.pgen.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes and eukaryotes. Ann NY Acad Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 8.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–326. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogenaru M, de Vos MG, Tans SJ. Revealing evolutionary pathways by fitness landscape reconstruction. Crit Rev Biochem Mol Biol. 2009;44:169–174. doi: 10.1080/10409230903039658. [DOI] [PubMed] [Google Scholar]
- 11.Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165–1174. [PubMed] [Google Scholar]
- 12.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 13.Wright S. The roles of mutation, inbreeding, cross-breading and selection in evolution; Proceedings of The Sixth International Congress on Genetics; 1932. pp. 356–366. [Google Scholar]
- 14.Povolotskaya IS, Kondrashov FA. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–926. doi: 10.1038/nature09105. [DOI] [PubMed] [Google Scholar]
- 15.Colgin LM, Hackmann AF, Emond MJ, Monnat RJ., Jr The unexpected landscape of in vivo somatic mutation in a human epithelial cell lineage. Proc Natl Acad Sci USA. 2002;99:1437–1442. doi: 10.1073/pnas.032655699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Gonzalez KD, Scaringe WA, Tsai K, Liu N, Gu D, et al. Evidence for mutation showers. Proc Natl Acad Sci USA. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PLoS One. 2008;3:3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Feng J, Saldivar JS, Gu D, Bockholt A, Sommer SS. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008;27:4336–4343. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 19.Kashuba VI, Pavlova TV, Grigorieva EV, Kutsenko A, Yenamandra SP, Li J, et al. High mutability of the tumor suppressor genes RASSF1 and RBSP3 (CTDSPL) in cancer. PLoS One. 2009;4:5231. doi: 10.1371/journal.pone.0005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordenin DA, Resnick MA. Yeast ARMs (DNA atrisk motifs) can reveal sources of genome instability. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassa PO, Hottiger MO. An epigenetic code for DNA damage repair pathways? Biochem Cell Biol. 2005;83:270–285. doi: 10.1139/o05-034. [DOI] [PubMed] [Google Scholar]
- 23.Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 25.Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 26.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, et al. Break-Induced Replication Is Highly Inaccurate. PLoS Biol. 2011;9:1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strandbreak-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 37.Unniraman S, Schatz DG. Strand-biased spreading of mutations during somatic hypermutation. Science. 2007;317:1227–1230. doi: 10.1126/science.1145065. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 39.Resnick MA, Setlow JK. Repair of pyrimidine dimer damage induced in yeast by ultraviolet light. J Bacteriol. 1972;109:979–986. doi: 10.1128/jb.109.3.979-986.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 41.Wheatcroft R, Cox BS, Haynes RH. Repair of UV-induced DNA damage and survival in yeast. I. Dimer excision. Mutat Res. 1975;30:209–218. [PubMed] [Google Scholar]
- 42.Pachkowski BF, Tano K, Afonin V, Elder RH, Takeda S, Watanabe M, et al. Cells deficient in PARP-1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP-1-proficient cells exposed to MMS. Mutat Res. 2009;671:93–99. doi: 10.1016/j.mrfmmm.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 44.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst) 2010;9:914–921. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. Regulation of ultraviolet light-induced gene expression by gene size. Proc Natl Acad Sci USA. 2004;101:6582–6586. doi: 10.1073/pnas.0308181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 49.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lydall D. Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways. J Cell Sci. 2003;116:4057–4065. doi: 10.1242/jcs.00765. [DOI] [PubMed] [Google Scholar]
- 51.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeifer GP. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 53.Chan GL, Doetsch PW, Haseltine WA. Cyclobutane pyrimidine dimers and (6-4) photoproducts block polymerization by DNA polymerase I. Biochemistry. 1985;24:5723–5728. doi: 10.1021/bi00342a006. [DOI] [PubMed] [Google Scholar]
- 54.Doetsch PW, Chan GL, Haseltine WA. T4 DNA polymerase (3′-5′) exonuclease, an enzyme for the detection and quantitation of stable DNA lesions: the ultraviolet light example. Nucleic Acids Res. 1985;13:3285–3304. doi: 10.1093/nar/13.9.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flibotte S, Edgley ML, Chaudhry I, Taylor J, Neil SE, Rogula A, et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics. 2010;185:431–441. doi: 10.1534/genetics.110.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci USA. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishant KT, Singh ND, Alani E. Genomic mutation rates: what high-throughput methods can tell us. Bioessays. 2009;31:912–920. doi: 10.1002/bies.200900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishant KT, Wei W, Mancera E, Argueso JL, Schlattl A, Delhomme N, et al. The Baker's Yeast Diploid Genome Is Remarkably Stable in Vegetative Growth and Meiosis. PLoS Genet. 2010;6:e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parkhomchuk D, Amstislavskiy V, Soldatov A, Ogryzko V. Use of high throughput sequencing to observe genome dynamics at a single cell level. Proc Natl Acad Sci USA. 2009;106:20830–20835. doi: 10.1073/pnas.0906681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zanders S, Ma X, Roychoudhury A, Hernandez RD, Demogines A, Barker B, et al. Detection of Heterozygous Mutations in the Genome of Mismatch Repair Defective Diploid Yeast Using a Bayesian Approach. Genetics. 2010;186:493–503. doi: 10.1534/genetics.110.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westmoreland JW, Summers JA, Holland CL, Resnick MA, Lewis LK. Blunt-ended DNA double-strand breaks induced by endonucleases PvuII and EcoRV are poor substrates for repair in Saccharomyces cerevisiae. DNA Repair (Amst) 2010;9:617–626. doi: 10.1016/j.dnarep.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 64.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 66.Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 67.Boulton SJ. DNA repair: Decision at the break point. Nature. 2010;465:301–302. doi: 10.1038/465301a. [DOI] [PubMed] [Google Scholar]
- 68.Hamdan SM, van Oijen AM. Timing, coordination and rhythm: acrobatics at the DNA replication fork. J Biol Chem. 2010;285:18979–18983. doi: 10.1074/jbc.R109.022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 70.Bjursell G, Gussander E, Lindahl T. Long regions of single-stranded DNA in human cells. Nature. 1979;280:420–423. doi: 10.1038/280420a0. [DOI] [PubMed] [Google Scholar]
- 71.Ma W, Panduri V, Sterling JF, Van Houten B, Gordenin DA, Resnick MA. The transition of closely opposed lesions to double-strand breaks during long-patch base excision repair is prevented by the coordinated action of DNA polymerase delta and Rad27/Fen1. Mol Cell Biol. 2009;29:1212–1221. doi: 10.1128/MCB.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- 76.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De S, Babu MM. A time-invariant principle of genome evolution. Proc Natl Acad Sci USA. 2010;107:13004–13009. doi: 10.1073/pnas.0914454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010.
- 82.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 83.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez C, Hadany L, Ponder RG, Price M, Hastings PJ, Rosenberg SM. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS Genet. 2008;4:1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.