Abstract
Stomach cancer is the second most frequent cause of cancer-related death worldwide. Thus, it is important to elucidate the properties of gastric stem cells, including their regulation and transformation. To date, such stem cells have not been identified in Drosophila. Here, using clonal analysis and molecular marker labeling, we identify a multipotent stem-cell pool at the foregut/midgut junction in the cardia (proventriculus). We found that daughter cells migrate upward either to anterior midgut or downward to esophagus and crop. The cardia functions as a gastric valve and the anterior midgut and crop together function as a stomach in Drosophila; therefore, we named the foregut/midgut stem cells as gastric stem cells (GaSC). We further found that JAK-STAT signaling regulates GaSCs' proliferation, Wingless signaling regulates GaSCs' self-renewal, and hedgehog signaling regulates GaSCs' differentiation. The differentiation pattern and genetic control of the Drosophila GaSCs suggest the possible similarity to mouse gastric stem cells. The identification of the multipotent stem cell pool in the gastric gland in Drosophila will facilitate studies of gastric stem cell regulation and transformation in mammal.
Key words: gastric stem cells, foregut/midgut junction, cardia, proventriculus, stomach, Drosophila
Introduction
The mammalian gastrointestinal (GI) tract is required for digestion, nutrient absorption, and homeostasis. It is composed of histologically distinct organs, including the oral cavity, pharynx, esophagus, stomach, small intestine and colon. An epithelial luminal lining with an underlying vascular lamina propria forms the GI mucosa, and the large numbers of epithelial cells are replenished throughout the GI tract by stem cells.1–3 Stomach cancer is the second most frequent cause of cancer-related death worldwide.4 Thus, it is fundamental to elucidate the properties of gastric stem cells, including their regulation and transformation.
In the mouse small intestine, two types of stem cells have been identified.5 One type is located at the +4 position from the crypt bottom; the other type is located below the +4 position in the stem cell zone. In the large intestine, stem cells appear to be directly located at the crypt bottom of the descending colon.5 The stem cells of both the small intestine and colon express a marker, Lgr5 (leucine-rich-repeat-containing G-protein-coupled receptor-5).6 In the gastric gland, multipotent stem cells have been identified to reside in the isthmus region, based on circumstantial morphological and cell kinetic evidence and in combination with 3H-thymidine-labeling.7–9 The stem cells first give rise to three progenitor cells: the pre-pit cells, pre-neck and pre-parietal cells. The pre-pit cells migrate up towards the lumen to become terminally differentiated pit cells. The pre-neck and pre-parietal cells migrate downwards and differentiate into one of the cell types of the fundic gland (mucous neck cell, parietal cell, zymogenic/chief cell, endocrine cell and caveolated cell).7–10 Recently, LGR5+ stem cells have been reported at the base of the antrum region of the gastric gland.11 Further, by lineage tracing, Lgr5+ve cells have been functionally characterized as self-renewing, multipotent stem cells, which are responsible for the long-term renewal of the gastric epithelium.10–12 Recently, it has been found that the two regions (fundic and antral) of the gastric gland vary because of differences in proliferation and differentiation, as well as in expression profiles.13
In Drosophila, the posterior midgut corresponds to the mammalian small intestine, the hindgut corresponds to the mammalian large intestine, and multipotent adult stem cells have been identified in both the posterior midgut and the hindgut.14–17 Notch (N) and other signaling pathways are known to regulate the self-renewal, differentiation, and regeneration of posterior midgut stem cells,15,16,18–28 while Wingless (Wg) and Hedgehog (Hh) signaling controls the self-renewal or differentiation of the hindgut stem cells.17 However, no stem cells have been identified in the location corresponding to the gastric gland in Drosophila.
The Drosophila cardia (proventriculus) is a structure at the junction of the foregut and midgut (referred to as F/M in the following text), where the esophagus, midgut, and crop (a bi-lobed structure in adult flies capable of considerable extension and storage of liquid ingested food) merge.29–31 The cardia functions as a valve and regulates the passage of food into the anterior midgut and crop, which together perform the functions of the mammalian stomach. The cardia is composed of three layers: the outer layer, derived from the anterior-most region of the endodermal midgut, and forming the gastric valve; the middle layer, derived from the ectodermal foregut; and the inner layer, derived from the posterior region of the esophagus (see Sup. Fig. 1).29–31 During embryogenesis, the multilayered cardia structure is formed from a simple epithelial tube through regulated epithelial cell sheet movement. Several signal transduction pathways, including those of Hh, Wg, Decapentaplegic (Dpp), N, and Janus kinase-signal transducer and activator of transcription (JAK-STAT), are required for this epithelial cell sheet movement.32–34 In this study, we identified a multipotent stem cell pool at the F/M junction in adult Drosophila. These cells differentiated into gastric and stomach cells. We further demonstrate that JAK-STAT signaling in cooperation with Wg and Hh signaling regulates the proliferation and self-renewal of these stem cells.
Figure 1.
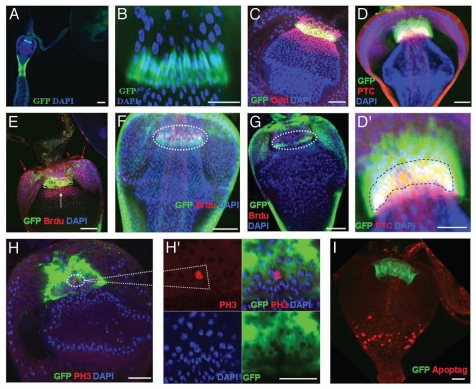
The molecular markers expression and proliferation in cardia. (A,B) The cardia of a Stat92E-GFP fly was stained with GFP (green) and DAPI (blue). (B) The high magnification of (A). (C) The cardia of a Stat92E-GFP fly was stained with GFP (green) and Odd (red) antibodies. (D, D′) the cardia of a wg-Gal4 UAS-GFP fly was stained with GFP (green) and Ptc (red) antibodies and DAPI (blue). (D′) A high magnification picture of (D), in which a part of wg-Gal4 UAS-GFP and anti-Ptc co-localize (yellow—highlighted by blue dashed line). (E) The cardia of a Stat92E-GFP fly was analyzed after five days of BrdU pulse for cell proliferation. F/M junction cells outlined by a white dotted line; anti-GFP (green), anti-BrdU (red), and DAPI (blue). The red arrows show the cell proliferation at anterior midgut region, white arrow shows the cell proliferation at foregut region (F) The Stat92E-GFP-positive cells are proliferating at the F/M junction after five days of BrdU pulse and five days chase period. Only the Stat92E-GFP-positive F/M junction cells retain BrdU, outlined by a white dotted line; anti-GFP (green), anti-BrdU (red), and DAPI (blue). (G) The cardia of a Stat92E-GFP fly was analyzed after five days BrdU pulse and 17 days chase. All BrdU-labeled cells at the F/M junction were gone. The cardia was stained with anti-GFP (green), anti-BrdU (red), and DAPI (blue). The F/M junction cells are outlined by a white dotted line (H, H′) the wg-Gal4 UAS-GFP flies were stained with phospho-histone H3 antibody, which labels only a rare population of cell with small nuclei (highlighted in white box; anti-PH3, red; anti-GFP, green; and DAPI, blue). (I) wg-Gal4 UAS-GFP flies were stained with anti-GFP and Apoptag kit to detect dead cells. Anterior is at the top in all panels. Scale bars: 50 µm (A); 5 µm ( B, D′ H′); 10 µm (C, D, E, H, I); 20 µm (F, G).
Results
The F/M junction of adult Drosophila contains unique cell types.
The adult Drosophila GI system and cardia are illustrated in Sup. Fig. 1,29 which shows the midgut and foregut joining at the junction of zones 3 and 4. We observed that during the third instar larvae stage, the cardia has four gastric caeca and contains a pool of small nuclei cells at the F/M junction, as well as big nuclei cells scattered in another region of the cardia (Sup. Fig. 2A and A′). Cardia does not contain crop at this stage. These small pools of nuclei may be the adult progenitor cells, because during metamorphosis, gastric caecas are degenerated and crop is formed in the adult. Further, we observed that the foregut portion of the adult cardia (zones 3 and part of zone 4) contained a population of cells with small nuclei (Sup. Fig. 2B and 2C′) that clearly differed from the anterior midgut cells, which had larger nuclei (Sup. Fig. 2B and 2C′ zones 4, 5 and 6) and lower foregut zones 1 and 2 (Sup. Fig. 2B and 2C′). A GFP-reporter of JAKSTAT signaling (Stat92E-GFP)35 is specifically expressed at the F/M junction cells (Fig. 1A–C and Sup. Fig. 3A). Furthermore, a transcription factor, Odd-Skipped (Odd) is expressed in the Stat92E-GFP domain, in nearby cells on both sides of it, and in both small and large nuclei (Fig. 1C). wg-Gal4/UAS-GFP (green in Fig. 1D and D′) and Patched (Ptc), a regulator of the Hh signal transduction pathway (red in Fig. 1D and D′) are also expressed in the Stat92E-GFP domain. Stat92E-GFP is a stem-cell marker in several other organs.17,36 Ptc (ptc-lacz) has been shown to mark the hub and cyst progenitor cells (CPCs) in Drosophila testis.37 We found that in addition to testis CPCs, ptc-lacz also express in the F/M junction in the cardia (Sup. Fig. 3B). This cellular organization and expression of markers at the F/M junction are very similar to those that have been reported at the junction of the posterior midgut and hindgut.17 Stem cells have been identified at the junction of the posterior midgut and hindgut and in nearby tissues.15–17,36 These findings led us to examine whether the F/M junction also contains stem cells.
Figure 2.
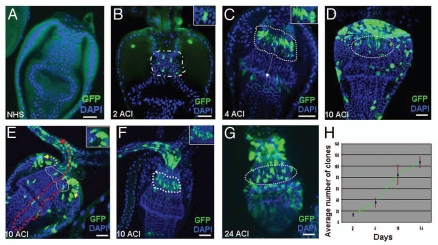
F/M junction cells are multipotent stem cells. (A) MARCM control flies without heat shock (NHS, no heat shock). (B–G) MARCM clones (green) induced in adult cardia. MARCM clone imaged two days after clone induction (ACI) (B), four days (C), ten days (D–F; E, outer view; F, inner view), and 24 days (G) after clonal induction. Clones are highlighted in insets. White arrow in C points to the migration direction of GFP-marked cells from the F/M junction to foregut. Red dotted arrow in (E) points to the migration direction of GFP-marked cells from the F/M junction to foregut and crop. Yellow dotted arrow in (E) points to the migration direction of GFP-marked cells from the F/M junction to midgut. (H) The average number of GFP clones per cardia at the indicated times ACI. Anterior is at the top in all panels. Scale bars: 10 µm (A–G).
Figure 3.
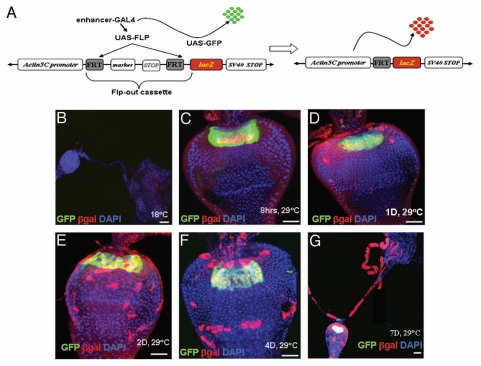
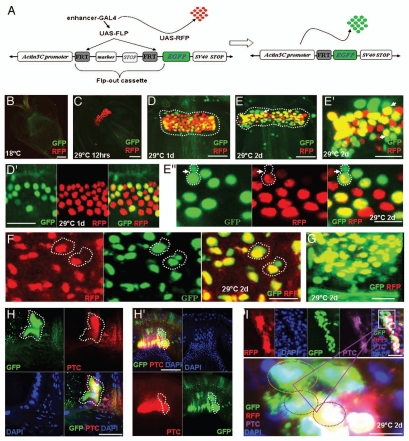
F/M junction cells are multipotent stem cells of the gastric and stomach organs. (A) Schematic diagram of the cell lineage marking system. After shifting the flies with genotype UAS-FLP/+; wg-Gal4 UAS-GFP/actin5C-FRT-draf-FRT-tau-lacZ; tub-Gal80ts/+ to 29°C, the Gal4 was activated and drove GFP (green) and FLP recombinase expression. The FLP recombinase then removed the ‘FLP-out’ cassette so that the constitutive actin5C promoter drove lacZ expression (red) permanently within all subsequent daughter cells. Flies with the genotype UAS-FLP/+; wg-Gal4 UAS-GFP/actin5C-FRT-draf-FRT-tau-lacZ; tub-Gal80ts/+ (B–G) were cultured at the permissive temperature (18° C, B) and then shifted to the restricted temperature (29°C) for 8 hours (C), 1 day (D), 2 days (E), 4 days (F), and 7 days (G). The cardia was stained with anti-GFP (green), anti-β-gal (red), and DAPI (blue). Anterior is at the top in all panels. Scale bars: 50 βm (B, G); 20 βm (C–F).
The F/M junction cells are proliferating.
In order to look for the F/M cell activity, we performed 5-bromodeoxyuridine (BrdU) incorporation experiments. Cells immediately next to the Stat92E-GFP zone in both foregut (Fig. 1E, white arrow) and anterior midgut (Fig. 1E, red arrows) showed strong BrdU incorporation. In contrast, cells in the Stat92E-GFP zone (Fig. 1E, outlined by a white dotted line) showed weak BrdU incorporation. We further analyzed the stability of BrdU-labeled cells at the F/M junction, using the “pulse-chase” technique. After a 5 day “chase,” the BrdU label was retained only in the Stat92E-GFP-expressing cells (Fig. 1F). We also chased the flies for 17 days and found that all BrdU-labeled cells at the F/M junction were gone (Fig. 1G). In invertebrates, BrdU is incorporated into many differentiated cells that endoreplicate.38 We further stained the tissue for phospho-histone H3 (PH3) to distinguish endoreplicating from dividing cells in the cardia. Although BrdU labeled several types of cells in the cardia, the PH3 staining was only detected in a very rare population of cells with small nuclei within the STAT92E-GFP and wg-Gal4/UAS-GFP regions of the cardia (8 PH3+ve cells were observed out of 127 tissues scored) (wg-Gal4/UAS-GFP shown in Fig. 1H and H′). These observations indicate that the Stat92E-expressing and part of wg-Gal4/UAS-GFP cells might be stem cells and the nearby cells at both foregut and anterior midgut sides are their proliferative progenitors.
We further examined the cell death by using an Apoptag kit. We detected very few dying cells in the Stat92E-GFP zone (Sup. Fig. 3E) or wg-Gal4/UAS-GFP (Fig. 1I) zone. However, a significant number of dying cells were detected in the esophagus, anterior midgut, and crop (Fig. 1H; also see Sup. Fig. 3E), indicating that the stem cells in the Stat92E-GFP zone and their proliferating progenitors are generating replacement cells for the dead cells in the esophagus, anterior midgut and crop.
The F/M junction cells are multipotent stem cells.
To determine whether the F/M junction cells included functional stem cells, we conducted three types of lineage tracing experiments. In the first experiment, we used the FLP/FRT system, a simple and efficient method for generating marked clones of random dividing cells.39 In this method, two complementary transgenes were introduced into the same genomic locus in two homologous chromosomes. One transgene bore a ubiquitously activated promoter, the Drosophila α1-tubulin promoter, followed by an FRT sequence. The homologous chromosome contained a reporter gene, β-galactosidase (lacZ), immediately after another FRT site. During mitotic division, when the chromosomes pair up at metaphase, the induced expression of FLP (in this case, by heat shock) can facilitate homologous recombination between the two FRT sites. This leads to the construction of a functional gene cassette that drives lacZ gene expression in a broad region. This technique is very sensitive, because the marker gene is turned on immediately after recombination. We briefly heat-shocked 3–5-day-old adult female flies that had the proper transgenic constructs and stained their guts with specific antibodies for β-gal and Odd 2 days after clone induction (ACI; Sup. Fig. 3F and F′, N = 15). We found that the β-gal-positive clones were mostly restricted to the F/M junction (outlined by a white dotted line in Sup. Fig. 1A), and that some of the labeled cells also expressed Odd (N = 20). No labeled cells were found without heat shock in the controls (data not shown).
In the second experiment, we used the MARCM (mosaic analysis with a repressible cell marker) system40 to trace the labeled cells for a longer time. In this system, a ubiquitously expressed Gal80 transgene (tubP-Gal80), which encodes a Gal4 repressor, is on one FRT chromosome, and a characterized mutation is on the complementary FRT chromosome. In addition, tubP-Gal4 and UAS-GFP are ubiquitously expressed. The induced FLP recombinase promotes recombination between the two FRT sites, and after the completion of cell division, a daughter cell is homozygous for the mutation and does not contain tubP-Gal80. In these cells, the active Gal4 will drive UAS-GFP expression to mark the mutant clones. No clones were detected without heat-shock in the controls (Fig. 2A, N = 15). We briefly heat-shocked 5- or 14-day-old adult female flies carrying the appropriate transgenic constructs and stained their gut with specific antibodies for GFP and DAPI. In cardias fixed two days ACI, the clones were smaller (mostly 1 to 2 cells) and primarily restricted to the F/M junction (outlined by a white dotted line in Fig. 2B, N = 25). However, four days ACI, the GFP-labeled cells had expanded at the F/M junction and some of the GFP-labeled cells started to move into the foregut (Fig. 2C, N = 19). 10 days ACI, the GFP-labeled cells remain at the F/M junction (Fig. 3D–F, N = 17) and some of the GFP-labeled cells migrate into both antier midgut (Fig. 3E, yellow dotted arrow, N = 21) and foregut/crops (Fig. 3E, red dotted arrow, N = 21). 24 days ACI, the GFP-labeled cells were continually expanded and migrate into midgut and foregut/crops, possibly by symmetric or asymmetric division (Fig. 2G, N = 22). From two days to ten days ACI, the total numbers of GFP-labeled cells increased ∼6 times (Fig. 2H); from two days to 24 days ACI, the total numbers of GFP-labeled cells increased ∼7.5 times (Fig. 2H). The number of tracing cells in the cardia did not significantly change over the 24-day period (Fig. 2H), indicating that the stem cells in the cardia are long-term, self-renewing adult stem cells.
To analyze the F/M junction cells further, we specifically traced them using the FLP-out method, modified from Jung et al.41 In this experiment, wg-Gal4 (expressed specifically in the F/M junction cells) was used to initiate the permanent marking of all the daughter cell lineages arising from them (Fig. 3A). In this system, the Wg-Gal4 initially drove the expression of both UAS-GFP and UAS-FLP. The FLP recombinase then excised an intervening FRT-flanked ‘Stop cassette,’ allowing the constitutive expression of lacZ under the control of the actin5C promoter.42 We also included a temperature-sensitive Gal4 inhibitor, Gal80ts,43 to manipulate the Wg-Gal4 activity. In this system, at any time point, GFP is expressed in cells where Gal80 is inactive and Wg-Gal4 is active, whereas lacZ is expressed in all the subsequent daughter cells, regardless of their continued expression of wg-Gal4.
Gal80ts suppresses Gal4 activity at the permissive temperature (18°C). When cultured at 18°C, these flies grow to adulthood with no obvious phenotype and no GFP or β-gal expression (Fig. 3B). We then shifted the adult flies to the restrictive temperature (29°C). After eight hours, GFP and a certain amount of β-gal were expressed at the F/M junction (Fig. 3C, N = 17). After one day, the GFP expression was still limited to the F/M junction, but the β-gal expression had extended to the esophagus and anterior midgut (Fig. 3D, N = 15). After two days, the β-gal expression had extended to the esophagus, anterior midgut, and crop (Fig. 3E, N = 18). After four days (Fig. 3F, N = 19) and seven days (Fig. 3G, N = 12), many cells in the esophagus, anterior midgut, and crop were permanently marked with β-gal, even though they do not express wg-Gal4 (as assessed by GFP) in these cells, indicating that these cells are derived from a wg-Gal4-positive precursor, and that the wg-Gal4 UAS-GFP-positive cells at the F/M junction were multipotent stem cells. The cardia functions as a gastric valve, and the crop and anterior midgut together perform the functions of the mammalian stomach. Since the F/M stem cells produce replacement cells for the gastric and stomach organs, we named them gastric stem cells (GaSCs).
As controls for the above experiment, we examined the β-gal expression patterns of three other Gal4 lines: byn-Gal4 (Sup. Fig. 3G), ppl-Gal4 (Sup. Fig. 3H), and esg-Gal4 (Sup. Fig. 3I), after shifting the adult flies to the restrictive temperature (29°C) for three days. The byn-Gal4 reporter was not expressed at the F/M junction or in the crop, and no specific β-gal was detected in the esophagus, anterior midgut, or crop (Sup. Fig. 3G, N = 10). The ppl-Gal4 reporter was expressed throughout the crop, and the entire crop was permanently marked by β-gal, but no specific β-gal was detected in the esophagus or anterior midgut (Sup. Fig. 3H, N = 12). The esg-Gal4 reporter was not expressed in the cardia or crop, and no specific β-gal was detected in the esophagus, anterior midgut or crop (Sup. Fig. 3I, N = 10).
The GaSCs are multipotent gastric stem cells.
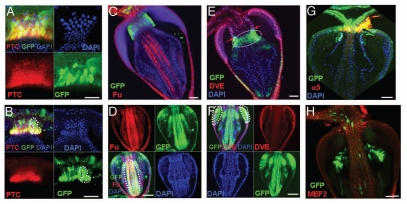
To demonstrate the multi-lineage potential of the GaSCs, we searched and identified several molecular markers that are expressed in specific epithelial cell types of the adult Drosophila gastric organ. Patched (Ptc), a regulator of the Hh signal transduction pathway, expresses in the wg-Gal4/UAS-GFP and Stat92E-GFP zones. We further found that a subset of MARCM clones with small nuclei express Ptc (Fig. 4A and B), which suggests that these cells are stem cells. Ptc does not express in the differentiated daughter cells. Fused (Fu) is a component of the Hh signal transduction pathway, specifically expressed in the differentiated foregut cells (Fig. 4C and D). Defective Proventriculus (Dve) is a transcription factor, specifically expressed in differentiated anterior midgut cells (Fig. 4E and F). Fu-positive cells (white dotted line in Fig. 4D) and Dve-positive cells (white dotted lines in Fig. 4F) were readily visible within the GFP-positive MARCM clones derived from the GaSCs cells in the cardia.
Figure 4.
The GaSCs are multipotent gastric stem cells. (A and B) MARCM clones (4 days) to see the expression of Ptc in the clones. Ptc expressing cells are stem cells in (A and B). Cardia was stained with Ptc (red), GFP (green) and DAPI (blue). White dotted line in (B) shows the expression of Ptc is limited in stem cells (C) The cardia of wg-Gal4 UAS-GFP flies were stained with GFP (green) and Fu (red in C). (D) The flies with MARCM clones were dissected and stained with anti-GFP (green) and anti-Fu (red in D). (E) The Stat92E-GFP flies were stained with GFP (green) and Dve (red in E) antibodies. (F–H) The flies with MARCM clones were dissected and stained with anti-GFP or anti-Dve (red in F) or anti-Na/K ATPase α-subunit α5 (red in G) or anti-MEF2 (red in H). White line highlighted Fu-positive (D) or Dve-positive (F) cells. Black arrow in G points to Na/K ATPase α-subunit α5-positive cells. Anterior is at the top in all panels. Scale bars: 5 µm (A, B); 10 µm (C–H).
In mammals, the gastric stem cells generate four main cell lineages: parietal, zymogen, enteroendocrine, and mucous cells.8 The parietal cells can be labeled by antibodies to the H+,K+-ATPase α-subunit,8 which is evolutionarily conserved. We stained fly gut bearing MARCM clones with antibodies against the Drosophila H+, K+-ATPase α-subunit (α5) and found that some GFP-positive cells were also labeled by the α5 antibodies (Fig. 4G), suggesting that these cells may be the fly parietal cells. We also stained cardia bearing MARCM clones with anti-Mef2 (a marker of muscle cells) and found that none of the GFP-positive cells was Mef2-positive (Fig. 4H).
In summary, the above results suggest that the stem cells at the F/M junction are multipotent gastric stem cells and their daughter cells either migrate up toward anterior midgut or migrate downward to become esophagus and crop cells. This may be similar to the mammalian system, where the gastric stem cell is located in the isthmus of the tubular unit and their daughter cells either migrate up toward the lumen and become pit cells or migrate downward to become one of the cell types of the fundic gland.9
The GaSCs are asymmetrically dividing.
To further analyze the self-renewal or division of GaSCs, we conducted two types of experiments. In the first experiment, we specifically traced them using the FLP-out method (Fig. 5A) similar to the technique used in Figure 3. Gal80ts suppresses Gal4 activity at a “permissive” temperature (18°C). When cultured at 18°C, these flies grow to adulthood with no obvious phenotype, and no RFP or EGFP expression was found (Fig. 5A, N = 20). We then shifted the adult flies to a “restrictive” temperature (29°C). After 12 h at 29°C, the first RFP appeared (Fig. 5C, N = 10). After one day, we could clearly see that EGFP-marked cells were budding out from RFP/EGFP, co-expressed parent cells (Fig. 5D and D′, N = 15). After two days, all of the RFP-positive cells were also EGFP-positive cells, and we could see a greater number of EGFP-marked cells budding out of RFP cells (white dotted line, Fig. 5E, E' and F, N = 20; and Fig. 5G, merged-z-stack). We also dissected the flies at six or ten days to two weeks and found the continued labeling of GFP+ cells in different regions of the cardia (data not shown).
Figure 5.
Asymmetric division of GaSCs. (A) Schematic diagram of the cell lineage marking system. After shifting the flies with genotype UAS-FLP/+; wg-Gal4 UAS-RFP/Act5C-FRT-y+-FRT-EGFP; tub-Gal80ts/+ to 29°C, the Gal4 was activated and drove RFP (red) and FLP recombinase expression. The FLP recombinase then removed the ‘FLP-out’ cassette so that the constitutive actin5C promoter drove EGFP expression (green) permanently within all subsequent daughter cells. Flies with the genotype UAS-FLP/+; wg-Gal4 UAS-RFP/ Act5C-FRT-y+-FRT-EGFP; tub-Gal80ts/+ (B–G, I, I′) were cultured at the permissive temperature (18°C, B) and then shifted to the restricted temperature (29°C, C–G, I, I′). At 18°C no EGFP/RFP expression (B). Flies shifted to 29°C for 12 hrs (C), 1 day (D, high magnification in D′), 2 days (E, E′, high magnification in E″, F, G). (G) 2 days (Z-stack merged). Division of GaSCs is highlighted in E′ (arrow) and E″ F (round dotted lines). In B–G the cardia was dissected and examined under confocal microscope without staining (live imaging). (H, H′) The flies with MARCM clones were dissected and stained with anti-GFP (green), anti-Ptc (red) and DAPI (blue). (I, and high magnification in I′) UAS-FLP/+; wg-Gal4 UAS-RFP/Act5C-FRT-y+-FRT-EGFP; tub-Gal80ts/+ flies (2 days) were stained with RFP (red), GFP (green), Ptc (purple) and DAPI (blue). Ptc is expressed in GaSCs as highlighted by dotted lines in H, H′. Anterior is at the top in all panels except in I, I′, where anterior is in right. Scale bars: 10 µm (B–E); 2 µm (E′, F, G, D′ E″, I′); 5 µm (H, H′ I).
In the second experiment, we use the MARCM system25 to trace the labeled cells and stained the cardia with specific antibodies for GFP, Ptc, and DAPI. In cardia fixed four days after clone induction, we can frequently detect a pair of partially connected GFP-marked cells (Fig. 5H and H′). In the pair, only one cell expresses the stem cell marker Ptc. Further, we also stained the flies of RFP/EGFP lines with Ptc antibody (Fig. 5I and I′) and found that asymmetric distribution of Ptc expression between stem cell and daughter cells. Cells with both RFP/EGFP express Ptc, and cells budding out of the stem cell zone do not express a stem cell marker Ptc (Fig. 5I and I′ and Sup. Fig. 3J). In summary, the above results suggest that GaSCs are dividing asymmetrically to produce one new GaSC and one differentiating daughter cell.
Wg signaling regulates GaSC self-renewal and maintenance.
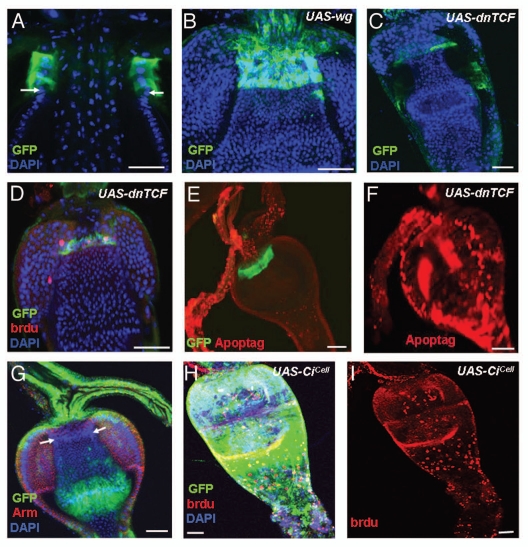
The wg-Gal4 UAS-GFP is expressed in the GaSCs and proliferating progenitor cells (Figs. 1D, D′, G, H, and 6A). To address the role of Wg signaling in regulating GaSCs, we overexpressed wg and a dominant-negative form of TCF (pangolin), a downstream transcription factor in the Wg signaling pathway,44 using the Gal4/UAS system45 combined with tubGal80ts.43 The overexpression of wg using actin5C-Gal4 resulted in a marked expansion of the number of GaSCs marked by Stat92E-GFP (compare Fig. 6B with Fig. 1A–C, N = 10). At 29°C after 2–4 days, the over- expression of dominant-negative TCF (dnTCF) strongly reduced the number of GaSCs (Fig. 6C, N = 27; 60% of the flies) as the numbers of Stat92E-GFP cells were decreased. When the BrdU incorporation assay described above was used in the dnTCF-overexpressing flies (N = 15), we found decreased proliferation of the GaSCs (compare Fig. 6D with Fig. 2F). Meanwhile, the significantly higher numbers of Apoptag-positive cells among the GaSCs in the dnTCF-overexpressing flies (N = 10), suggesting an increase in the apoptosis (compare Fig. 6E with 6F). These data together suggest that Wg signaling is required for GaSCs' self-renewal and maintenance.
Figure 6.
Wg and Hh signaling regulate GaSCs maintenance and differentiation, respectively. The cardia of a wg-Gal4 UAS-GFP fly was stained with anti-GFP (green) and DAPI (blue). White arrows point to the F/M junction. (B) Flies with the genotype Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts/UAS-wg were cultured at the restricted temperature (29°C) for 4 days. The cardia was stained with anti-GFP (green), and DAPI (blue). (C) Flies with the genotype Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts/UAS-dnTCF were cultured at the restricted temperature (29°C) for 4 days. The cardia was stained with anti-GFP (green) and DAPI (blue). (D) Flies with the genotype Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts/UAS-dnTCF were given food containing BrdU for 5 days, followed by a 5-day chase at the restricted temperature (29°C). The cardia was stained with anti-GFP (green), anti-BrdU (red), and DAPI (blue). (E) The cardia of a wg-Gal4 UAS-GFP flies (as control) were stained with Apoptag (red) GFP (green) for dying cells. (F) Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts/ UAS-dnTCF flies were stained for Apoptag (red) for dying cells. (G) The cardia of an hh-Gal4 UAS-GFP fly stained with anti-GFP (green), anti-Arm (red), and DAPI (blue). White arrows point to the F/M junction. (H, I) Flies with the genotype Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts//UAS-CiCell were given food containing BrdU for 5 days, followed by a 5-day chase at the restricted temperature (29°C). The cardia was stained with anti-GFP (green), anti-BrdU (red), and DAPI (blue). Anterior is at the top in all panels. Scale bars: 10 µm (A–D, G–I); 20 µm (E, F).
Hh signaling regulates GaSCs' differentiation.
Hh-Gal4-UAS-GFP and Fu are specifically expressed in the differentiated cells in the foregut side of the cardia (outside of the Stat92E-GFP zone; Figs. 4C, D and 6G) while Ptc is specifically expressed in the GaSCs and some of the immediate progenitor cells (Figs. 1D and D′; 4A and B; 5G and H). We therefore examined the role of Hh signaling in GaSC regulation. We overexpressed a repressor isoform of Cubitus interruptus (Cicell), a transcription factor that acts downstream in the Hh signaling pathway.46,47 The overexpression of Cicell resulted in a significantly enlarged cardia, in two weeks at 29°C (compare Sup. Fig. 4B with C). The cardia in the wild-type flies was 32.34 µM wide on average (Sup. Fig. 4B, N = 20), while that in flies overexpressing Cicell was 57.67 µM wide on average (Sup. Fig. 4C, N = 23). The enlarged cardia was almost entirely filled with small Stat92E-GFP-positive GaSCs (Fig. 6H and Sup. Fig. 4A, N = 20). The Cicell-expressing flies actively incorporated BrdU into all of the cells of the cardia (Fig. 6H and I, N = 17), suggesting that blocking the Hh signal stimulates GaSC proliferation and prevents their differentiation.
JAK-STAT signaling regulates GaSC proliferation.
Drosophila JAK-STAT signal transduction pathway regulates stem cell self-renewal in several other systems.36,48–51 These findings and our observation that Stat92E-GFP is specifically expressed in the GaSCs (Fig. 1A–C) prompted us to examine the function of JAK-STAT signaling in GaSC regulation. The Drosophila JAK-STAT signaling pathway has four major components.52 A secreted glycoprotein, unpaired (upd), is an in vivo ligand that initiates signaling by binding its receptor,53 a transmembrane protein, Domeless [Dome; also called Master of Marelle (MOM)].54–58 Dome signals through the only Drosophila JAK kinase homologue, Hopscotch (Hop),59 and the only Drosophila STAT homologue, Stat92E.53,54 The activated Stat92E then enters the nucleus to activate its target genes' transcription.52
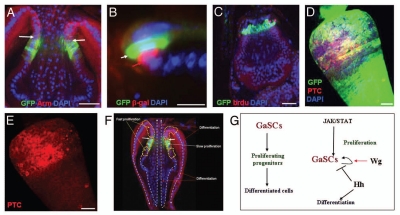
We first examined the expression of JAK-STAT pathway components in the cardia and found that upd (as assessed upd-Gal4 UAS-GFP) was expressed in zone 3, immediately anterior to the Stat92E-GFP cells (Fig. 7A; more clearly shown in Fig. 7B), while its receptor dome (as assessed by dome-Gal4 UAS-GFP) is expressed in the Stat92E-GFP cells (Sup. Fig. 4C). We then used a temperature-sensitive allele of Stat92E (Stat92EF)54 to study the function of JAK-STAT signaling in the regulation of the GaSCs. Because this allele has a closely linked background lethal mutation, we used flies that were Stat92EF heterozygous with Stat92E06346. The Stat92EF/Stat92E06346 (referred to as Stat92Ets from now on) genotype is lethal at 29°C, but viable and fertile at 18°C and at room temperature (RT, ∼22°C.60 data not shown). Adult Stat92E-GFP/+; Stat92Ets female flies were first cultured at 29°C for three days and then fed a BrdU-containing diet for five days, followed by a five day chase. Although the number of GFP-positive cells were relatively normal, the amount of BrdU incorporation was dramatically lower in the Stat92E-GFP/+; Stat92Ets flies (N = 17) than in wild-type flies (N = 20) (Fig. 7C versus Fig. 1F). This result suggests that JAK-STAT signaling regulates GaSC proliferation. We next overexpressed upd using the Gal4/UAS system44 in combination with tubGal80ts43. The overexpression of upd resulted in a significant expansion of the number of GaSCs marked by Stat92E-GFP and Ptc (Fig. 7D and E, N = 16), compared with wild-type flies (Fig. 1A, N = 18). These data further support the idea that JAK-STAT signaling is required for GaSCs proliferation.
Figure 7.
JAK-STAT signaling regulates GaSCs proliferation. (A) The cardia of a upd-Gal4 UAS-GFP fly was stained with anti-GFP (green), anti-Arm (red), and DAPI (blue). White arrows point to the F/M junction. (B) The cardia of upd-Gal4 UAS-GFP/ STAT92E-lacZ flies were stained with anti-GFP (green), anti-β-gal (red), and DAPI (blue). White arrows point to the upd-Gal4 UAS-GFP (green) and red arrow point to the STAT92E-lacZ (red). (C) Flies with the genotype Stat92E-GFP/+; Stat92E06346/Stat92EF were given food containing BrdU for five days, followed by a five day chase at the restricted temperature (29°C). The cardia was stained with anti-GFP (green), anti-BrdU (red), and DAPI (blue). (D,E) Flies with the genotype Stat92E-GFP/+; Act-Gal4/+; tub-Gal80ts/UAS-upd were cultured at the restricted temperature (29°C) for four days. The cardia was stained with anti-GFP (green), anti-Ptc (red in D–E), and DAPI (blue). (F,G) Diagram showing behavior and regulation of GaSCs in Drosophila. (F) The slow proliferative GaSCs first give rise to the fast proliferative progenitors in both foregut and anterior midgut. The progenitors then migrate and differentiate into the terminally differentiated cell types in both crop and midgut. (G) Schematic diagram summarizing the functions of JAK-STAT, Wg, and Hh pathways in regulating GaSCs′ behaviors in Drosophila. Anterior is at the top in all panels. Scale bars: 10 µm (A, C–F); 5 µm (B).
Discussion
The gastric stem cells may have similar behavior in Drosophila and mouse.
In this study, we identified multipotent gastric stem cells (GaSCs) at the junction of the adult Drosophila foregut and midgut. The GaSCs express the Stat92E-GFP reporter, wg-Gal4 UAS-GFP, and Ptc, and are slowly proliferating. The GaSCs first give rise to the fast proliferative progenitors in both foregut and anterior midgut. The foregut progenitors migrate downward and differentiate into crop cells. The anterior midgut progenitors migrate upward and differentiate into midgut cells (Fig. 7F). However, at this stage because of limited markers availability, complex tissues system at cardia location, we are not sure how many types of cells are produced and how many progenitor cells are in the cardia. Our clonal and molecular markers analysis suggest that cardia cells are populated from gastric stem cells at the F/M junction; however, we cannot rule out that there may be other progenitor cells with locally or limited differential potential may also take part in cell replacement of cardia cells. Nevertheless, the observed differentiation pattern of GaSCs in Drosophila may be similar to that of the mouse gastric stem cells.10,12 The gastric stem cell in mouse is located at the neck-isthmus region of the tubular unit. They produce several terminally differentiated cells with bidirectional migration, in which, upward migration towards lumen become pit cells, and downward migration results in fundic gland cells.10,12
Signal transduction pathways that regulate gastric stem cell activity.
We found that three signal transduction pathways differentially regulate the GaSC self-renewal or differentiation (Fig. 7G). The loss of JAK-STAT signaling resulted in quiescent GaSCs; that is, the stem cells remained but did not incorporate BrdU or rarely incorporated BrdU. In contrast, the amplification of JAK-STAT signaling resulted in GaSCs' expansion. These observations indicate that JAK-STAT signaling regulates GaSCs' proliferation. On the other hand, the loss of Wg signaling resulted in GaSC loss, while the amplification of Wg resulted in GaSC expansion, indicating that Wg signaling regulates GaSC self-renewal and maintenance. Finally, the loss of Hh signaling resulted in GaSC expansion at the expense of differentiated cells, indicating that Hh signaling regulates GaSC differentiation.
The JAK-STAT signaling has not been directly connected to gastric stem cell regulation in mammal. However, the quiescent gastric stem cells/progenitors are activated by interferon γ (an activator of the JAK-STAT signal transduction pathway),61 indicating that JAK-STAT pathways may also regulate gastric stem cell activity in mammal. Amplification of JAK-STAT signaling resulted in expansion of stem cells in germline, posterior midgut and malpighian tubules of adult Drosophila.22,36,49 In the mammalian system, it has been reported that activated STAT contributes to gastric hyperplasia and that STAT signaling regulates gastric cancer development and progression.62 Wnt signaling has an important function in the maintenance of intestinal stem cells and progenitor cells in mice63,64 and hindgut stem cells in Drosophila,17 and its activation results in gastrointestinal tumor development.63 Tcf plays a critical role in the maintenance of the epithelial stem cell.64 Mice lacking Tcf resulted in depletion of epithelial stem-cell compartments in the small intestine64 as well as being unable to maintain long-term homeostasis of skin epithelia.65 A recent study even demonstrates that the Wnt target gene Lgr5 is a stem cell marker in the pyloric region and at the esophagus border of the mouse stomach.11 Further, it has been found that overactivation of the Wnt signaling can transform the adult Lgr5+ve stem cells in the distal stomach,11 indicating that Wnt signaling may also regulate gastric stem cell self-renewal and maintenance in the mammal. Sonic Hedgehog (Shh) and its target genes are expressed in the human and rodent stomach.12 Blocking Shh signaling with cyclopamine in mice results in an increase in the cell proliferation of gastric gland, suggesting that Shh may also regulate the gastric stem cell differentiation in mice.11,12 These data together suggest that the genetic control of the Drosophila GaSCs may be similar to that of the mammalian gastric stem cells.
The potential GaSCs niche.
In most stem cell systems that have been well characterized to date, the stem cells reside in a specialized microenvironment, called a niche.66 A niche is a subset of neighboring stromal cells and has a fixed anatomical location. The niche stromal cells often secrete growth factors to regulate stem cell behavior, and the stem cell niche plays an essential role in maintaining the stem cells, which lose their stem-cell status once they are detached from the niche.67
Loss of the JAK-STAT signaling results in the GaSCs being quiescent; the stem cells remain but do not proliferate or rarely proliferate (Fig. 7C). The Dome receptor is expressed in GaSCs (Sup. Fig. 3C), while the ligand Upd is expressed in adjacent cells (Fig. 7B). Upd-positive hub cells function as a germline stem-cell niche in the Drosophila testis.49,50 Further, we demonstrated that overexpression of upd results in GaSC expansion (Fig. 7D), suggesting that the Upd-positive cells may function as GaSCs niche. Furthermore, while the Stat92E-GFP expression is regulated by the JAK-STAT signaling in other systems,35 its expression at the F/M junction seems independent of the JAK-STAT signaling because the Stat92E-GFP expression is not significantly disrupted in the Stat92Ets mutant flies (Fig. 7C), suggesting that the GaSCs may have unique properties.
The stomach epithelium undergoes continuous renewal by gastric stem cells throughout adulthood. Disruption of the renewal process may be a major cause of gastric cancer, the second leading cause of cancer-related death worldwide,35 yet the gastric stem cells and their regulations have not been fully characterized. A more detailed characterization of markers and understanding of the molecular mechanisms control gastric stem cell behavior will have a major impact on future strategies for gastric cancer prevention and therapy. The information gained from this report may facilitate studies of gastric stem cell regulation and transformation in mammal.
Materials and Methods
Drosophila stocks.
Oregon R was used as the wild-type strain. The other fly stocks used in this study, described either in FlyBase or as otherwise specified, were as follows: Stat92E reporter-GFP (provided by G. Baeg),35 hh-Gal4 (provided by K. Basler), wg-Gal4 (provided by JP. Vincent and V. Hartenstein), esg-Gal4 (provided by S. Hyashi), stat92E-lacZ68, byn-Gal4 (provided by J. Merriam), ppl-Gal4 (provided by M. Pankratz), UAS-Cicell (provided by K. Basler), Act5C-FRT-Draf-FRT-tau-lacZ (provided by J. Skeath), and X-15-29 and X-15-33, used for clonal analysis (provided by T. Xie).39 Act5C-FRT-y+-FRT-EGFP was generated in our laboratory. UAS-upd and stat92E06346 were described previously;53,58 stat92EF, a temperature-sensitive stat92E allele55 was provided by S. DiNardo; and ptc-lacZ; Act-Gal4, UAS-Flp, UAS-wg, UAS-dnTCFΔN, UAS-RFP, tub-GAL80ts, AyGal4 UAS-GFP, SM6, hs-Flp, FRT82B;-tub-Gal80 and FRT82B were obtained from the Bloomington stock center.
Flies were raised on standard Drosophila medium at room temperature (22°C) or in incubators at 18°, 25° or 29°C. Genes and mutations that are not described in the text can be found at Flybase (flybase.bio.indiana.edu).
Temporal overexpression of genes and generation of clones using the Lacz, MARCM and flip-out techniques.
The tubulin (tub)-GAL80ts in combination with the conventional Gal4/UAS system was used for the temporal overexpression of genes. Adult flies, which had been raised at 18°C, were shifted to 29°C to inactivate GAL80ts, which inhibits the Gal4-driven expression of genes following the UAS. The genotypes of the flies used for the temporal overexpression of genes were Stat92E-GFP/+; ActGal4/+; tub-GAL80ts/UAS-wg, Stat92E-GFP/+; Act-Gal4/+; tubGAL80ts/UAS-Cicell, Stat92E-GFP/+; Act-Gal4/+; tub-GAL80ts/UAS-dTCFΔN, and Stat92E-GFP/+; Act-Gal4/+; tub-GAL80ts/UAS-upd. The genotype of LacZ lineage labeling, hsFLP;X-15-29/X15-33.39 3–5-day-old adult females were heat-shocked for 40 min in a 37°C water bath. After heat shock, flies were transferred to fresh food every day, and the gut was processed for staining at the indicated times. The genotype for MARCM clone induction was AyGal4 UAS-GFP/SM6, hs-Flp; FRT82B tub-GAL80/FRT82B. 3–5-day-old adult females were subjected to heat shock twice (37°C, 60 min each) with an interval of 8–12 h between heat shocks. The flies were transferred to fresh food once a day after the heat-shock treatments, and the gut was processed for staining at the indicated times. For lineage tracing using the Flp-out technique, adults with the following genotypes were generated: UAS-Flp/+; Act5C-FRT-Draf-FRT-tau-lacZ/wg-Gal4 UAS-GFP; tub-GAL80ts/+or UAS-Flp/+; Act5C-FRT-y+-FRT-EGFP/wg-Gal4 UAS-RFP; tub-GAL80ts/+. Crosses were established and cultured at 18°C, the permissive temperature, until adulthood. The progeny were divided into two equal pools; the control group was cultured at 18°C and the experimental group was shifted to 29°C. Flies kept at 29°C were dissected and stained after the indicated times.
BrdU labeling.
Female flies were starved at 25°C and then fed 100 mM BrdU (Invitrogen) in a paste of yeast granules, sucrose, and water, for 5 days. For the chase experiment, flies were given normal fly food without BrdU for another 5 or 17 days before dissection. The gut was dissected, fixed with 4% formaldehyde, treated for 30 min at 37°C with DNase, and stained with anti-BrdU (Invitrogen).
Immunofluorescence staining and microscopy.
The gut was dissected and stained as described previously.36,67,69–72 Confocal images were obtained using the Zeiss LSM510 system and processed with Adobe Photoshop CS2. The following antibodies were used: rabbit anti-β-gal (1:1000; Cappel), mouse anti-β-gal (1:100; Promega), rabbit anti-Odd (1:400; gift from J. Skeath), rabbit anti-MEF2 (1:1000; gift from B. Patterson), anti-Dve (1:1000; gift from F. Matsuzaki), mouse anti-Armadillo N7A1 [1:4; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Na/K ATPase α-subunit α5 (1:50; DSHB), mouse anti-Fu (1:100; DSHB), mouse anti-Ptc (1:100; DSHB), mouse anti-BrdU (1:100; BD Biosciences or Invitrogen), rabbit anti-GFP (1:200; Invitrogen), and mouse anti-GFP (1:100; Invitrogen), chicken anti-GFP (Abcam), and rabbit anti-RFP (Invitrogen). Secondary antibodies were goat anti-mouse and goat anti-rabbit IgG conjugated to Alexa 488 or Alexa 568 (1:400; Invitrogen). DAPI (Sigma) was used to stain DNA.
Detection of apoptosis.
We used an Apoptag Red in Situ Detection Kit (Chemicon) to detect cell death in the cardia.
Acknowledgements
We thank G. Baeg, K. Basler, J.P. Vincent, V. Hartenstein, S. Hyashi, J. Merriam, M. Pankratz, J. Skeath, T. Xie, S. DiNardo, and the Bloomington stock center for fly stocks; J. Skeath, F. Matszaki, and B. Patterson for antibodies; Dr. Stephen Lockett and Optical Microscopy and Analysis Laboratory (OMAL) staff for help with the confocal microscopy; Dr. David King for help in identifying cell types in the cardia; and Maritta Perry Grau for editing the manuscript. This research was supported by the Intramural Research Program, National Cancer Institute of the National Institutes of Health.
Supplementary Material
References
- 1.Brittan M, Wright NA. Stem cell origin of cell lineages, proliferative units, and cancer in the gastrointestinal tract. In: Sell S, editor. Stem Cells Handbook Totowa. NJ: Humana Press; 2002. pp. 329–343. [Google Scholar]
- 2.McDonald SA, Preston SL, Lovell MJ, Wright NA, Jankowski JA. Mechanisms of disease: from stem cells to colorectal cancer. Nat Clin Pract Gastroenterol Hepatol. 2006;3:267–274. doi: 10.1038/ncpgasthep0473. [DOI] [PubMed] [Google Scholar]
- 3.Wright NA. Epithelial stem cell repertoire in the gut: clues to the origin of cell lineages, proliferative units and cancer. Int J Exp Path. 2000;81:117–143. doi: 10.1046/j.1365-2613.2000.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 7.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 8.Bjerknes M, Cheng H. Multipotential stem cells in adult mouse gastric epithelium. Am J Physiol: Gastrointest Liver Physiol. 2002;283:G767–G777. doi: 10.1152/ajpgi.00415.2001. [DOI] [PubMed] [Google Scholar]
- 9.Karam SM, Straiton T, Hassan WM, Leblond CP. Defining epithelial cell progenitors in the human oxyntic mucosa. Stem Cells. 2003;21:322–336. doi: 10.1634/stemcells.21-3-322. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann W. Regeneration of the gastric mucosa and its glands from stem cells. Curr Med Chem. 2008;15:3133–3144. doi: 10.2174/092986708786848587. [DOI] [PubMed] [Google Scholar]
- 11.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es J H, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 12.van den Brink G R, Hardwick JC, Tytgat GNJ, Brink MA, ten Kate FJ, van Deventer SJH, Peppelenbosch MP. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology. 2001;121:317–328. doi: 10.1053/gast.2001.26261. [DOI] [PubMed] [Google Scholar]
- 13.Kouznetsova I, Kalinski T, Meyer F, Hoffmann W. Self-renewal of the human gastric epithelium: new insights from expression profiling using laser microdissection. Mol BioSyst. 2011;7:1105–1112. doi: 10.1039/C0MB00233J. [DOI] [PubMed] [Google Scholar]
- 14.Fox DT, Spradling AC. The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell. 2009;5:290–297. doi: 10.1016/j.stem.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;39:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 16.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 17.Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signaling. Nature. 2008;454:651–655. doi: 10.1038/nature07156. [DOI] [PubMed] [Google Scholar]
- 18.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 19.Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Singh SR, Hou SX. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. 2010;109:992–999. doi: 10.1002/jcb.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development. 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- 26.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 28.Lin G, Xu N, Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of Drosophila intestinal stem cells. J Mol Cell Biol. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- 29.King DG. Cellular organization and peritrophic membrane formation in the cardia (proventriculus) of Drosophila melanogaster. J Morphol. 1988;196:253–282. doi: 10.1002/jmor.1051960302. [DOI] [PubMed] [Google Scholar]
- 30.King DG. The origin of an organ: Phylogenetic analysis of evolutionary innovation in the digestive tract of flies (Insecta: Diptera) Evolution. 1991;45:568–588. doi: 10.1111/j.1558-5646.1991.tb04330.x. [DOI] [PubMed] [Google Scholar]
- 31.Skaer H. The alimentary canal. In: Bate M, Martinez A, editors. The Development of Drosophila melanogaster. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 941–1012. [Google Scholar]
- 32.Fuss B, Josten F, Feix M, Hoch M. Cell movements controlled by the Notch signalling cascade during fore-gut development in Drosophila. Development. 2004;131:1587–1595. doi: 10.1242/dev.01057. [DOI] [PubMed] [Google Scholar]
- 33.Jung S, Evans CJ, Uemura C, Banerjee U. Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev Biol. 2004;267:181–189. doi: 10.1016/j.ydbio.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Pankratz MJ, Hoch M. Control of epithelial morphogenesis by cell signaling and integrin molecules in the Drosophila foregut. Development. 1995;121:1885–1898. doi: 10.1242/dev.121.6.1885. [DOI] [PubMed] [Google Scholar]
- 35.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Singh SR, Liu W, Hou SX. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell. 2007;1:191–203. doi: 10.1016/j.stem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci USA. 2006;103:8402–8407. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 39.Harrison DA, Perrimon N. A simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3:424–433. doi: 10.1016/0960-9822(93)90349-s. [DOI] [PubMed] [Google Scholar]
- 40.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 41.Jung S, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 42.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 43.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 44.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 45.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 46.Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9:501–510. doi: 10.1016/j.devcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 50.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 51.Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arbouzova NI, Zeidler MP. JAK/STAT signaling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:1605–1616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 53.Hou XS, Melnick MB, Perrimon N. marelle acts downstream of the Drosophila hop/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 54.Yan R, Small S, Desplan C, Dearolf CR, Darnell JJ. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 55.Baksa K, Parke T, Dobens LL, Dearolf CR. The Drosophila STAT protein, Stat92E, regulates follicle cell differentiation during oogenesis. Dev Biol. 2002;243:166–175. doi: 10.1006/dbio.2001.0539. [DOI] [PubMed] [Google Scholar]
- 56.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 58.Chen HW, Chen X, Oh S, Marinissen MJ, Gutkind JS, Hou XS. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- 60.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 61.Qiao XT, Ziel JW, McKimpson W, Madison BB, Todisco A, Merchant JL, et al. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology. 2007;133:1989–1998. doi: 10.1053/j.gastro.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong W, Wang L, Yao JC, Ajani JA, Wei D, Aldape KD, et al. Expression of activated signal transducer and activator of transcription 3 predicts expression of vascular endothelial growth factor in and angiogenic phenotype of human gastric cancer. Clin Cancer Res. 2005;11:1386–1393. doi: 10.1158/1078-0432.CCR-04-0487. [DOI] [PubMed] [Google Scholar]
- 63.Oguma K, Oshima H, Aoki M, Uchio R, Naka K, Nakamura S, et al. Activated macrophages promote Wnt signalling through tumour necrosis factor-alpha in gastric tumour cells. EMBO J. 2008;27:1671–1681. doi: 10.1038/emboj.2008.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morrison SJ, Spradling AC. Stem cell and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Singh SR, Zheng Z, Oh SW, Chen X, Edwards K, Hou SX. A Rap-GEF/Rap GTPase signaling controls stem cell maintenance through regulating adherens junction positioning and cell adhesion in Drosophila testis. Dev Cell. 2006;10:117–126. doi: 10.1016/j.devcel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Zeidler MP, Perrimon N, Strutt DI. The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr Biol. 1999;9:1363–1372. doi: 10.1016/s0960-9822(00)80081-0. [DOI] [PubMed] [Google Scholar]
- 69.Singh SR, Zhen W, Zheng Z, Wang H, Oh SW, Liu W, et al. The Drosophila homologue of the human tumor suppressor gene BHD regulates male germline stem cell maintenance and functions downstream of Dpp. Oncogene. 2006;25:5933–5941. doi: 10.1038/sj.onc.1209593. [DOI] [PubMed] [Google Scholar]
- 70.Zeng X, Singh SR, Hou D, Hou SX. Tumor suppressors Sav/Scrib and oncogene Ras regulate stem-cell transformation in adult Drosophila malpighian tubules. J Cell Physiol. 2010;224:766–774. doi: 10.1002/jcp.22179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh SR, Zheng Z, Wang H, Oh SW, Chen X, Hou SX. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J Cell Physiol. 2010;223:500–510. doi: 10.1002/jcp.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng X, Chauhan C, Hou SX. Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis. 2010;48:607–611. doi: 10.1002/dvg.20661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.