Abstract
Cytochrome P450 reductase (POR) is a microsomal electron transport protein essential to cytochrome P450-mediated drug metabolism and sterol and bile acid synthesis. The conditional deletion of hepatic POR gene expression in mice results in a marked decrease in plasma cholesterol levels counterbalanced by the accumulation of triglycerides in lipid droplets in hepatocytes. To evaluate the role of cholesterol and bile acid synthesis in this hepatic lipidosis, as well as the possible role of lipid transport from peripheral tissues, we developed a stable, small interfering RNA (siRNA)-mediated cell culture model for the suppression of POR. POR mRNA and protein expression were decreased by greater than 50% in McArdle-RH7777 rat hepatoma cells 10 days after transfection with a POR-siRNA expression plasmid, and POR expression was nearly completely extinguished by day 20. Immunofluorescent analysis revealed a marked accumulation of lipid droplets in cells by day 15, accompanied by a nearly 2-fold increase in cellular triglyceride content, replicating the lipidosis seen in hepatic POR-null mouse liver. In contrast, suppression of CYP51A1 (lanosterol demethylase) did not result in lipid accumulation, indicating that loss of cholesterol synthesis is not the basis for this lipidosis. Indeed, addition of cholesterol to the medium appeared to augment the lipidosis in POR-suppressed cells, whereas removal of lipids from the medium reversed the lipidosis. Oxysterols did not accumulate in POR-suppressed cells, discounting a role for liver X receptor in stimulating triglyceride synthesis, but addition of chenodeoxycholate significantly repressed lipid accumulation, suggesting that the absence of bile acids and loss of farnesoid X receptor stimulation lead to excessive triglyceride synthesis.
Introduction
Cytochrome P450 reductase (POR) is the obligate electron donor to the cytochromes P450 (P450s) found on the endoplasmic reticulum, which includes most P450s involved in drug metabolism but also P450s essential to physiological processes such as cholesterol, steroid, and bile acid synthesis. Indeed, the genomic inactivation of POR in mice results in embryonic lethality by day 14 (Shen et al., 2002), and developmental defects associated with POR deletion seem to be due in large part to the loss of cholesterol synthesis (Otto et al., 2003; Schmidt et al., 2009). To explore the role of microsomal P450s in drug metabolism and circumvent the embryonic lethality associated with genomic deletion of POR, the conditional, liver-specific deletion of P450 reductase was reported by two groups in 2003 (Gu et al., 2003; Henderson et al., 2003). Unexpectedly, these hepatic POR-null mice developed a severe hepatic lipidosis characterized by the accumulation of diglycerides, triglycerides, and monounsaturated fatty acids (Weng et al., 2005; Mutch et al., 2007), even while serum triglyceride and cholesterol levels were decreased. Since POR was not known to be involved in triglyceride synthesis or secretion the basis for the lipidosis was not readily apparent.
Disruption of POR activity would be expected to prevent cholesterol synthesis as well as its catabolism to bile acids. The net result in hepatic POR-null mice is a dramatic decrease in circulating cholesterol and bile acids. Because the liver serves as a depot for lipids from the peripheral tissues, it is unclear to what extent extrahepatic lipids contribute to the accumulation of triglycerides in hepatic POR-null mice. Weng et al. (2005) found an increase in the expression of a gene responsible for fatty acid uptake (Cd36) and a decrease in the expression of genes involved in fatty acid oxidation (Cpt1a, Ascl1). Mutch et al. (2007) made similar observations, concluding that although hepatic fatty acid synthesis was neither increased nor decreased in POR-null liver, lipid secretion was impaired and that this, coupled with continual uptake from the circulation, lead to the lipidosis. Indeed, Finn et al. (2009) showed that placing these mice on a fat-deficient diet completely prevented the hepatic lipidosis and the fall in circulating cholesterol and triglycerides. However, why the disruption of POR activity, manifested as a block to cholesterol and bile acid synthesis, would result in this impaired secretion of triglycerides was not elucidated. As a first step toward answering this question, we have developed a cellular model of the hepatic POR-null mouse by using cultured hepatoma cells and small interfering RNA (siRNA) to suppress POR expression. This model appears to faithfully replicate the lipidosis seen in whole animals and has allowed us to explore the role of extracellular lipids, cholesterol, and bile acids in the accumulation of triglycerides in these cells.
Materials and Methods
Construction of a POR siRNA Expression Plasmid.
siRNA sequences for the rat POR mRNA were identified using a commercial program (Integrated DNA Technologies, San Diego, CA). Three sequences were identified, and each was synthesized by Integrated DNA Technologies, along with its complementary sequence, to generate “hairpin”-formation oligonucleotides. BamHI and HindIII restriction sites were included at the ends to allow cloning into the pSilencer4.1-CMVneo siRNA expression vector (Applied Biosystems, Foster City, CA) after annealing the complementary pairs. Each plasmid was prepared and transfected into McA-RH7777 rat hepatoma cells, and POR mRNA expression was tested after 2, 4, 6, 8, and 10 days by reverse transcriptase-polymerase chain reaction (RT-PCR). The plasmid yielding the best suppression was selected for further use; the siRNA sequence corresponds to nucleotides 2372 to 2390 in the 3′-nontranslated region of the rat POR mRNA. A plasmid containing a scrambled siRNA sequence served as the wild-type control.
Transfection and Culture of Hepatoma Cells.
McArdle-RH7777 (McA-RH7777; American Type Culture Collection, Manassas, VA) rat hepatoma cells between passages 12 and 22 were cultured in six-well plates in Dulbecco's modified Eagle's medium supplemented with 1× penicillin-streptomycin mixture and 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) at 37°C under a humidified atmosphere of 5% CO2. For plasmid transfection, best results were obtained by mixing the plasmid DNA with Fugene 6 transfection reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions at a ratio of 1 μg of DNA/5 μl of reagent, allowing complexes to form for a minimum of 30 min at room temperature. The culture medium then was replaced with antibiotic-free medium containing 10% FBS, and the transfection mixture was added dropwise to the cells with gentle swirling. The medium was replaced after 24 h with standard medium; cells were split as necessary and assayed for expression beginning after 5 days. For cholesterol-supplemented medium, cholesterol was added dropwise to FBS in ethanol (5 mg/ml) with vortexing to give a final medium concentration of 50 μg/ml (Meyer et al., 1990). For the preparation of delipidated medium, FBS was mixed with an equal volume of diethyl ether with vigorous mixing, after which the ether was removed and the delipidated serum was sterile filtered for use (Slater and Robertson, 1979).
siRNA Suppression of CYP51.
Four plasmids for the suppression of rat CYP51A1 (CYP51, lanosterol demethylase) were purchased (SureSilencing shRNA; SABiosciences, Frederick, MD), and two were tested for their ability to suppress CYP51 expression in McA-RH7777 cells, following the transfection protocol described above. A plasmid containing a scrambled siRNA sequence served as the negative control. Cells transfected with the “yellow” plasmid were selected and maintained in the presence of 1500 μg/ml geneticin (Invitrogen).
Expression Analysis by RT-PCR.
Total RNA was isolated from cells grown to confluence in six-well plates, with the use of TRIzol reagent (Invitrogen), and the concentration of RNA was determined spectrophotometrically with the use of a Nanodrop instrument (Thermo Fisher Scientific, Waltham, MA). RT-PCR was performed with Superscript ΙΙ Reverse Transcriptase (Invitrogen) and quantified on an Applied Biosystems 7000 instrument using gene-specific primer sets as follows: POR, tctcacgaagacaccagtgc (forward), cgttgtttggatcttgctga (reverse); CYP51, gtgccaaatgcagttttcct (forward), tcagacagcgcttcaaacac (reverse). Dissociation curves were established for each polymerase chain reaction product. The detection and quantitation of nucleic acid levels was done by using the comparative Ct method (Livak and Schmittgen, 2001) with β-actin as the reference gene.
Immunoblot Analysis of Protein Expression.
Thirty micrograms of protein from cell lysates was fractionated by SDS-polyacrylamide gel electrophoresis on 8% gels and electroblotted to nitrocellulose (Bio-Rad, Hercules, CA). The membrane was blocked with 0.05% Tween 20 and 5% defatted milk for 1 h at room temperature and incubated in this same buffer with a rabbit polyclonal antibody to rat POR (Stressgen/Enzo Life Sciences, Plymouth Meeting, PA) or human CYP51 (Proteintech Group, Inc., Chicago, IL) overnight at 4°C with gentle shaking. The immunoblot was developed with a secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature, and the chemiluminescent image (SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher Scientific) was captured and quantified by autoradiography on a Kodak Image Station. β-Actin served as a loading control and was detected with a mouse monoclonal antibody (Sigma-Aldrich, St. Louis, MO).
Immunofluorescence Analysis.
Hepatoma cells transfected with an siRNA expression plasmid were plated on coverslips coated with collagen in 35-mm dishes at 0.5 × 106 cells per 1 ml in Dulbecco's modified Eagle's medium containing 5% FBS and 1% penicillin-streptomycin. At 5, 10, 15, and 20 days after transfection, the cells were immunostained as follows: coverslip cultures were rinsed twice with ice-cold phosphate-buffered saline (PBS) and fixed by incubation with 4% paraformaldehyde for 30 min at room temperature. The cells were washed three times for 5 min with ice-cold PBS, treated with 0.1% Triton X-100 for 8 min at room temperature, washed with PBS, and quenched with 100 mM ammonium chloride for 20 min. The cells were washed with PBS, blocked in 1% bovine serum albumin (BSA) in PBS for 30 min, and incubated for 10 min with the Nile Red stain (Sigma-Aldrich) for lipids (1:5000). Stained cells were then washed and mounted on glass slides with use of ProLong antifade reagent (Invitrogen) containing DAPI (4,6-diamidino-2-phenylindole) nuclear stain for visualization on a Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss Inc., Thornwood, NY). For detection of POR expression, coverslips were incubated with the antibody to POR (1:100) described above in 1% BSA in PBS overnight, washed with PBS containing 0.1% BSA, and incubated in the dark with a secondary antibody conjugated to a fluorochrome (Alexa Fluor488; Invitrogen) for 1 h. Images were captured at 400× unless otherwise indicated.
Gas Chromatography-Mass Spectrometry Analysis of Cellular Sterols.
For oxysterol analysis, hepatoma cells cultured for 12 days in standard medium were homogenized at 4°C for 5 min in 1 ml of absolute ethanol containing 10 μg/ml butylated hydroxytoluene. The homogenates were subjected to centrifugation at 12,000g for 20 min, the supernatant was set aside, and the pellets were resuspended and reextracted with sonication and centrifugation in a methylene chloride/methanol mixture (1:1, v/v, 1 ml) containing 10 μg/ml butylated hydroxytoluene. This supernatant fraction was combined with the ethanol extract, and the solvents were removed by evaporation under N2 gas. The extracted sterols were resuspended in 50 μl of methylene chloride/hexane (8:2, v/v) and derivatized with trimethylsilane. Mass spectra were acquired by the University of Kentucky Mass Spectrometry Facility on a Trace gas chromatograph with a DB5-ms (Agilent Technologies, Santa Clara, CA) 30 m × 0.25 mm (0.25 μm df) column coupled to a Thermo-Finnigan PolarisQ ion-trap mass spectrometer (Thermo Fisher Scientific), using selective ion monitoring. The following sterols were run as standards and monitored in the cell extracts: 22β-hydroxycholesterol, 24,25-dihydrolanosterol, 25-hydroxycholesterol, and 27-hydroxycholesterol. For measurement of lanosterol in CYP51-suppressed cells, lipids were extracted twice with 2 ml of hexane, the solvent was evaporated under N2 gas, and the samples were derivatized by resuspension in 50 μl of bis(trimethylsilyl)trifluoroacetamide/pyridine (1:1).
Analysis of Triglycerides.
Cells were cultured and treated in triplicate in 96-well plates and pooled and assayed in duplicate. Triglyceride content was determined with 30 μg of cell lysate protein using the LabAssay Triglyceride kit from Wako Pure Chemical Industries (Richmond, VA).
Results
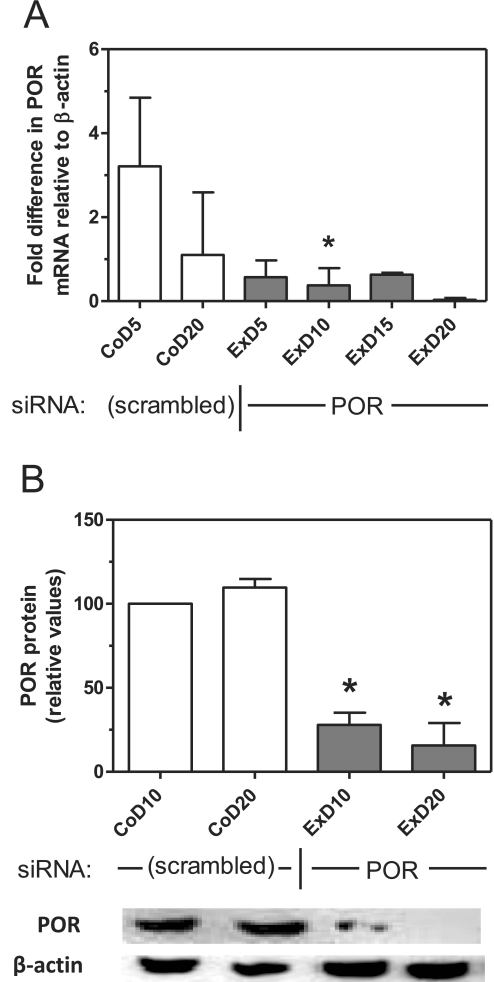
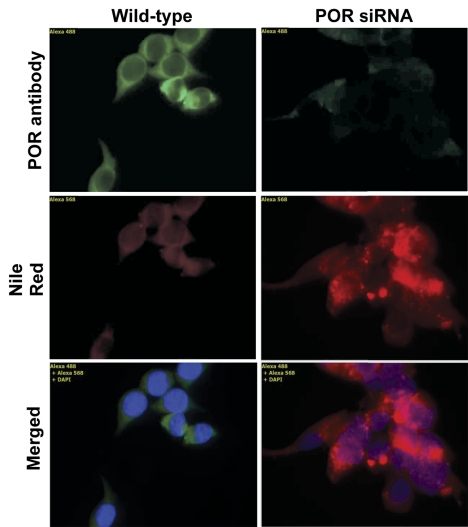
Transfection of a plasmid that expresses siRNA complementary to rat cytochrome P450 reductase mRNA into McA-RH7777 rat hepatoma cells decreased POR mRNA in these cells by greater than 80% by day 5, and this suppression was maintained through 20 days of culture (Fig. 1A). POR protein expression, measured by immunoblot analysis at days 10 and 20, was decreased to 28 and 16% of control cell levels, respectively (Fig. 1B). Immunocytochemical analysis of these cells at 20 days post-transfection with an antibody to cytochrome P450 reductase confirmed the near-complete loss of POR expression (Fig. 2). Staining of these cells with Nile Red stain for lipids demonstrated a diffuse increase in lipid content throughout the cell with numerous and prominent intracellular lipid vesicles of various sizes. Triglyceride content in POR-suppressed cells was increased at 5 and 10 days and was statistically significant at 15 days (Table 1). This pattern of lipidosis closely resembles that observed in hepatocytes of hepatic POR-null mice (Gu et al., 2003; Henderson et al., 2003).
Fig. 1.
Suppression of POR mRNA and protein in hepatoma cells. Hepatoma cells were transfected with the POR siRNA expression plasmid or a control vector (scrambled), and POR mRNA (A) and protein (B) expression were measured at various time points by RT-PCR and immunoblot analysis, respectively. An immunoblot image is shown at the bottom corresponding to the time points in B. Values represent the mean and S.D. of two determinations done in duplicate; values that are statistically different from the corresponding control are indicated by asterisks, as determined by one-way analysis of variance with Bonferroni's multiple comparisons post hoc test (p < 0.05). CoD5, control cells, day 5; ExD5, experimental (POR-suppressed) cells, day 5.
Fig. 2.
Immunofluorescence analysis of POR siRNA-transfected hepatoma cells. Hepatoma cells were transfected with the POR siRNA expression plasmid or a control vector, and cytochrome P450 reductase expression in these cells was visualized with a POR antibody followed by a fluorochrome-coupled secondary antibody. Lipid content was assessed by staining with Nile Red; nuclei are stained with DAPI nuclear stain (blue). Cells were stained 20 days after transfection; original magnification, 630×.
TABLE 1.
Triglyceride content of hepatoma cells
Data are means ± S.D., n = 2, in milligram per milligram cell lysate protein.
| Control | POR siRNA | POR siRNA DLPS | POR siRNA CDCA | CYP51 siRNA | |
|---|---|---|---|---|---|
| 5 Days | 132.9 ± 19.8 | 184.8 ± 26.2 | 139.5 ± 23.1 | ||
| 10 Days | 143.8 ± 23.1 | 184.8 ± 30.8 | 125.3 ± 21.3 | 62.5 ± 11.6* | |
| 15 Days | 154.8 ± 34.1 | 247.0 ± 39.6* | 72.9 ± 12.6* | 59.7 ± 9.3* | 34.1 ± 5.0* |
DLPS, delipidated serum.
p < 0.05, Bonferroni's multiple comparisons post hoc test with comparison to the control value for that time point.
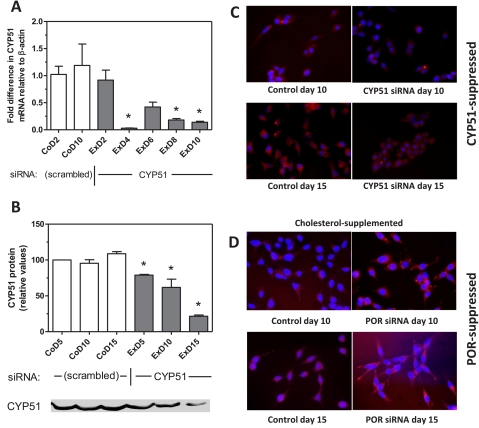
To determine whether the increase in lipid content in POR-suppressed cells was due to the loss of cholesterol synthesis, CYP51 expression was suppressed in McA-RH7777 cells by siRNA. CYP51 uniquely catalyzes the demethylation of lanosterol in the cholesterol biosynthetic pathway and has no other known biochemical roles. Moreover, it requires cytochrome P450 reductase for activity, and thus its suppression blocks cholesterol synthesis at the same step as does POR suppression. CYP51 mRNA was decreased by 86% at 10 days (Fig. 3A), and CYP51 protein was decreased by 79% at 15 days (Fig. 3B); lanosterol content in these cells increased by up to 8-fold, similar to the 10-fold increase seen in POR-suppressed cells (data not shown). However, CYP51-suppressed cells had significantly lower triglyceride content than did control cells or POR-suppressed cells (Table 1) and displayed no evidence of lipidosis (Fig. 3C), arguing that the lipidosis observed in POR-suppressed cells does not result solely from the loss of cholesterol synthesis.
Fig. 3.
Lack of cholesterol does not lead to lipid accumulation. A and B, hepatoma cells were transfected with a CYP51 siRNA expression plasmid or a control vector (scrambled), and CYP51 mRNA (A) and protein (B) expression were measured at various time points by RT-PCR and immunoblot analysis, respectively. An immunoblot image is shown at the bottom corresponding to the time points in B. C and D, lipid accumulation in CYP51-suppressed cells (C) is shown with Nile Red staining of cells after 10 and 15 days; the effect of cholesterol supplementation on POR-suppressed cells (D) is also shown. Cells were cultured in standard medium supplemented from day 2 post-transfection with 50 μg/ml cholesterol and stained with Nile Red lipid stain at days 10 and 15. Values in A and B represent the mean and S.E., n = 3, of duplicate determinations; values that are statistically different from the control group (CoD2; A) or the corresponding control (B) are indicated by asterisks, as determined by one-way analysis of variance with Dunnett's multiple comparisons post hoc test (A) or Bonferroni's multiple comparisons post hoc test (B) (p < 0.05). CoD5, control cells, day 5; ExD5, experimental (CYP51-suppressed) cells, day 5.
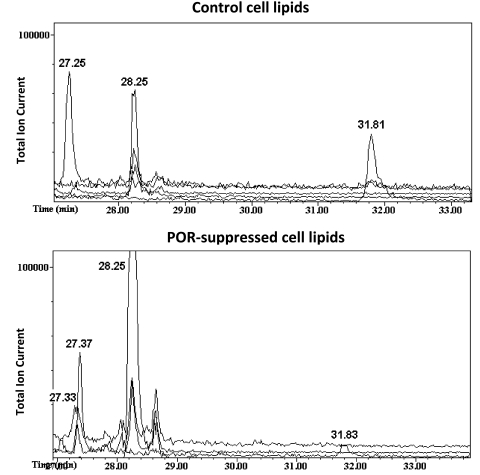
The conclusion that loss of cholesterol synthesis alone is not responsible for the lipidosis is further supported by the observation that addition of cholesterol to the culture medium appeared to augment the accumulation of lipids in POR-suppressed cells (Fig. 3D). Because cholesterol addition had no effect on the lipid content of control hepatoma cells, this observation raised the possibility that the inability of POR-suppressed cells to eliminate cholesterol might lead to the generation of oxysterols in these cells. Oxysterols serve as potent ligands for the liver X receptor (LXR), which stimulates triglyceride synthesis, accumulation, and hepatic steatosis (Kalaany and Mangelsdorf, 2006; Moya et al., 2010). To test this hypothesis, the abundance of three oxysterols whose formation does not require cytochrome P450 reductase (22β-hydroxycholesterol, 25-hydroxycholesterol, and 27-hydroxycholesterol) was determined by gas chromatography-mass spectrometry in control and POR-suppressed hepatoma cells. As shown in Fig. 4, these three oxysterols were less abundant in POR-suppressed cells than in control cells, indicating that the loss of cholesterol catabolism does not result in the accumulation of cholesterol oxidation products that might otherwise lead to an LXR-mediated stimulation of triglyceride accumulation.
Fig. 4.
Oxysterols do not accumulate in POR-suppressed cells. Control and POR-suppressed hepatoma cells were grown in normal medium for 12 days, after which lipids were extracted and gas chromatography-mass spectrometry with selective ion monitoring was used to identify 22β-hydroxycholesterol (eluting at ∼27.3 min) and 27-hydroxycholesterol (31.8 min). 25-Hydroxycholesterol could not be detected in either lipid sample (uppermost baseline tracing); lanosterol was eluted at 28.25 min, and 24,25-dihydrolanosterol was eluted at 27.37 min.
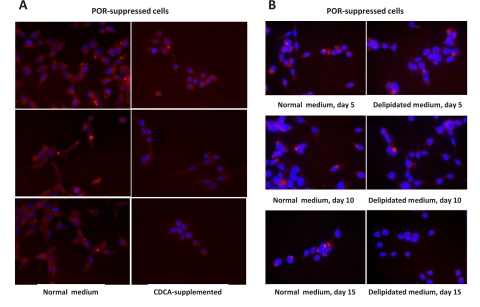
The catabolism of cholesterol to bile acids is dependent upon multiple P450 oxidations, several of which, including the regulatory step, are dependent on cytochrome P450 reductase. As bile acids act through the farnesoid X receptor (FXR) to suppress the synthesis of fatty acids, triglycerides, and bile acids (Zhang and Edwards, 2008), the loss of bile acids in POR-suppressed cells might be expected to derepress the synthesis of fatty acids and lead to the accumulation of triglycerides, as has been found in FXR-null mice (Sinal et al., 2000). To test this hypothesis, chenodeoxycholic acid (CDCA), a strong FXR ligand, was added to POR-suppressed cells. As shown in Fig. 5A, the addition of 50 μM CDCA largely prevented the accumulation of lipids in these cells and decreased triglyceride content below that of control and POR-suppressed cells (Table 1).
Fig. 5.
Bile acid supplementation or depletion of serum lipids prevents lipid accumulation. A, POR-suppressed hepatoma cells were cultured for 15 days in normal medium (left images) or medium supplemented with 50 μM CDCA (right images). B, cells were cultured in normal medium (left images) or delipidated medium (right images) for 5, 10, or 15 days. All cells were stained for lipids with Nile Red.
Although the results obtained with bile acid supplementation suggest that triglyceride synthesis is up-regulated in POR-suppressed cells, studies with hepatic POR-null mice indicated that genes involved in fatty acid and triglyceride synthesis are not up-regulated in the livers of these mice (Wang et al., 2005; Weng et al., 2005). These studies instead suggested that the accumulating triglycerides are derived largely from the circulation. To test the role of lipid uptake in POR-suppressed hepatoma cells, lipids were extracted from FBS and cells were cultured for up to 15 days in medium prepared with this delipidated serum. POR-suppressed cells initially accumulated lipids to the same extent as cells cultured in normal medium, and triglyceride levels were not statistically different from POR-suppressed or control hepatoma cells cultured in normal medium after up to 10 days of culture (Table 1). By 15 days, however, triglyceride levels were significantly elevated in POR-suppressed cells cultured in normal medium but were greatly decreased in POR-suppressed cells maintained in delipidated serum. Lipid staining of these cells demonstrated marked lipidosis in cells cultured in normal medium for 15 days and a notable lack of lipids in cells cultured in the delipidated serum (Fig. 5B). Although the viability of POR-suppressed cells was limited to 15 days in delipidated serum, these results suggest that lipid uptake contributes to the lipidosis.
Discussion
The present studies were undertaken to determine whether suppression of cytochrome P450 reductase expression in an in vitro hepatoma cell culture model would replicate the lipidosis observed in hepatic POR-null mice. The intent was to remove the whole animal physiological and compensatory components that might contribute to, or diminish the effect of, loss of cytochrome P450 reductase on lipid homeostasis in hepatocytes. Ultimately, the goal was, and is, to define the mechanism by which loss of this enzyme leads to triglyceride lipidosis; this goal is all the more interesting given that cytochrome P450 reductase is not known to play a role in triglyceride uptake, synthesis, catabolism, or secretion and all the more challenging given the several pathways in which cytochrome P450 reductase participates in lipid metabolism in the cell.
siRNA-mediated suppression of cytochrome P450 reductase in hepatoma cells was effective in generating a lipidosis that closely resembles that seen in hepatic POR-null mice. In this cell culture model, POR mRNA was decreased to 20% of control levels at 5 days post-transfection, POR protein was similarly decreased by day 10, and lipid accumulation was clearly evident by day 15. These changes match the results obtained by Finn et al. (2009) with an inducible hepatic POR-null mouse in which hepatic triglyceride accumulation was evident 5 days after loss of POR expression. Notably, plasma cholesterol levels fell in concert with POR protein levels in these mice, whereas plasma and liver triglyceride levels took an additional 5 days to reach a new equilibrium. Their results, and ours, suggest that changes in triglycerides are secondary to changes in cholesterol synthesis or catabolism.
To address the possibility that the lipidosis resulted from the loss of cholesterol synthesis in POR-suppressed cells (or from the fall in plasma cholesterol levels in hepatic POR-null mice), we supplemented the medium with cholesterol, but this appeared to augment, rather than attenuate, the lipidosis. Because hepatic cholesterol levels are normal in hepatic POR-null mice (Finn et al., 2009), it seems unlikely that a cholesterol deficit leads to the triglyceride accumulation; the cells and liver appear able to compensate for the loss of cholesterol synthesis by increasing cholesterol uptake from the medium or circulation. This conclusion is further supported by our observation that CYP51 suppression does not lead to lipidosis and, indeed, lowers triglyceride levels. Suppression of this enzyme is complementary and equivalent to POR suppression in the cholesterolgenic pathway but, unlike POR suppression, does not affect other biochemical pathways. This provides strong evidence that loss of cholesterol synthesis per se is not sufficient to cause triglyceride accumulation.
Oxysterols are well established agonists for the liver X receptor, a potent transcriptional inducer of genes involved in fatty acid synthesis. LXR agonists readily induce hepatic triglyceride accumulation (Kalaany and Mangelsdorf, 2006). However, studies with hepatic POR-null mice do not show a strong transcriptional response characteristic of LXR stimulation (Wang et al., 2005; Weng et al., 2005), indicating that oxysterols are unlikely to be elevated in POR-null mice despite the inability of these liver cells to catabolize excess cholesterol to bile acids. Our cell culture studies support this conclusion, because three of the most common oxysterols, none of which are dependent on cytochrome P450 reductase for their synthesis, were less abundant in POR-suppressed cells than in control cells. However, there is at least one additional sterol that could contribute to LXR stimulation: 24,25-dihydrolanosterol. Dihydrolanosterol was recently shown to be a moderate LXR agonist (Zhu et al., 2010); this sterol is formed from lanosterol when CYP51 is inhibited or absent. Interestingly, Zhu et al. (2010) showed that inhibition of CYP51 in 3T3L1 fibroblasts results in a triglyceride lipidosis, presumably by activation of LXR. Dihydrolanosterol is not abundant in POR-suppressed or CYP51-suppressed McA-RH7777 hepatoma cells (our data) but is abundant in the livers and hepatocytes of hepatic POR-null mice (Li and Porter, 2007) and may generate some level of LXR activation in these animals. However, overall, the evidence from both POR-null mice and POR-suppressed cells does not support a significant role for oxysterols and other LXR agonists in the triglyceride lipidosis that follows loss of POR expression.
FXR is a bile acid receptor that down-regulates bile acid synthesis in the liver in the presence of excess bile acids. In the absence of FXR signaling, triglyceride synthesis is greatly increased through derepression of sterol regulatory element binding protein-1c signaling, which increases transcription of several lipogenic genes. The addition of a bile acid, CDCA, to POR-suppressed cells largely prevented the lipidosis and decreased triglycerides to below normal levels, arguing that FXR signaling is absent or greatly decreased in these cells; this would be consistent with an inability of these cells to synthesize bile acids in the absence of cytochrome P450 reductase. FXR signaling also appears to be decreased in the livers of hepatic POR-null mice, consistent with a 90% reduction in the bile acid pool in these animals (Henderson et al., 2003). Expression of CYP7A1, a gene down-regulated by FXR, is increased in the livers of these mice by at least 3.5-fold (Wang et al., 2005; Weng et al., 2005), and similar increases in other genes negatively regulated by FXR are also seen. However, genes involved in fatty acid and triglyceride synthesis in the livers of these mice are not significantly up-regulated, leading the investigators in these studies to conclude that the lipidosis is not the result of increased triglyceride synthesis, but rather due to increased triglyceride uptake and decreased catabolism and secretion.
We tested the role of lipid uptake from the medium in our cell culture model by using delipidated medium. Although in normal medium triglyceride content trended upward in POR-suppressed cells, reaching significance at 15 days, triglyceride content in cells grown in delipidated medium trended downward, also reaching significance at 15 days. At 15 days, lipid vesicles were clearly absent in cells grown in delipidated medium, demonstrating that extracellular lipids are required for lipid content to increase in POR-suppressed cells. Because we did not measure fatty acid or triglyceride synthesis or uptake in these cells, we cannot determine to what extent synthesis and uptake contribute to the lipidosis, but it would appear that both play a role in this system. We do note that POR-suppressed cells grown in delipidated medium do not survive to 20 days, whereas control cells are still dividing at 20 days despite the absence of extracellular lipids. Thus, an external source of lipid is essential to the survival of POR-suppressed cells; we suspect that this required lipid is cholesterol. Interestingly, POR-suppressed cells supplemented with CDCA also die after approximately 15 days, whereas control cells remain viable in this medium; because cholesterol and the normal complement of serum lipids are present in this medium, the toxicity must not be due to the lack of a lipid but rather to presence of CDCA. Why POR-suppressed cells are sensitive to this concentration of a bile acid, whereas control cells are not, remains to be determined.
McA-RH7777 rat hepatoma cells retain a number of characteristics of liver parenchymal cells and are widely used in lipid metabolism studies, including cholesterol and triglyceride synthesis (Singh and Porter, 2006; Ko et al., 2009) and lipoprotein secretion (Rasouli et al., 2004). They more closely resemble the intact liver with regard to sterol regulatory element binding protein expression than does the also widely used HepG2 cell line (DeBose-Boyd et al., 2001) and thus are very attractive for studies on the regulation of hepatic lipid synthesis. They do not appear to be efficient at bile acid synthesis (Labonte et al., 2000), although we have not tested this hypothesis. Further studies on the expression level of regulatory and catalytic genes in cholesterol, fatty acid, and bile acid synthesis in these cells would clearly help elucidate which pathways are being activated in the absence of POR expression in this defined in vitro system.
We have established a hepatoma cell culture system that replicates the hepatic lipidosis observed in hepatic POR-null mice. Studies with this model suggest that a loss of bile acid synthesis leading to enhanced triglyceride synthesis, coupled with uptake and perhaps altered metabolism of triglycerides and fatty acids, is responsible for the lipidosis observed in these cells. The extent to which these complementary pathways contribute to the lipidosis remains to be determined, and potential mechanisms by which triglyceride metabolism and secretion are altered in these cells have not been defined. Further studies with this model, as well as with hepatic POR-null mice, will be needed to delineate fully the interplay between cytochrome P450 reductase and triglyceride metabolism.
Acknowledgments
We thank Melissa Douglas for assistance with these studies.
This work was supported by the National Institutes of Health National Center for Complementary and Alternative Medicine [Grants AT003488, AT005235].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.111.038562.
- POR
- cytochrome P450 reductase
- P450
- cytochrome P450
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- LXR
- liver X receptor
- CDCA
- chenodeoxycholic acid
- siRNA
- small interfering RNA
- FXR
- farnesoid X receptor
- DAPI
- 4,6-diamidino-2-phenylindole.
Authorship Contributions
Participated in research design: Porter, Banerjee, Stolarczyk, and Zou.
Conducted experiments: Banerjee, Stolarczyk, and Zou.
Performed data analysis: Porter, Banerjee, Stolarczyk, and Zou.
Wrote or contributed to the writing of the manuscript: Porter, Banerjee, and Stolarczyk.
References
- DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. (2001) Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci USA 98:1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Henderson CJ, Scott CL, Wolf CR. (2009) Unsaturated fatty acid regulation of cytochrome P450 expression via a CAR-dependent pathway. Biochem J 417:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278:25895–25901 [DOI] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278:13480–13486 [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. (2006) LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol 68:159–191 [DOI] [PubMed] [Google Scholar]
- Ko KW, Erickson B, Lehner R. (2009) Es-x/Ces1 prevents triacylglycerol accumulation in McArdle-RH7777 hepatocytes. Biochim Biophys Acta 1791:1133–1143 [DOI] [PubMed] [Google Scholar]
- Labonté ED, Li Q, Agellon LB. (2000) Expression of cholesterol 7alpha-hydroxylase restores bile acid synthesis in McArdle RH7777 cells. Arch Biochem Biophys 381:273–277 [DOI] [PubMed] [Google Scholar]
- Li L, Porter TD. (2007) Hepatic cytochrome P450 reductase-null mice reveal a second microsomal reductase for squalene monooxygenase. Arch Biochem Biophys 461:76–84 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Meyer R, Malewicz B, Baumann WJ, Johnson RG. (1990) Increased gap junction assembly between cultured cells upon cholesterol supplementation. J Cell Sci 96 (Pt 2):231–238 [DOI] [PubMed] [Google Scholar]
- Moya M, Gómez-Lechón MJ, Castell JV, Jover R. (2010) Enhanced steatosis by nuclear receptor ligands: a study in cultured human hepatocytes and hepatoma cells with a characterized nuclear receptor expression profile. Chem Biol Interact 184:376–387 [DOI] [PubMed] [Google Scholar]
- Mutch DM, Klocke B, Morrison P, Murray CA, Henderson CJ, Seifert M, Williamson G. (2007) The disruption of hepatic cytochrome p450 reductase alters mouse lipid metabolism. J Proteome Res 6:3976–3984 [DOI] [PubMed] [Google Scholar]
- Otto DM, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, Adams RH, Tickle C, Wolf CR. (2003) Identification of novel roles of the cytochrome p450 system in early embryogenesis: effects on vasculogenesis and retinoic acid homeostasis. Mol Cell Biol 23:6103–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli M, Trischuk TC, Lehner R. (2004) Calmodulin antagonist W-7 inhibits de novo synthesis of cholesterol and suppresses secretion of de novo synthesized and preformed lipids from cultured hepatocytes. Biochim Biophys Acta 1682:92–101 [DOI] [PubMed] [Google Scholar]
- Schmidt K, Hughes C, Chudek JA, Goodyear SR, Aspden RM, Talbot R, Gundersen TE, Blomhoff R, Henderson C, Wolf CR, et al. (2009) Cholesterol metabolism: the main pathway acting downstream of cytochrome P450 oxidoreductase in skeletal development of the limb. Mol Cell Biol 29:2716–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, O'Leary KA, Kasper CB. (2002) Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem 277:6536–6541 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- Singh DK, Porter TD. (2006) Inhibition of sterol 4alpha-methyl oxidase is the principal mechanism by which garlic decreases cholesterol synthesis. J Nutr 136:759S–764S [DOI] [PubMed] [Google Scholar]
- Slater HR, Robertson FW. (1979) A comparison of delipidated sera used in studies of sterol synthesis by human mononuclear leukocytes. J Lipid Res 20:413–416 [PubMed] [Google Scholar]
- Wang XJ, Chamberlain M, Vassieva O, Henderson CJ, Wolf CR. (2005) Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (POR) null mice. Biochem J 388:857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, DiRusso CC, Reilly AA, Black PN, Ding X. (2005) Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase gene. Mechanistic implications for the regulation of microsomal cytochrome P450 and the fatty liver phenotype. J Biol Chem 280:31686–31698 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Edwards PA. (2008) FXR signaling in metabolic disease. FEBS Lett 582:10–18 [DOI] [PubMed] [Google Scholar]
- Zhu J, Mounzih K, Chehab EF, Mitro N, Saez E, Chehab FF. (2010) Effects of FoxO4 overexpression on cholesterol biosynthesis, triacylglycerol accumulation, and glucose uptake. J Lipid Res 51:1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]