Abstract
Molecular chaperones are proteins that assist the folding, unfolding, and remodeling of other proteins. In eukaryotes, heat shock protein 90 (Hsp90) proteins are essential ATP-dependent molecular chaperones that remodel and activate hundreds of client proteins with the assistance of cochaperones. In Escherichia coli, the activity of the Hsp90 homolog, HtpG, has remained elusive. To explore the mechanism of action of E. coli Hsp90, we used in vitro protein reactivation assays. We found that E. coli Hsp90 promotes reactivation of heat-inactivated luciferase in a reaction that requires the prokaryotic Hsp70 chaperone system, known as the DnaK system. An Hsp90 ATPase inhibitor, geldanamycin, inhibits luciferase reactivation demonstrating the importance of the ATP-dependent chaperone activity of E. coli Hsp90 during client protein remodeling. Reactivation also depends upon the ATP-dependent chaperone activity of the DnaK system. Our results suggest that the DnaK system acts first on the client protein, and then E. coli Hsp90 and the DnaK system collaborate synergistically to complete remodeling of the client protein. Results indicate that E. coli Hsp90 and DnaK interact in vivo and in vitro, providing additional evidence to suggest that E. coli Hsp90 and the DnaK system function together.
Keywords: Hsp40, DnaJ, CbpA, CbpM, GrpE
Heat shock protein 90 (Hsp90) is a highly conserved molecular chaperone that is essential in eukaryotes, where it is involved in many cellular functions, such as protein trafficking, signal transduction, and receptor maturation (1, 2). It acts by facilitating the folding and activation of client proteins with the assistance of cochaperones. At least 20 cochaperones have been identified (1). Hsp90 of Escherichia coli, which is encoded by the htpG gene and referred to here as Hsp90Ec, is a very abundant protein, and levels of Hsp90Ec are further induced during heat stress. Although not essential for growth, deletion of Hsp90Ec results in slower growth at high temperatures (3) and a slight increase in protein aggregation in heat-stressed cells (4). To date no Hsp90Ec cochaperones have been identified and only one Hsp90Ec client protein has been reported, ribosomal protein L2 (5). L2 was identified in a screen for Hsp90Ec interacting partners, and in vitro it stimulates Hsp90Ec ATPase activity. However, the physiological implication of this association is unknown. In cyanobacteria, Hsp90 is important during stress conditions (6) and has been shown in an ATP-independent reaction to stabilize a linker polypeptide of phycobilisome, a large structure involved in photosynthesis (7).
Prokaryotic and eukaryotic Hsp90 proteins are dimeric, with each protomer containing three conserved domains (Fig. 1A): an N-terminal domain (NTD) that binds and hydrolyzes ATP, a middle domain, and a C-terminal domain that is important for dimerization (2, 8–10). In addition to the three conserved domains, eukaryotic Hsp90s have a charged linker that connects the NTD with the middle domain and a C-terminal extension of approximately 35 amino acids. This C-terminal extension contains the conserved MEEVD amino acid sequence that is involved in the binding of several of the cochaperones (1, 2).
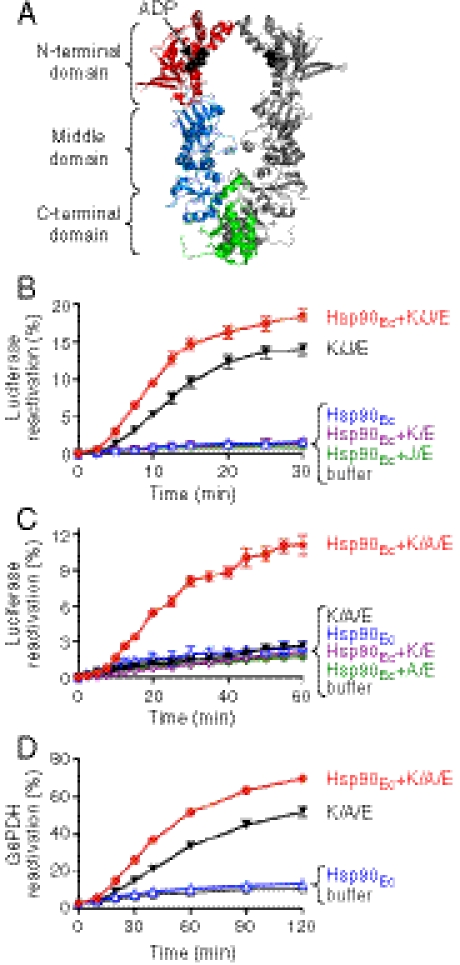
Fig. 1.
Hsp90Ec reactivates denatured substrates in collaboration with the DnaK chaperone system. (A) Model of the Hsp90Ec dimer made from the X-ray structure of Hsp90Ec in the ADP-bound form (Protein Data Bank ID 2iop) using PyMOL. In one protomer, the N-terminal domain, middle domain, and C-terminal domain are red, blue, and green, respectively. ADP is black and represented as Corey–Pauling–Koltun models. (B) Reactivation of heat-denatured luciferase was measured over time as described in Methods in the presence of Hsp90Ec and DnaK, DnaJ, and GrpE (K/J/E); K/J/E; Hsp90Ec; Hsp90Ec and K/E; Hsp90Ec and J/E; or buffer. (C) Reactivation of heat-denatured luciferase was measured as in B except that CbpA was substituted for DnaJ in the DnaK system, referred to as K/A/E. (D) Reactivation of denatured G6PDH was measured over time as described in Methods in the presence of Hsp90Ec and K/A/E; K/A/E; Hsp90Ec; or buffer. For all panels, data from at least three replicates are presented as mean ± SEM. Some error bars are covered by plot symbols.
Hsp90 undergoes large conformational rearrangements that are modulated by ATP binding and hydrolysis; several distinguishable conformational states of Hsp90 have been identified through structural analyses (11–16). Hsp90 dimers transition from an extended open conformation to a compact closed conformation. ATP binding promotes the conversion from the open to closed conformation (1, 2, 9, 10). During this transition, a conserved region in the N terminus, known as the active site lid, closes over the nucleotide-binding pocket, and the N-terminal domains transiently interact. Following ATP hydrolysis, dissociation of ADP restores the open conformation. These conformational rearrangements are conserved in bacteria, yeast, and human Hsp90 family members, however, the equilibrium between these conformational states varies across species (14).
In vitro, both eukaryotic and prokaryotic Hsp90s have weak ATPase activity (17–19). In addition, they protect proteins from heat-induced aggregation in vitro (20, 21), but the lack of an ATP requirement for this activity suggests that it may not be physiologically relevant. Although ATP-dependent protein remodeling activity by prokaryotic Hsp90 has not been previously reported, eukaryotic Hsp90 has been shown to reactivate proteins in vitro. For example, eukaryotic Hsp90 reactivates denatured luciferase in vitro in conjunction with a cochaperone, Hop/Sti1, and the Hsp70 chaperone system (22–24). This reaction involves ATP hydrolysis by Hsp90 (23). Similarly, eukaryotic Hsp90 promotes the maturation of steroid hormone receptors in vitro by a mechanism that involves the ATP-dependent protein remodeling activity of Hsp90, along with the assistance of the Hsp70 system and several cochaperones (25).
In this paper, we show that Hsp90Ec reactivates inactive proteins in collaboration with the DnaK chaperone system. Reactivation requires ATP hydrolysis by Hsp90Ec, demonstrating the ATP-dependent molecular chaperone activity of Hsp90Ec.
Results
Hsp90Ec and the DnaK System of E. coli Act Synergistically in Protein Remodeling.
To explore the mechanism of the molecular chaperone activity of Hsp90Ec, we developed an in vitro protein reactivation assay. Firefly luciferase was used as a model substrate. We heat-inactivated luciferase and then monitored reactivation by Hsp90Ec alone or in combination with the E. coli DnaK chaperone system. The DnaK system was chosen for these experiments because DnaK is the prokaryotic homolog of Hsp70, which is known to participate in luciferase reactivation in vitro with eukaryotic Hsp90 and cochaperones (22–24). The DnaK chaperone system is comprised of DnaK and two cochaperones: DnaJ, a bacterial homolog of Hsp40, and GrpE, a nucleotide exchange factor (26). We observed that, in the presence of ATP, Hsp90Ec alone did not reactivate luciferase, whereas DnaK, DnaJ, and GrpE reactivated luciferase as previously reported (27) (Fig. 1B). Importantly, when Hsp90Ec and the DnaK system were added together with heat-denatured luciferase in the presence of ATP, we observed a 1.6-fold increase in luciferase reactivation compared to reactivation by the DnaK system alone (Fig. 1B). Reactivation required both DnaK and DnaJ (Fig. 1B), whereas GrpE stimulated reactivation twofold in the presence or absence of Hsp90Ec (Fig. S1A). Hsp90Ec and the DnaK system, as well as the DnaK system alone, reactivated the soluble, but not the insoluble, heat-inactivated luciferase* (Fig. S2 A and B).
Although Hsp90Ec stimulated luciferase reactivation above that seen by the DnaK system alone, we wanted to optimize conditions to more clearly observe the Hsp90Ec effect in vitro. By varying the concentrations of the components or the reaction conditions, we were unable to obtain a greater dependence on Hsp90Ec. Because E. coli, like many organisms, has several DnaJ proteins that function with DnaK, we substituted DnaJ with a homolog, CbpA. In vivo, high-level expression of CbpA suppresses the temperature sensitive growth phenotype of a dnaJ deletion strain (28). In vitro, CbpA can replace DnaJ in DnaK-dependent protein remodeling reactions (29). We found that CbpA substituted for DnaJ in luciferase reactivation in combination with Hsp90Ec, DnaK, and GrpE (Fig. 1C). However, there was insignificant reactivation by CbpA, DnaK, and GrpE in the absence of Hsp90Ec (Fig. 1C), showing that Hsp90Ec was necessary for reactivation in combination with DnaK, CbpA, and GrpE. The reaction depended upon both DnaK and CbpA (Fig. 1C), whereas GrpE stimulated the reaction by 15% (Fig. S1B). Hsp90Ec, DnaK, CbpA, and GrpE reactivated only the soluble inactivated luciferase (Fig. S2C). These results demonstrate that Hsp90Ec plays an essential role in luciferase reactivation in collaboration with DnaK, CbpA, and GrpE.
We also tested whether Hsp90Ec could reactivate another model substrate, denatured glucose-6-phosphate dehydrogenase (G6PDH), in combination with the DnaK system. We found that Hsp90Ec stimulated G6PDH reactivation in combination with DnaK, CbpA, and GrpE by twofold (Fig. 1D). In this reactivation reaction, DnaK, CbpA, and GrpE were active alone.† We also observed synergistic effects of Hsp90Ec and the combination of DnaK, CbpA, and GrpE when chemically denatured luciferase was used as the substrate (Fig. S3). In contrast, when we tested several other model substrates, including heat-inactivated GFP and malate dehydrogenase, we observed no stimulation by Hsp90Ec in conjunction with DnaK, DnaJ, and GrpE or with DnaK, CbpA, and GrpE. Hsp90Ec alone did not reactivate any of the substrates tested. Our experiments indicate that Hsp90Ec and the DnaK system function together on some but not all inactive substrates.
Taken altogether, these data demonstrate that Hsp90Ec and the DnaK chaperone system, with either DnaJ or CbpA, act collaboratively to reactivate inactive protein substrates.
Protein Remodeling by Hsp90Ec in Collaboration with the DnaK System Requires the ATPase Activity of Hsp90Ec.
Next, we tested whether ATP hydrolysis by Hsp90Ec is essential for client protein reactivation by Hsp90Ec and the DnaK system. Hsp90 proteins contain a highly conserved ATP binding site (1). In Saccharomyces cerevisiae Hsp90, Hsp82, a Glu to Ala substitution in the ATP binding site at amino acid position 33 abolishes Hsp82 function in vivo and the mutant protein is defective in ATPase activity in vitro (18, 19). An Hsp90Ec mutant, Hsp90Ec(E34A), with a Glu to Ala substitution in the homologous residue is also defective in ATP hydrolysis, possessing 10% of the wild-type ATPase activity (15) (Fig. 2 A and B). When we measured luciferase reactivation by Hsp90Ec(E34A), DnaK, CbpA, and GrpE, we found that the mutant was inactive (Fig. 2C). Hsp90Ec(E34A) was unable to stimulate luciferase reactivation by DnaK, DnaJ, and GrpE (Fig. 2D). Similarly, Hsp90Ec(E34A) was unable to stimulate reactivation of denatured G6PDH by DnaK, CbpA, and GrpE (Fig. S4).
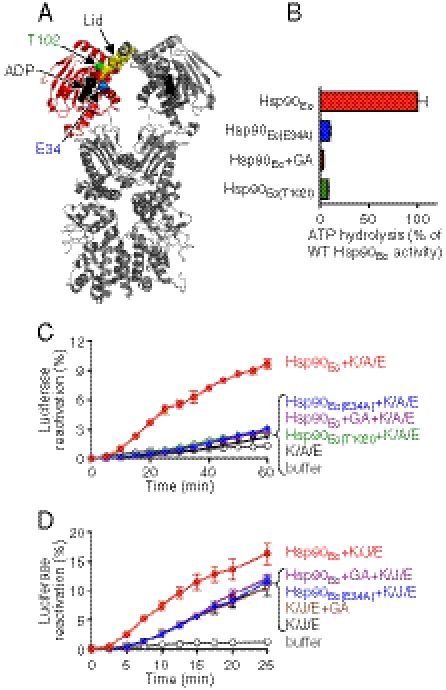
Fig. 2.
Hsp90Ec ATPase activity is required for substrate remodeling in collaboration with the DnaK system. (A) Model of the Hsp90Ec dimer created as in the legend to Fig. 1A showing one N-terminal domain of the dimer in red with E34 and T102 shown in blue and green, respectively, as Corey–Pauling–Koltun models. The active site lid is gold. (B) Hsp90Ec ATPase activity was measured as described in Methods for wild-type Hsp90Ec with or without GA, and for Hsp90Ec(E34A) and Hsp90Ec(T102I). ATPase activities are represented as the percentage of wild-type Hsp90Ec activity. (C) Reactivation of heat-denatured luciferase was measured over time as described in Methods in the presence of DnaK, CbpA, and GrpE (K/A/E); Hsp90Ec and K/A/E with or without GA; Hsp90Ec(E34A) and K/A/E; Hsp90Ec(T102I) and K/A/E; or buffer. (D) Reactivation of heat-denatured luciferase was measured as described in C except that DnaJ was substituted for CbpA in the DnaK system, referred to as K/J/E. GA was added where indicated. In B–D, data from three replicates are presented as mean ± SEM. Some error bars are covered by plot symbols.
To further assess the ATP requirement in protein reactivation by Hsp90Ec, we tested the effect of an Hsp90 inhibitor, geldanamycin (GA). GA binds to the nucleotide-binding site of eukaryotic Hsp90 (30–32) and inhibits the ATPase and ATP-dependent chaperone activity (17–19, 33). It also binds to Hsp90Ec and inhibits ATP hydrolysis (19) (Fig. 2B). When GA was added to luciferase reactivation reactions containing Hsp90Ec, DnaK, CbpA, and GrpE, luciferase reactivation was inhibited (Fig. 2C). GA similarly inhibited Hsp90Ec-stimulated luciferase reactivation in conjunction with DnaK, DnaJ, and GrpE (Fig. 2D). GA had no effect on reactivation by DnaK, DnaJ, and GrpE in the absence of Hsp90Ec (Fig. 2D). GA also prevented the Hsp90Ec-dependent stimulation of G6PDH reactivation by DnaK, CbpA, and GrpE (Fig. S4).
Hsp90 proteins contain a conserved structural element in the N-terminal domain called the active site lid that is involved in ATP hydrolysis and in transient dimerization of the N domains during the ATP hydrolytic cycle (11, 30, 31) (Fig. 2A). A substitution mutation in this region (T101I) in S. cerevisiae Hsp82 is temperature sensitive for growth (34). In vitro the ATPase activity and N-domain dimerization of Hsp82(T101I) are impaired (35). We constructed the corresponding Thr to Ile substitution in Hsp90Ec and observed that Hsp90Ec(T102I) had 10% of the wild-type Hsp90Ec ATPase activity (Fig. 2 A and B). We found that Hsp90Ec(T102I) did not reactivate heat-denatured luciferase in collaboration with DnaK, CbpA, and GrpE (Fig. 2C). It was also unable to stimulate luciferase reactivation by DnaK, DnaJ, and GrpE (Fig. S5). Thus, this active site lid residue is important for ATPase activity and for the chaperone activity of Hsp90Ec in collaboration with the DnaK system.
Altogether the results using Hsp90Ec mutants and the specific Hsp90 inhibitor show that ATP hydrolysis by Hsp90Ec is essential for client protein remodeling in collaboration with the DnaK system.
ATP-Dependent Chaperone Activity of the DnaK System Is Required for Collaboration with Hsp90Ec.
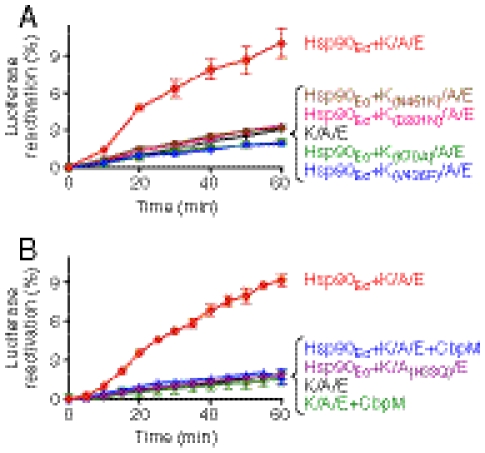
We analyzed two DnaK substitution mutants, DnaK(K70A) and DnaK(D201N), that have impaired ATP hydrolysis (36, 37) to test if ATP hydrolysis by DnaK is required for DnaK to function with Hsp90Ec. Neither mutant reactivated heat-denatured luciferase in conjunction with Hsp90Ec, CbpA, and GrpE (Fig. 3A). Thus ATP hydrolysis by DnaK is required for the synergistic action of the two chaperones. We also tested two DnaK mutants defective for peptide binding, DnaK(V436F) and DnaK(N451K) (38, 39). Both mutants failed to reactivate luciferase in combination with Hsp90Ec, CbpA, and GrpE (Fig. 3A), suggesting that substrate binding by DnaK is also necessary for substrate reactivation. All four of the DnaK mutants were defective in protein remodeling with DnaK, DnaJ, and GrpE in the presence or absence of Hsp90Ec (Fig. S6). These data provide evidence that the ATP-dependent chaperone activity of DnaK is critical for protein remodeling with Hsp90Ec.
Fig. 3.
The chaperone function of the DnaK system is required for luciferase remodeling in collaboration with Hsp90Ec. (A) ATP hydrolysis and substrate binding by DnaK are required for luciferase reactivation. Reactivation of heat-denatured luciferase was measured over time as described in Methods in the presence of wild-type DnaK, CbpA, and GrpE (K/A/E) or in reactions containing Hsp90Ec, CbpA, and GrpE in the presence of wild-type DnaK, DnaK(N451K), DnaK(D201N), DnaK(K70A), or DnaK(V436F). (B) The activity of the CbpA cochaperone is required for luciferase reactivation. Reactivation of heat-denatured luciferase was measured over time as described in Methods in the presence of wild-type DnaK, CbpA, and GrpE (K/A/E) without or with CbpM; Hsp90Ec and K/A/E without or with CbpM; or Hsp90Ec and DnaK, CbpA(H33Q), and GrpE (K/A(H33Q)/E). For both panels, data from three replicates are presented as mean ± SEM. Some error bars are covered by plot symbols.
We next evaluated the importance of CbpA in the collaboration between Hsp90Ec and the DnaK system by testing a CbpA mutant with a His to Gln substitution at position 33 in the conserved HPD amino acid sequence (40). This signature motif of J proteins is essential for interaction with Hsp70/DnaK and for stimulation of Hsp70/DnaK ATPase, which is in turn coupled to polypeptide binding and release (26). We found that CbpA(H33Q) was defective in luciferase reactivation with Hsp90Ec, DnaK, and GrpE, indicating a requirement for an active J protein (Fig. 3B). We also took advantage of a small protein inhibitor of CbpA, CbpM. CbpM specifically interacts with the J domain of CbpA (29, 40, 41), thereby inhibiting the chaperone activity of DnaK (29). When CbpM was added to reaction mixtures containing Hsp90Ec, DnaK, CbpA, and GrpE, luciferase reactivation was prevented (Fig. 3B). CbpM also inhibited G6PDH reactivation by DnaK, CbpA, and GrpE in the presence or absence of Hsp90Ec (Fig. S7A). CbpM had no effect on Hsp90Ec ATPase activity (Fig. S7B) and did not inhibit luciferase reactivation by DnaK, DnaJ, and GrpE in the presence or absence of Hsp90Ec (Fig. S7C). These results demonstrate that the cochaperone activity of CbpA is required for the collaboration between Hsp90Ec and DnaK in substrate remodeling. Taken altogether, our results show that the synergistic action of Hsp90Ec and the DnaK system in protein remodeling requires the ATP-dependent chaperone activity of both the DnaK system as well as Hsp90Ec.
The DnaK System Acts On the Inactivated Client Protein Prior to Hsp90Ec.
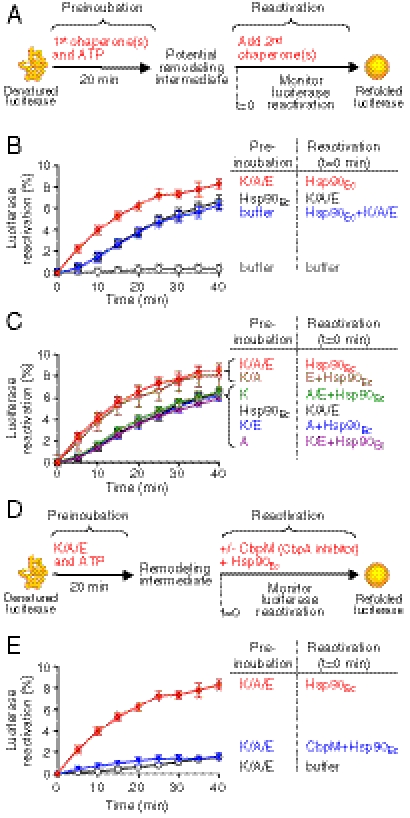
To explore the synergistic action of Hsp90Ec and the DnaK system in protein reactivation, we tested if one chaperone acts on the client protein before the other chaperone. Heat-denatured luciferase was first preincubated for 20 min at 24 °C with the DnaK system, Hsp90Ec, or buffer alone. After the preincubation step, the full reactivation reaction was reconstituted by adding the remaining components, and luciferase reactivation was measured with time (Fig. 4A). The expectation was that if one chaperone acts alone in the first step, then the lag time observed in reactivation, as seen in Fig. 1C, would decrease when the chaperones were added sequentially. We observed that preincubation of inactive luciferase with the DnaK system prior to the addition of Hsp90Ec eliminated the lag phase (Fig. 4B). In contrast, preincubation of luciferase with Hsp90Ec prior to the addition of the DnaK system did not reduce the duration of the lag phase of the luciferase reactivation reaction (Fig. 4B).
Fig. 4.
DnaK system acts first on the denatured luciferase but both chaperones collaborate in protein reactivation. (A) Schematic of experiments shown in B and C. (B) Heat-denatured luciferase and ATP were preincubated with DnaK, CbpA, and GrpE (K/A/E); Hsp90Ec; or buffer for 20 min. After preincubation, the remaining reaction component(s) were added at t = 0 and luciferase reactivation was monitored over time as described in Methods. (C) During the preincubation step, Hsp90Ec or DnaK system components were included in the reactions as indicated. After preincubation, the omitted component(s) of the DnaK system were added with Hsp90Ec at t = 0 and luciferase reactivation was measured. (D) Schematic of experiment shown in E. (E) Heat-denatured luciferase was preincubated for 20 min with DnaK, CbpA, and GrpE (K/A/E). At t = 0, CbpM was added where indicated to inhibit the DnaK system followed by the addition of Hsp90Ec. In B, C, and E, data from at least three replicates are presented as mean ± SEM.
To determine if both of the essential components of the DnaK system, DnaK and CbpA, were required to eliminate the lag phase, heat-inactivated luciferase was preincubated with various combinations of proteins comprising the DnaK system. After 20 min at 24 °C, the full reaction was reconstituted by adding the omitted DnaK system component(s) and Hsp90Ec, and luciferase reactivation was monitored (Fig. 4C). We found that both DnaK and CbpA were required in the preincubation step to eliminate the lag phase of reactivation following the addition of Hsp90Ec (Fig. 4C). Additional observations support the suggestion that the chaperone activity of the DnaK system is required in the first step: (i) ATP was required during the preincubation with the DnaK system (Fig. S8A); (ii) preincubation at 4 °C did not reduce the duration of the lag phase (Fig. S8B); and (iii) the duration of the lag phase decreased as the time of preincubation at 24 °C increased (Fig. S8C). Together, these results suggest that the DnaK chaperone system acts first on heat-inactivated luciferase, followed by the action of Hsp90Ec.
The Coupled Activity of the DnaK System and Hsp90Ec Is Required for Protein Reactivation.
We next wanted to understand whether the actions of the DnaK system and Hsp90Ec on denatured luciferase were independent of one another or if the two activities were coupled. We first incubated heat-denatured luciferase with DnaK, CbpA, and GrpE for 20 min at 24 °C. Then CbpM was added to inhibit the action of the DnaK system. Finally we added Hsp90Ec and monitored luciferase reactivation (Fig. 4D). If the two chaperones act independently and sequentially on the substrate, then inhibition of the DnaK system prior to the addition of Hsp90Ec should have no effect on the extent of luciferase reactivation. We observed no reactivation of luciferase in reactions where DnaK, CbpA, and GrpE were first incubated with heat-denatured luciferase followed by the addition of CbpM and then Hsp90Ec (Fig. 4E). These results show that the successive action of the DnaK system and Hsp90Ec is not sufficient to reactivate heat-denatured luciferase.
Altogether, these data suggest that the DnaK system acts on the inactive substrate prior to Hsp90Ec, possibly performing partial remodeling. Hsp90Ec is then recruited and together the two chaperones collaborate to complete the remodeling and reactivation process.
Hsp90Ec Interacts with DnaK in Vivo and in Vitro.
Based on our observations that Hsp90Ec and the DnaK system collaborate and on a previous report that Hsp90Ec and DnaK were seen to interact in a genome-wide screen (42), we examined if Hsp90Ec interacts directly with DnaK. We used a bacterial two-hybrid method to test for an in vivo interaction (43). The coding regions of Hsp90Ec and DnaK were fused to the two fragments of the Bordetella pertussis adenylate cyclase gene, T18 and T25. We cotransformed plasmids carrying these fusions into an E. coli strain deleted for the adenylate cyclase gene and induced expression. If Hsp90Ec and DnaK interact, then the T18 and T25 domains reconstitute active adenylate cyclase, and β-galactosidase, the cAMP-dependent reporter protein, is expressed.
When we expressed one combination of the Hsp90Ec and DnaK fusion proteins, we observed a 30-fold higher level of β-galactosidase activity than the vector control (Fig. 5A). In control experiments, cells expressing the other combination of the Hsp90Ec and DnaK fusion proteins produced an eightfold higher level of β-galactosidase activity than the vector control (Fig. 5A). Cells expressing either fragment of adenylate cyclase in combination with an Hsp90Ec or DnaK fusion protein produced a low level of β-galactosidase activity similar to the vector control (Fig. 5A). Cells expressing both Hsp90Ec fusion proteins had high levels of β-galactosidase activity, as expected, because Hsp90Ec is a dimeric protein (Fig. 5A). Together these data suggest that DnaK and Hsp90Ec interact in vivo.
Fig. 5.
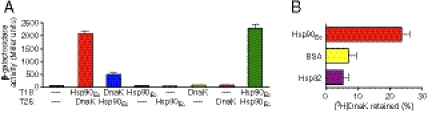
Hsp90Ec interacts with DnaK. (A) A bacterial two-hybrid assay was used to detect interactions between Hsp90Ec and DnaK in vivo as described in Methods. Interactions were detected by measuring β-galactosidase activity in E. coli extracts. Data from three replicates are presented as mean ± SEM. (B) In vitro interaction between DnaK and Hsp90Ec was monitored by measuring retention of [3H]DnaK on filters with a 100-kDa exclusion limit in the presence of Hsp90Ec, BSA, or S. cerevisiae Hsp82 as described in Methods. Data from at least six replicates are presented as mean ± SEM.
We also measured binding of Hsp90Ec to DnaK using purified proteins in vitro. Hsp90Ec was incubated with radiolabeled DnaK and complex formation was monitored by ultrafiltration. We observed specific retention of radiolabeled DnaK (approximately 25%) in the presence of Hsp90Ec (Fig. 5B). Two proteins that would not be expected to interact with DnaK, BSA, and Hsp82, the S. cerevisiae Hsp90 homolog, retained approximately 7% and 5%, respectively, of the DnaK over the background level of DnaK alone (Fig. 5B). Taken altogether, the in vivo and in vitro results suggest Hsp90Ec and DnaK interact directly.
Discussion
Our results show that Hsp90Ec has ATP-dependent protein remodeling activity and acts collaboratively with the DnaK chaperone system. These conclusions suggest a possible model for the action of Hsp90Ec and the DnaK system (Fig. 6). First, the DnaK chaperone system interacts with the substrate, likely being targeted to the substrate by DnaJ (or CbpA), and begins the initial remodeling process. Hsp90Ec is then recruited to the substrate through its interaction with DnaK. The coupled ATP-dependent remodeling activities of Hsp90Ec and the DnaK system complete the protein reactivation process. The nature of the protein remodeling performed by Hsp90Ec remains to be elucidated.
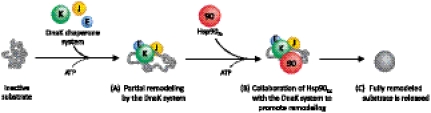
Fig. 6.
Model illustrating client protein reactivation by the collaborative activities of Hsp90Ec and the DnaK system. (A) First, the DnaK system (DnaK, DnaJ, or CbpA and GrpE) interacts with an inactive or denatured substrate and initiates the ATP-dependent remodeling process. (B) Then, Hsp90Ec interacts with DnaK and the substrate, and together in a nucleotide-dependent manner the two chaperones promote remodeling. (C) The substrate is released by the chaperones.
In contrast to protein reactivation by Hsp90Ec and the DnaK system that we describe here, the action of the eukaryotic Hsp90 homolog often requires not only the Hsp70 system, but also specific cochaperones. For example, eukaryotic Hsp90 requires the Sti1/Hop cochaperone to collaborate with the Hsp70 system in reactivation of denatured luciferase (22–24). Studies of the yeast Hsp82 reaction in vitro suggest that Hsp70 binds to denatured luciferase first, then through its interaction with Sti1, Hsp70 transfers luciferase to Hsp82 (24). In this reaction, Sti1 simultaneously binds Hsp82 and Hsp70 forming a bridge linking the two chaperones. Although Hsp90Ec and the DnaK system reactivate proteins in vitro without additional cochaperones and the two chaperones appear to interact in vivo and in vitro, it is possible that as yet unidentified cochaperones could be involved.
Hsp90Ec and the DnaK system reactivated several denatured model substrates, however, many other denatured proteins were not reactivated, indicating that the Hsp90Ec-DnaK collaboration has substrate specificity. Only one potential Hsp90Ec client protein, ribosomal protein L2, has been reported to date (5). It is possible that Hsp90Ec may act on a limited number of proteins to regulate activity or under particular stress conditions to reactivate specific inactivated proteins.
We observed that, when the DnaJ homolog, CbpA, was substituted for DnaJ, Hsp90Ec was essential for luciferase reactivation in combination with DnaK and GrpE. Although the difference between DnaJ and CbpA is not understood, the difference is independent of Hsp90Ec. It is likely that CbpA and DnaJ differ in client protein recognition or the transfer of the client protein to DnaK.
In summary, our results demonstrate that Hsp90Ec has ATP-dependent protein remodeling activity. Moreover, Hsp90Ec acts synergistically with the DnaK chaperone system. The in vitro dependence on Hsp90Ec and the DnaK system for client protein reactivation provides a much needed model system to elucidate the ATP-dependent chaperone action of Hsp90Ec and collaboration with the DnaK system.
Methods
Plasmids, Proteins, and Reagents.
The htpG gene coding for Hsp90Ec was amplified from E. coli DH5α and cloned into pET24b (Novagen) via NdeI/HindIII restriction sites to yield pET-HtpG. Single substitution mutations of Hsp90Ec (E34A and T102I), DnaK (K70A, D201N, V436F, and N451K), and CbpA (H33Q) were introduced into pET-HtpG, pREdnaK, and pCU60 (44), respectively, using the QuikChange mutagenesis system (Stratagene) as described by the manufacturer. Plasmid pRSETA-Hsp82 for the expression of His6-tagged S. cerevisiae Hsp82 (35) was a gift from Len Neckers [National Cancer Institute (NCI)]. Hsp90Ec wild type and mutants were purified as described in SI Methods. DnaK wild type and mutants (45), DnaJ (45), GrpE (45), CbpA wild type and mutant (28), and CbpM (29) were prepared as described. Hsp82 was purified as described (19) with some modifications (SI Methods). [3H]DnaK was prepared as described (46). Luciferase from Photinus pyralis was purchased from Promega. G6PDH from Leuconostoc mesenteroides and BSA were purchased from Sigma. GA was a gift from Len Neckers (NCI). Protein concentrations were determined by Bradford assay and are given for monomeric luciferase, G6PDH, DnaK, CbpM, and BSA; dimeric Hsp90Ec, DnaJ, CbpA, GrpE, and Hsp82.
Assays.
Heat-denatured luciferase reactivation assays were performed as described (27) with some modifications (SI Methods) using 80 nM luciferase, 0.75 μM DnaK, 0.15 μM DnaJ or CbpA, 0.05 μM GrpE, and 0.5 μM Hsp90Ec. Where indicated, GA (15 μM) and CbpM (1.6 μM) were added. G6PDH reactivation assays were performed as described (47, 48) with modifications (SI Methods) using 1.3 μM DnaK, 0.2 μM CbpA, 0.1 μM GrpE, and 1.5 μM Hsp90Ec. ATPase activity assay was performed as described (17) with modifications (SI Methods) using 1 μM Hsp90Ec. The bacterial two-hybrid assay (43) and the in vitro protein binding assay (49) were performed as described with some modifications (SI Methods).
Supplementary Material
Acknowledgments.
We thank Len Neckers (NCI) for Hsp90 inhibitors, plasmid, and discussions, Aurelia Battesti for lending her expertise with the two-hybrid system, and Marika Miot for plasmids. We are also grateful to Danielle Johnston and Marika Miot for critical reading of the manuscript and helpful discussions. This research was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104703108/-/DCSupplemental.
*Approximately 20% of the total heat-inactivated luciferase was soluble and therefore available for reactivation by chaperones (Fig. S2 A and B); 80–90% of the soluble inactive luciferase was reactivated by Hsp90Ec with DnaK, DnaJ, and GrpE (Fig. S2B).
†In another protein remodeling reaction, monomerization of plasmid P1 RepA, CbpA and DnaJ are similarly active in combination with DnaK and GrpE (29).
References
- 1.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 2.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell JC, Craig EA. Ancient heat shock gene is dispensable. J Bacteriol. 1988;170:2977–2983. doi: 10.1128/jb.170.7.2977-2983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas JG, Baneyx F. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol Microbiol. 2000;36:1360–1370. doi: 10.1046/j.1365-2958.2000.01951.x. [DOI] [PubMed] [Google Scholar]
- 5.Motojima-Miyazaki Y, Yoshida M, Motojima F. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem Biophys Res Commun. 2010;400:241–245. doi: 10.1016/j.bbrc.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka N, Nakamoto H. HtpG is essential for the thermal stress management in cyanobacteria. FEBS Lett. 1999;458:117–123. doi: 10.1016/s0014-5793(99)01134-5. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H. HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol Microbiol. 2010;76:576–589. doi: 10.1111/j.1365-2958.2010.07139.x. [DOI] [PubMed] [Google Scholar]
- 8.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 9.Mayer MP. Gymnastics of molecular chaperones. Mol Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011 doi: 10.1017/S0033583510000314. 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 12.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: Insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 17.Scheibel T, Weikl T, Buchner J. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc Natl Acad Sci USA. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperones protein folding in vitro. Nature. 1992;358:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- 21.Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 23.Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 24.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275:18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- 26.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: A paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 27.Schroder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ, and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chae C, Sharma S, Hoskins JR, Wickner S. CbpA, a DnaJ homolog, is a DnaK co-chaperone, and its activity is modulated by CbpM. J Biol Chem. 2004;279:33147–33153. doi: 10.1074/jbc.M404862200. [DOI] [PubMed] [Google Scholar]
- 30.Stebbins CE, et al. Crystal structure of an Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 31.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 32.Grenert JP, et al. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 33.Neckers L. Using natural product inhibitors to validate Hsp90 as a molecular target in cancer. Curr Top Med Chem. 2006;6:1163–1171. doi: 10.2174/156802606777811979. [DOI] [PubMed] [Google Scholar]
- 34.Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prodromou C, et al. The ATPase cycle of Hsp90 drives a molecular “clamp” via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barthel TK, Zhang J, Walker GC. ATPase-defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP-induced conformational change and peptide release. J Bacteriol. 2001;183:5482–5490. doi: 10.1128/JB.183.19.5482-5490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath-Loeb AS, Lu CZ, Suh WC, Lonetto MA, Gross CA. Analysis of three DnaK mutant proteins suggests that progression through the ATPase cycle requires conformational changes. J Biol Chem. 1995;270:30051–30059. doi: 10.1074/jbc.270.50.30051. [DOI] [PubMed] [Google Scholar]
- 38.Laufen T, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 40.Bird JG, Sharma S, Roshwalb SC, Hoskins JR, Wickner S. Functional analysis of CbpA, a DnaJ homolog and nucleoid-associated DNA-binding protein. J Biol Chem. 2006;281:34349–34356. doi: 10.1074/jbc.M603365200. [DOI] [PubMed] [Google Scholar]
- 41.Sarraf NS, et al. Structural basis of the regulation of the CbpA co-chaperone by its specific modulator CbpM. J Mol Biol. 2010;398:111–121. doi: 10.1016/j.jmb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Arifuzzaman M, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueguchi C, Shiozawa T, Kakeda M, Yamada H, Mizuno T. A study of the double mutation of dnaJ and cbpA, whose gene products function as molecular chaperones in Escherichia coli. J Bacteriol. 1995;177:3894–3896. doi: 10.1128/jb.177.13.3894-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skowyra D, Wickner S. The interplay of the GrpE heat shock protein and Mg2+ in RepA monomerization by DnaJ and DnaK. J Biol Chem. 1993;268:25296–25301. [PubMed] [Google Scholar]
- 46.Hoskins JR, Pak M, Maurizi MR, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnett ME, Nagy M, Kedzierska S, Zolkiewski M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J Biol Chem. 2005;280:34940–34945. doi: 10.1074/jbc.M505653200. [DOI] [PubMed] [Google Scholar]
- 48.Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 49.Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.