Abstract
Chromosome 14 was transferred into tumorigenic nasopharyngeal carcinoma and esophageal carcinoma cell lines by a microcell-mediated chromosome transfer approach. Functional complementation of defects present in the cancer cells suppressed tumor formation. A candidate tumor-suppressor gene, cysteine-rich intestinal protein 2 (CRIP2), located in the hot spot for chromosomal loss at 14q32.3, was identified as an important candidate gene capable of functionally suppressing tumor formation. Previous studies have shown that CRIP2 is associated with development. To date, no report has provided functional evidence supporting a role for CRIP2 in tumor development. The present study provides unequivocal evidence that CRIP2 can functionally suppress tumorigenesis. CRIP2 is significantly down-regulated in nasopharyngeal carcinoma cell lines and tumors. CRIP2 reexpression functionally suppresses in vivo tumorigenesis and angiogenesis; these effects are induced by its transcription-repressor capability. It interacts with the NF-κB/p65 to inhibit its DNA-binding ability to the promoter regions of the major proangiogenesis cytokines critical for tumor progression, including IL6, IL8, and VEGF. In conclusion, we provide compelling evidence that CRIP2 acts as a transcription repressor of the NF-κB–mediated proangiogenic cytokine expression and thus functionally inhibits tumor formation and angiogenesis.
Keywords: transcription regulator, antiangiogenesis
Functional complementation of internal defects present in cancer cells can be used to identify candidate tumor-suppressor genes (TSGs) contributing to tumor development. The microcell-mediated chromosome transfer (MMCT) approach can be used to transfer a chromosome to a cancer cell. The resulting hybrid cells containing the exogenous transferred chromosome, known as microcell hybrids (MCHs), can be used to investigate the ability of that particular chromosome to induce tumor suppression and identify TSGs (1). Using a panel of tumor-suppressive chromosome 14 MCH cell lines established in a previous study (1), a novel candidate TSG, cysteine-rich intestinal protein 2 (CRIP2), was identified and shown to induce tumor suppression in nasopharyngeal carcinoma (NPC). CRIP2 is located in the chromosome 14q32.3 region, which often shows high allelic loss in many cancers, including NPC (2, 3) and esophageal, renal, and colon carcinomas (4–7). In fact, because of its location in a hot spot for chromosome truncation in tumor development, it was reported to be a candidate for leukemic translocation (8). However, there have been no follow-up studies to examine the functional role of this gene in tumor development. Our earlier chromosome 14 studies indicate that CRIP2 may be a potential TSG and provide the impetus for further in-depth study of a new functional role for CRIP2 in NPC.
CRIP2 is a member of the LIM domain protein family (9). Members in this family encode different proteins, including transcription factors, adhesion molecules, and cytoskeleton proteins (10, 11). CRIP2 belongs to the cysteine-rich intestine protein family l and specifically to the second class of LIM domain proteins, which contains between one and three LIM domains but usually lacks DNA-binding homeodomains (9). Interestingly, the mouse CRIP2 protein was found to interact with a protein tyrosine phosphatase, PTP-BL, which is important in cancer development (12).
CRIP2 is highly expressed in the heart as well as in the ovaries, brain, skeletal muscle, spleen, prostate, small intestine, pancreas, testis, and neuronal ganglia (9, 13). CRIP2 expression has been detected in the heart endothelium during development and in the adult heart (14). CRIP2 also has been identified as a heart vascular marker (15).
The present study shows that CRIP2 acts as a transcriptional repressor of NF-κB. NF-κB is an important and well-studied transcription factor for various genes involved in the regulation of cancer development and angiogenesis. Loss of regulation of this gene is commonly seen in various types of cancer. The dominant NF-κB complex is p50/p65. This complex is mainly controlled by IκB, which binds to NF-κB to inactivate its transcription function. Phosphorylation of IκB proteins by the upstream IκB kinase results in degradation of IκB protein. NF-κB is then translocated to the nucleus and activates proangiogenesis and cell proliferation target gene transcription. Inactivation of NF-κB transcription factor regulation is an important event contributing to tumor suppression (16, 17).
We have investigated the possible role of CRIP2 in regulating angiogenesis in cancer and associated molecular pathways for CRIP2-inhibited angiogenesis. This report investigates the functional role of CRIP2 in cancer development and provides critical evidence of its crucial role in regulating angiogenesis during tumor development.
Results
Identification of a Candidate TSG Using a Chromosome 14 MMCT Approach.
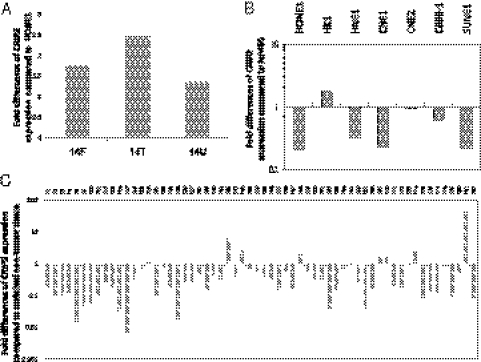
Our earlier study of NPC chromosome 14 MCH cell lines established by the MMCT approach demonstrated these cell lines’ ability to functionally suppress tumor formation in vivo (1, 18). Critical regions were delineated. One gene, CRIP2, was identified and investigated for its importance in NPC. Interestingly, up-regulation of CRIP2 expression was observed in three tumor-suppressive chromosome 14 MCHs (14F, 14T, and 14U), compared with the tumorigenic recipient cell line HONE1 (Fig. 1A), suggesting an important role for CRIP2.
Fig. 1.
CRIP2 gene expression analysis. (A) qPCR analysis of CRIP2 expression in three chromosome 14 MCHs. The expression fold changes were compared with the recipient HONE1 cell line. (B) qPCR analysis of CRIP2 expression in seven NPC cell lines. The expression fold changes of the NPC cell lines were compared with that of the immortalized nontumorigenic cell line NP460. (C) qPCR analysis of CRIP2 expression in 60 NPC paired biopsy specimens. The fold changes of the NPC tumor tissues were compared with their corresponding nontumor tissues.
Significant CRIP2 down-regulation was observed by qPCR analysis in five out of seven NPC cell lines (HONE1, HNE1, CNE1, C666-1, and SUNE1), compared with the nontumorigenic immortalized nasopharyngeal epithelial cell line NP460 (Fig. 1B). The clinical relevance of CRIP2 was investigated in 60 NPC tumors; 42 (70%) showed CRIP2 mRNA down-regulation compared with their corresponding nontumor tissues (Fig. 1C). This suggests that CRIP2 down-regulation is likely an important event during NPC tumor progression.
CRIP2 Reexpression Is Associated with in Vivo Tumor Suppression.
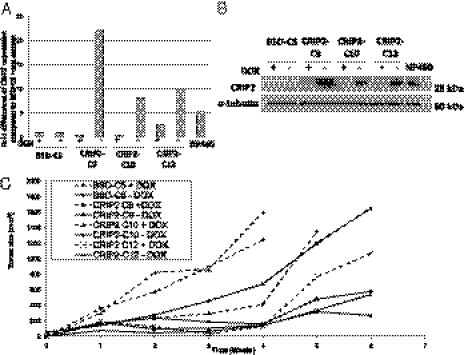
The functional role of CRIP2 was investigated by reexpressing CRIP2 in a HONE1-2 NPC cell line in a previously described tetracycline-regulated inducible system (19). Three CRIP2-expressing clones (CRIP2-C9, -C10, and -C12) expressed increased levels of CRIP2 at both the mRNA and protein levels compared with the vector-alone control (BSD-C5) in the absence of doxycycline (−dox) (Fig. 2 A and B). The expression levels were similar to or higher than those of the immortalized NP460 cell line. In the presence of dox (+dox), transgene expression levels were switched off and CRIP2 expression levels were reduced (Fig. 2 A and B).
Fig. 2.
CRIP2 expression levels in the CRIP2-expressing clones in an in vivo tumorigenicity assay. (A) qPCR analysis of CRIP2 expression in the vector-alone BSD-C5 (±dox) and the three CRIP2-expressing clones CRIP2-C9, -C10, and -C12 (±dox). The expression levels were compared with that of BSD-C5 (+dox). The NP460 cell line was used as a control for endogenous CRIP2 expression. (B) Western blot analysis of BSD-C5 and CRIP2-C9, -C10, and -C12 (±dox). α-tubulin served as a loading control. (C) In vivo tumorigenicity assay of CRIP2-expressing clones and vector-alone controls (+dox). The curves represent an average tumor volume of all sites inoculated for each cell population. Vector-alone BSD-C5 (±dox) formed large tumors by 6 wk postinjection. Small tumors were induced after 5–6 wk postinjection in all of the CRIP2-expressing clones. Tumorigenicity was restored when CRIP2 gene expression was switched off (+dox).
The CRIP2 in vivo tumor-suppressive ability was studied in the nude mouse tumorigenicity assay. All six injection sites of the vector-alone control, BSD-C5 (±dox), formed tumors within 14–21 d postinjection (Fig. 2C and Table S1). The three CRIP2-expressing clones (−dox) suppressed tumor formation. A longer tumor formation latency period was observed compared with that for the vector-alone control. The tumorigenicity of all three CRIP2-expressing clones was restored when CRIP2 expression was switched off by dox. These findings confirm the potent tumor-suppressive ability of CRIP2. The differences in tumor growth kinetics among the vector-alone, CRIP2-expressing clones, and their corresponding +dox controls were statistically significant (P < 0.05) (Table S1).
CRIP2 Suppresses in Vitro and in Vivo Angiogenesis.
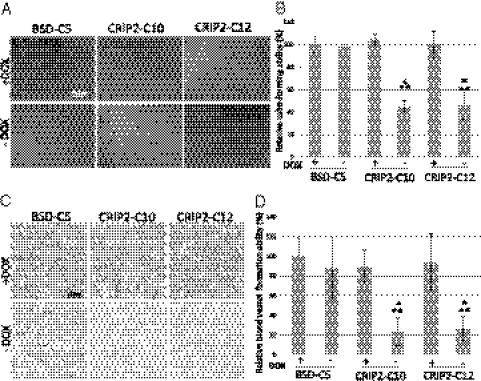
CRIP2 has been reported to regulate blood vessel formation during heart development (14). We studied the functional role of CRIP2 in cancer angiogenesis using in vitro human umbilical vein endothelial cell (HUVEC) tube formation and in vivo Matrigel plug assays. In the in vitro HUVEC tube formation assay, conditioned media from the vector-alone and CRIP2-expressing clones (±dox) were added to the HUVEC cells. The conditioned media from transfectants CRIP2-C10 and -C12 significantly reduced HUVEC cell tube-like formation ability to 44.5% and 45.6%, respectively, compared with the vector-alone (BSD-C5) (±dox) and corresponding +dox controls when transgene expression was repressed (Fig. 3 A and B). The differences in relative tube-forming ability between the vector-alone and CRIP2-stable transfectants (±dox) were statistically significant (P < 0.05).
Fig. 3.
In vitro and in vivo angiogenesis assays of CRIP2-expressing clones. (A) Representative images of the HUVEC tube formation assay of vector-alone (BSD-C5) and CRIP2-expressing clones. (B) Summary of the relative tube-forming ability of the vector-alone (BSD-C5) and CRIP2-expressing clones (±dox). Relative tube-forming ability was calculated by comparing the total tube length of each sample to that of BSD-C5 (+dox). (C) Representative images of the in vivo Matrigel plug CD34 IHC staining of vector-alone (BSD-C5) and CRIP2-expressing clones. The brown color represents positive staining of the blood vessels. (D) Summary of the relative blood vessel formation ability of vector-alone (BSD-C5) and CRIP2-expressing clones (±dox). Relative blood vessel formation ability was calculated by comparing the total tube length of each sample with that of the BSD-C5 (+dox) control. *P < 0.05 and **P < 0.05, statistically significant differences compared with the vector-alone control and corresponding +dox control, respectively.
We further studied the antiangiogenesis effect of CRIP2 in an in vivo nude mouse model Matrigel plug assay. After CD34 staining, we analyzed the density of blood vessels observed inside the Matrigel plug. The blood vessel formation observed with two CRIP2-expressing clones, CRIP2-C10 and -12, decreased dramatically, to 23.2% and 25.6%, respectively, of the vector-alone levels (Fig. 3 C and D). The tumor-forming ability was restored when the transgene was switched off. This further confirms the antiangiogenesis effect of CRIP2.
CRIP2 Reexpression Inhibits Angiogenesis Through Transcriptional Inhibition of Proangiogenesis Cytokines.
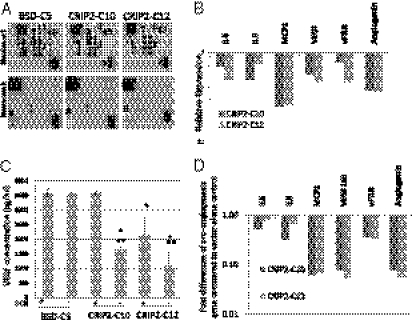
We investigated the functional pathway through which CRIP2 inhibits tumorigenesis and angiogenesis using an angiogenesis-specific antibody protein array. Significant down-regulation of six angiogenesis-related cytokines—IL-6, IL-8, MCP1, VEGF, uPAR, and angiogenin—was observed with the conditioned media from CRIP2-expressing clones (Fig. 4 A and B).The secreted form of VEGF, an important proangiogenic protein, in conditioned media was quantitated by ELISA to confirm the antibody array results. The vector-alone BSD-C5 (±dox) showed a high concentration of VEGF (∼3,500 pg/mL). When CRIP2 was expressed in the CRIP2-C10 and -C12 clones, VEGF protein concentration was reduced to 1,616 and 1,079 pg/mL, respectively (Fig. 4C). VEGF protein concentration was restored when the transgene expression was switched off by dox treatment of the CRIP2-C10 and -C12 clones, further confirming the antibody array results.
Fig. 4.
Angiogenesis-related protein expression in CRIP2-expressing clones. (A) Results of the angiogenesis protein array of the vector-alone BSD-C5 and CRIP2-expressing clones. Duplicate spots: 1, angiogenin; 2, IL-6; 3, IL-8; 4, MCP1; 5, PDGF-BB; 6, RANTES; 7, VEGF; 8, uPAR. (B) Summary of the relative expression of angiogenesis-related proteins in the CRIP2-expressing clones. Relative expression was calculated by comparing the intensity on the protein array of CRIP2-expressing clones vs. the vector-alone. (C) VEGF ELISA of the vector-alone and CRIP2-expressing clones. The total VEGF protein concentration (pg/mL) in the conditioned media of the vector-alone and CRIP2-expressing clones (+dox) was determined. (D) qPCR analysis verifying mRNA transcription levels of candidate genes identified by angiogenesis protein array in the CRIP2-expressing clones. The fold changes were compared with those of the vector-alone BSD-C5. *P < 0.05 and **P < 0.05, statistically significant differences compared with the vector-alone control and corresponding +dox control, respectively.
We studied the mRNA transcription levels of candidate genes IL6, IL8, MCP1, VEGF165, uPAR, and angiogenin by qPCR. All demonstrated decreased mRNA levels in the CRIP2-expressing clones (Fig. 4D), suggesting that reduction of these proangiogenic proteins occurs at the mRNA transcription level. These findings were further substantiated by the discovery of CRIP2 transiently expressed in three CRIP2-down-regulated NPC cell lines, HONE1, HNE1, and SUNE1. When CRIP2 was overexpressed (Fig S1A), down-regulation of IL6, IL8, and VEGF165 was observed (Fig S1B). Thus, this CRIP2 overexpression can functionally down-regulate these proangiogenic proteins at the transcriptional level. In contrast, down-regulation of NF-κB–mediated antiapoptotic factors BclXL and Survivin was not observed when CRIP2 was expressed (Fig S1C).
CRIP2 Suppresses the Transcription of NF-κB–Mediated Proangiogenesis Cytokines IL-6, IL-8, and VEGF.
Because the down-regulation of the proangiogenesis cytokines is regulated mainly by CRIP2 at the transcriptional level, we investigated the functional role of CRIP2 as a transcription repressor. We confirmed the subcellular localization of CRIP2 protein. It was detected mainly in the nuclear fraction, not in the cytoplasmic and membrane fractions, as observed after cell fractionation (Fig. 5A) followed by Western blot detection with antihistone and anti–α-tubulin antibodies as positive controls. This is consistent with a nuclear transcriptional-repressor role for CRIP2.
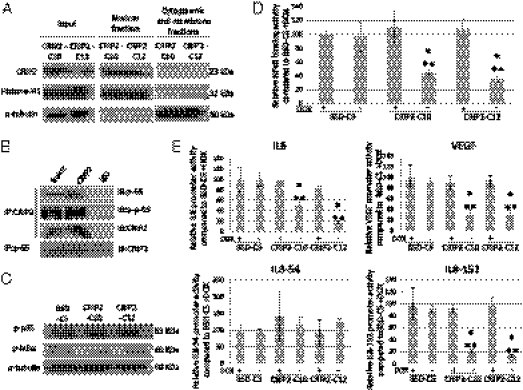
Fig. 5.
Transcriptional regulatory role of CRIP2. (A) Subcellular fractionation of CRIP2-expressing clones. Input protein, nuclear fraction, and cytoplasmic and membrane fraction were used for Western blot analysis. Anti–histone-1H and anti–α-tubulin antibodies were used as positive controls for nuclear and cytoplasmic and membrane fractions, respectively. (B) Co-IP assay using CRIP2 and NF-κB/p65 antibodies. CRIP2 and the total and phosphorylated NF-κB/p65 were detected in the CRIP2-expressing clones after co-IP. Anti-rabbit IgG was used as a negative control. (C) Phosphorylation status of p65 and IκBα in CRIP2-expressing clones. α-tubulin served as a loading control. (D) NF-κB–binding ability assay. The relative NF-κB–binding abilities of CRIP2-expressing clones were compared with those of the BSD-C5 (+dox) control. (E) Promoter assay results for IL-6, VEGF, and IL-8 promoters. Relative promoter activities were compared with the BSD-C5 (+dox) control. *P < 0.05 and **P < 0.05, statistically significant differences compared with the vector-alone control and corresponding +dox control, respectively.
We investigated the interaction between CRIP2 and a well-studied angiogenesis key regulator, NF-κB, a potential interacting partner. The interaction between CRIP2 and NF-κB/p65 subunit was confirmed by coimmunoprecipitation (co-IP) with the CRIP2 and p65 antibodies (Fig. 5B), providing strong evidence that CRIP2 interacts with NF-κB/p65 and subsequently inhibits its transcription factor capability.
We further investigated the mechanism for CRIP2 control of NF-κB function by studying one of the key regulators of NF-κB subcellular translocation, IκB. The phosphorylation status of the NF-κB regulator IκB was confirmed by Western blot analysis. There was no significant change in phosphorylated IκBα levels between the vector-alone and CRIP2-expressing clones (Fig. 5C). There was also no significant change in the phosphorylated NF-κB/p65 levels between the vector-alone and CRIP2-expressing clones (Fig. 5C). This suggests that CRIP2 does not regulate NF-κB function by controlling its phosphorylation status or subcellular localization.
We examined the functional role of CRIP2 in regulating NF-κB–binding affinity to a defined NF-κB–responsive DNA-binding element in CRIP2-expressing clones, to evaluate CRIP2’s ability to inhibit NF-κB's transcription activation capability. NF-κB DNA-binding activity was significantly reduced in the CRIP2-expressing clones compared with the vector-alone and their corresponding +dox controls. The NF-κB–binding activity dropped to 56% in the CRIP2-C10 cell lines and to 32% in the CRIP2-C12 cell lines (Fig. 5D). These findings suggest that CRIP2 regulates NF-κB's transcriptional activity by inhibiting its binding to the promoter region.
We studied the promoter activities of some proangiogenic cytokines to confirm the transcription-repressive effect of NF-κB. The promoter regions of IL-6 and VEGF were cloned in the pGL3 vector (20, 21). Our luciferase reporter assay found significantly reduced promoter activity of the proangiogenesis proteins IL-6 and VEGF by CRIP2 expression (Fig. 5E) and confirmed the role of CRIP2 in repressing their transcription. In the IL-8 promoter region, the IL-8-54 construct showed no significant change in promoter activity between the vector-alone and the two CRIP2-expression clones. In the IL-8-152 construct, which contains a deletion of the transcription factor-binding element, the promoter activity was significantly reduced in the two CRIP2-expressing clones compared with the vector-alone control and their corresponding +dox controls. These findings indicate the importance of the -152 to -55 region of the promoter, which contains the NF-κB–responsive site, for CRIP2 suppression of NF-κB–mediated IL-8 expression (Fig. 5E).
Discussion
In this study, we identified a novel candidate TSG, CRIP2, which induces significant in vivo tumor suppression and angiogenesis inhibition, after chromosome 14 MMCT. CRIP2 demonstrated up-regulation in the MCHs and down-regulation in both NPC cell lines and patient tumors, further validating its importance in cancer development and making it an important candidate for further in-depth study.
Three stably CRIP2-expressing clones were found to be potent inducers of tumor suppression in an in vivo nude mouse tumorigenicity assay, providing unequivocal evidence that CRIP2 can act as a tumor suppressor. We used the CRIP2-C10 and -C12 clones, which express near-physiological levels of CRIP2, for further functional studies.
CRIP2 is a group 2 LIM protein that lacks the DNA-binding LIM domain (9) and thus might not bind directly to the target promoters. Instead, it can interact with other transcription factors to suppress transcription or affect binding to the responsive DNA-binding elements for transcriptional inactivation. The interaction of CRIP2 protein with the NF-κB/p65 subunit shows that CRIP2 is a transcription regulator for NF-κB–mediated transcriptional activation of IL-6, IL-8, and VEGF to inhibit angiogenesis. NF-κB is an important transcription factor in cancer owing to its effective control of key genes promoting cell survival, antiapoptosis, angiogenesis, and invasion (16).
The expression of IL-6, IL-8, and VEGF can serve as an indicator of NF-κB activity, because they contain NF-κB–responsive elements in their promoter regions. These three proangiogenic proteins play important roles in regulating angiogenesis. VEGF is a key angiogenic stimulator, and its expression can directly induce angiogenesis. It is a prognostic factor and is highly expressed in metastatic cancers, including NPC (22, 23). IL-8 is proangiogenic (24) and contributes to a poor prognosis in primary NPC (25). IL-8 has been reported to increase expression of the antiapoptotic protein Bcl-2 in endothelial cells (26) and thus to maintain the angiogenic phenotype of endothelial cells (27). IL-6 promotes angiogenesis and also tumor growth in different cancers by inducing VEGF expression (28–30). CRIP2 induces transcriptional repression of these three proangiogenic proteins, resulting in reduction of stimulating signals to induce angiogenesis. CRIP2 has been detected in the heart endothelium during development and in the adult heart (14) and has been identified as a heart vascular marker (15), further supporting the importance of CRIP2 in regulating angiogenesis.
Although NF-κB is an important regulator in controlling antiapoptotic protein transcription, apoptosis was not observed in the CRIP2-expressing clones. Apoptosis appears to be controlled by different signaling pathways not involving CRIP2 in NPC.
Our findings confirm the mechanism for CRIP2 involvement in regulating NF-κB. The transcription factor activity of NF-κB is tightly regulated by IκB. IκB binds to the NF-κB complex in the cytoplasmic region and prevents subcellular translocation of NF-κB to the nucleus. Phosphorylation of I-κB releases the NF-κB complex and induces its subcellular translocation. CRIP2 expression does not affect I-κB phosphorylation, and thus CRIP2 does not regulate NF-κB activity by affecting NF-κB subcellular localization.
CRIP2 regulates NF-κB's transcription capability by blocking its DNA binding to target gene promoter regions. Previous studies have shown that 17 β-estradiol inhibits NF-κB DNA-binding ability and results in decreased IL-6 secretion in human retinal pigment epithelial cells and an endometrial adenocarcinoma cell line (31, 32). This demonstrates the importance of the inhibition of NF-κB DNA-binding ability in regulating NF-κB target gene promoter activities to critically affect target gene expression levels. This study provides valuable insight into the importance of CRIP2 in regulating NF-κB function. In addition to the NF-κB–responsive element, the promoter region of the IL-8 construct used in this study also contains the AP1- and NF-IL-6–binding elements (33); this suggests that CRIP2, like NF-κB, may have the ability to regulate AP1 and NF-IL-6 transcriptional activities. Taken together, our findings indicate that NF-κB is an important CRIP2 target responsible for its function.
In conclusion, we have provided irrefutable evidence that CRIP2 plays an essential role in suppressing tumor development through inhibiting angiogenesis. This interesting gene has hitherto been associated only with developmental processes. We have shown that CRIP2 plays a key role as a transcriptional repressor of NF-κB–mediated transcription, and that its subsequent functional ramifications in cell regulation mediate its critical role in cancer development.
Materials and Methods
See SI Materials and Methods for more detailed information. Table S2 contains qPCR primer sequences.
Gene Transfection.
The CRIP2 cDNA clone was purchased from the Mammalian Gene Collection (Invitrogen). The CRIP2 gene ORF was cloned into the pETE-BSD (19) vector to establish stable transfectants. Transfection into HONE1-2 was performed with Lipofectamine 2000 Reagent (Invitrogen), as described previously (34).
Luciferase Reporter Assay.
The promoter region of the proangiogenesis cytokines IL-6 and VEGF was cloned into pGL3-basic vector as described previously (20, 21), The IL-8 promoter plasmids pGL-IL-8-54 and -152 were described previously (33). Luciferase activity was measured 48 h after transfection with the Promega Dual-Luciferase Reporter assay system according to the manufacturer's instructions. The relative promoter expression was calculated after normalization with Renilla luciferase activity to eliminate the effect of differences in transfection efficiency.
NF-κB/p65 Transcription Factor Assay.
The NF-κB/p65 transcription factor assay was performed with the Millipore Universal EZ-TFA Transcription Factor Assay Chemiluminescent Kit according to the manufacturer's instructions and as described previously (35).
Supplementary Material
Acknowledgments
This work was supported by the Research Grants Council and the University Grants Council of the Hong Kong Special Administrative Region, People's Republic of China (Grants 6615/07M and AoE/M-06/08, to M.L.L.); the University of Hong Kong Small Project Fund (Grant 200907176081, to A.K.L.C.); and the Swedish Cancer Society, Swedish Research Council, Swedish Institute, Royal Swedish Academy of Sciences, and Karolinska Institute (E.Z.).
Footnotes
The authors declare no conflicts of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101747108/-/DCSupplemental.
References
- 1.Cheung AK, et al. Chromosome 14 transfer and functional studies identify a candidate tumor-suppressor gene, Mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 2009;106:14478–14483. doi: 10.1073/pnas.0900198106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui AB, et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int J Cancer. 1999;82:498–503. doi: 10.1002/(sici)1097-0215(19990812)82:4<498::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 3.Lo KW, et al. High-resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res. 2000;60:3348–3353. [PubMed] [Google Scholar]
- 4.Hoshi M, et al. Detailed deletion mapping of chromosome band 14q32 in human neuroblastoma defines a 1.1-Mb region of common allelic loss. Br J Cancer. 2000;82:1801–1807. doi: 10.1054/bjoc.2000.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohta M, et al. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102:220–224. doi: 10.1002/ijc.10705. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimoto T, et al. High-resolution analysis of DNA copy number alterations and gene expression in renal clear cell carcinoma. J Pathol. 2007;213:392–401. doi: 10.1002/path.2239. [DOI] [PubMed] [Google Scholar]
- 7.Mourra N, et al. High frequency of chromosome 14 deletion in early-onset colon cancer. Dis Colon Rectum. 2007;50:1881–1886. doi: 10.1007/s10350-007-9040-3. [DOI] [PubMed] [Google Scholar]
- 8.Tsui SK, et al. A novel cDNA encoding for a LIM domain protein located at human chromosome 14q32 as a candidate for leukemic translocation. Biochem Mol Biol Int. 1996;39:747–754. doi: 10.1080/15216549600201831. [DOI] [PubMed] [Google Scholar]
- 9.Karim MA, et al. Human ESP1/CRP2, a member of the LIM domain protein family: Characterization of the cDNA and assignment of the gene locus to chromosome 14q32.3. Genomics. 1996;31:167–176. doi: 10.1006/geno.1996.0028. [DOI] [PubMed] [Google Scholar]
- 10.Sadler I, Crawford AW, Michelsen JW, Beckerle MC. Zyxin and cCRP: Two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Lee G, Liebhaber SA, Cooke NE. Human cysteine-rich protein: A member of the LIM/double-finger family displaying coordinate serum induction with c-myc. J Biol Chem. 1992;267:9176–9184. [PubMed] [Google Scholar]
- 12.van Ham M, et al. Cloning and characterization of mCRIP2, a mouse LIM-only protein that interacts with PDZ domain IV of PTP-BL. Genes Cells. 2003;8:631–644. doi: 10.1046/j.1365-2443.2003.00660.x. [DOI] [PubMed] [Google Scholar]
- 13.Bourane S, et al. A SAGE-based screen for genes expressed in sub-populations of neurons in the mouse dorsal root ganglion. BMC Neurosci. 2007;8:97. doi: 10.1186/1471-2202-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu TS, Moctezuma-Anaya M, Kubo A, Keller G, Robertson S. The heart LIM protein gene (Hlp), expressed in the developing and adult heart, defines a new tissue-specific LIM-only protein family. Mech Dev. 2002;116:187–192. doi: 10.1016/s0925-4773(02)00139-9. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Hoffman JA, Ruoslahti E. Molecular profiling of heart endothelial cells. Circulation. 2005;112:1601–1611. doi: 10.1161/CIRCULATIONAHA.104.529537. [DOI] [PubMed] [Google Scholar]
- 16.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 17.Karin M. NF-κB and cancer: Mechanisms and targets. Mol Carcinog. 2006;45:355–361. doi: 10.1002/mc.20217. [DOI] [PubMed] [Google Scholar]
- 18.Ko JM, et al. Functional evidence of decreased tumorigenicity associated with monochromosome transfer of chromosome 14 in esophageal cancer and the mapping of tumor-suppressive regions to 14q32. Genes Chromosomes Cancer. 2005;43:284–293. doi: 10.1002/gcc.20190. [DOI] [PubMed] [Google Scholar]
- 19.Protopopov AI, et al. Human cell lines engineered for tetracycline-regulated expression of tumor-suppressor candidate genes from a frequently affected chromosomal region, 3p21. J Gene Med. 2002;4:397–406. doi: 10.1002/jgm.283. [DOI] [PubMed] [Google Scholar]
- 20.Watters KM, Dean J, Gautier V, Hall WW, Sheehy N. Tax 1-independent induction of vascular endothelial growth factor in adult T-cell leukemia caused by human T-cell leukemia virus type 1. J Virol. 2010;84:5222–5228. doi: 10.1128/JVI.02166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AJ, et al. Association of serum interleukin-6 concentration with a functional IL-6 -6331T>C polymorphism. Clin Chem. 2008;54:841–850. doi: 10.1373/clinchem.2007.098608. [DOI] [PubMed] [Google Scholar]
- 22.Krishna SM, James S, Balaram P. Expression of VEGF as prognosticator in primary nasopharyngeal cancer and its relation to EBV status. Virus Res. 2006;115:85–90. doi: 10.1016/j.virusres.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Guang-Wu H, et al. The relationship between microvessel density, the expression of vascular endothelial growth factor (VEGF), and the extension of nasopharyngeal carcinoma. Laryngoscope. 2000;110:2066–2069. doi: 10.1097/00005537-200012000-00017. [DOI] [PubMed] [Google Scholar]
- 24.Harada K, et al. Cepharanthine inhibits angiogenesis and tumorigenicity of human oral squamous cell carcinoma cells by suppressing expression of vascular endothelial growth factor and interleukin-8. Int J Oncol. 2009;35:1025–1035. doi: 10.3892/ijo_00000417. [DOI] [PubMed] [Google Scholar]
- 25.Xie LQ, Bian LJ, Li Z, Li Y, Liang YJ. Co-elevated expression of hepatocyte growth factor and interleukin-8 contributes to poor prognosis of patients with primary nasopharyngeal carcinoma. Oncol Rep. 2010;23:141–150. [PubMed] [Google Scholar]
- 26.Nör JE, et al. Up-regulation of Bcl-2 in microvascular endothelial cells enhances intratumoral angiogenesis and accelerates tumor growth. Cancer Res. 2001;61:2183–2188. [PubMed] [Google Scholar]
- 27.Strieter RM, et al. Cancer CXC chemokine networks and tumour angiogenesis. Eur J Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Adachi Y, et al. Interleukin-6 induces both cell growth and VEGF production in malignant mesotheliomas. Int J Cancer. 2006;119:1303–1311. doi: 10.1002/ijc.22006. [DOI] [PubMed] [Google Scholar]
- 29.Cohen T, Nahari D, Cerem LW, Neufeld G, Levi BZ. Interleukin 6 induces the expression of vascular endothelial growth factor. J Biol Chem. 1996;271:736–741. doi: 10.1074/jbc.271.2.736. [DOI] [PubMed] [Google Scholar]
- 30.Loeffler S, Fayard B, Weis J, Weissenberger J. Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int J Cancer. 2005;115:202–213. doi: 10.1002/ijc.20871. [DOI] [PubMed] [Google Scholar]
- 31.Paimela T, et al. The effect of 17beta-estradiol on IL-6 secretion and NF-κB DNA-binding activity in human retinal pigment epithelial cells. Immunol Lett. 2007;110:139–144. doi: 10.1016/j.imlet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Ray P, Ghosh SK, Zhang DH, Ray A. Repression of interleukin-6 gene expression by 17 beta-estradiol: Inhibition of the DNA-binding activity of the transcription factors NF-IL-6 and NF-κB by the estrogen receptor. FEBS Lett. 1997;409:79–85. doi: 10.1016/s0014-5793(97)00487-0. [DOI] [PubMed] [Google Scholar]
- 33.Kashima L, et al. CHFR, a potential tumor suppressor, down-regulates interleukin-8 through the inhibition of NF-κB. Oncogene. 2009;28:2643–2653. doi: 10.1038/onc.2009.123. [DOI] [PubMed] [Google Scholar]
- 34.Cheung AK, et al. Functional analysis of a cell cycle-associated tumor-suppressive gene, protein tyrosine phosphatase receptor type G, in nasopharyngeal carcinoma. Cancer Res. 2008;68:8137–8145. doi: 10.1158/0008-5472.CAN-08-0904. [DOI] [PubMed] [Google Scholar]
- 35.Yamini B, et al. Inhibition of nuclear factor-κB activity by temozolomide involves O6-methylguanine–induced inhibition of p65 DNA binding. Cancer Res. 2007;67:6889–6898. doi: 10.1158/0008-5472.CAN-06-4496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.