Abstract
In most mammals, the MHC class I molecules are polymorphic and determine the specificity of peptide presentation, whereas the transporter associated with antigen presentation (TAP) heterodimers are functionally monomorphic. In chickens, there are two classical class I genes but only one is expressed at a high level, which can result in strong MHC associations with resistance to particular infectious pathogens. However, the basis for having a single dominantly expressed class I molecule has been unclear. Here we report TAP1 and TAP2 sequences from 16 chicken lines, and show that both genes have high allelic polymorphism and moderate sequence diversity, with variation in positions expected for peptide binding. We analyze peptide translocation in two MHC haplotypes, showing that chicken TAPs specify translocation at three peptide positions, matching the peptide motif of the single dominantly expressed class I molecule. These results show that coevolution between class I and TAP genes can explain the presence of a single dominantly expressed class I molecule in common chicken MHC haplotypes. Moreover, such coevolution in the primordial MHC may have been responsible for the appearance of the antigen presentation pathways at the birth of the adaptive immune system.
Keywords: avian, evolution, bird
Compared with typical mammals, the chicken MHC has only a single classical class I and class II molecule expressed at high level (1–4). Moreover, the chicken MHC can determine decisive resistance and susceptibility to several infectious pathogens, as well as response to live and killed commercial vaccines (5–7). We developed the hypothesis of a “minimal essential MHC” to provide a molecular explanation for the infectious disease associations of chickens, in comparison with what is known for well characterized mammals (1–4, 8–11). In this view, multigene families of well expressed class I and class II molecules, as well as other disease resistance genes of the huge and complex mammalian MHC, confer more or less protection against most pathogens (leading to weak associations with infectious disease). In contrast, the properties of the single dominantly expressed class I and class II molecules of the small and simple classical chicken MHC (now referred to as the “core MHC” by some authors; ref. 12) confer resistance or susceptibility to particular pathogens (leading to strong genetic associations with infectious disease). Examining iconic chicken diseases, we found that chickens may indeed live or die by their dominantly expressed class I molecule (3, 13).
However, the chicken MHC has two class I genes, BF1 and BF2 (1–4), so why are both not equally well expressed? Based on our discovery that the transporter associated with antigen presentation (TAP) genes are located between the class I genes in chicken (1, 2), we proposed that the coevolution of linked polymorphic genes could be the explanation (1, 2, 8–11). This concept was first developed for mouse class II genes and then used to explain the relationship of rat class I and TAP2 genes (14–16).
The two TAP genes encode a heterodimer of TAP1 and TAP2, which, together, pump peptides from the cytoplasm to the lumen of the endoplasmic reticulum, where the peptides can be loaded onto class I molecules (17). In humans (and most placental mammals examined), the genes for TAPs (as well as other molecules involved in antigen processing and loading) are located in the class II region, separated by considerable physical and recombinational distance from the multigene family of classical class I genes located in the class I region (18, 19). Moreover, these TAPs are nearly monomorphic in sequence, supplying a wide variety of peptides that are used by all alleles of the classical class I multigene family (20, 21).
In contrast, all the classical class I genes of the rat are located in the extended class II region relatively close to the TAP genes (22). The rat TAP2 gene has two allelic lineages, one of which restricts the peptide C terminus to aliphatic amino acids whereas the other allows most amino acids (20, 23, 24). For most rat MHC haplotypes, a particular TAP2 allele is found together with class I molecule(s) of the same peptide-binding specificity (15, 16).
In chickens, we found that the class I genes flank the TAP genes (1, 2), all of which are highly polymorphic at the nucleotide level, with each MHC haplotype having a unique combination of TAP1, TAP2, and BF2 genes (25, 26). In our view (1, 2, 8–11), the large distance in the human MHC between the class I genes and the genes encoding the antigen processing machinery result in the evolution of monomorphic TAPs, which supply peptides to all class I alleles and loci, thus allowing a multigene family. The closer distance in the rat MHC allows just enough coevolution to support one or two class I genes with some specificity in one peptide position. However, the different organization and lack of recombination across the chicken MHC allows the coevolution of genes in stable haplotypes, so that the peptide-translocation specificity of polymorphic TAPs converge with the peptide-binding specificity of the dominantly expressed class I molecule BF2, with BF1 receiving few if any peptides.
Here, we show that the chicken TAP1 and TAP2 proteins do indeed fit our speculations: both TAP1 and TAP2 proteins are found in association with class I heavy chains, both chicken TAP genes are polymorphic and diverse at the amino acid level, and a functional assay for peptide translocation shows polymorphism, specificity, and coevolution with the dominantly expressed class I molecule in each haplotype examined.
Results
Chicken TAPs Form a Complex with Class I Molecules.
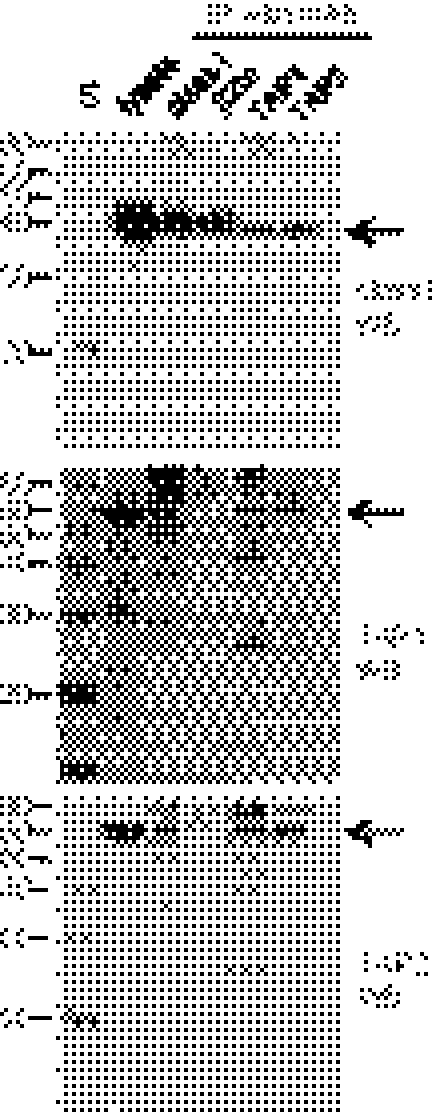
We produced mAbs against TAP1 and TAP2 peptides. By Western blot (of membrane preparations; Fig. S1), these mAbs recognized bands of appropriate mobility on SDS gels. Moreover, after solubilization in the detergent digitonin, immunoprecipitation with mAb to class I, TAP1, and TAP2 showed that all three components interact (Fig. 1), as has been shown for the so-called peptide-loading complex (PLC) in mammals (17). Surprisingly, the mAb F21-21 directed to chicken β2-microglobulin (β2m) did not immunoprecipitate TAP1 or TAP2, but rabbit antisera to chicken β2m did (Fig. 1), so it is likely that the epitope recognized by F21-21 is hidden in the PLC. Affinity isolation by ATP-agarose also identified all three components (Fig. S2). Thus, as in mammals, chicken TAP1 and TAP2 are part of a PLC.
Fig. 1.
Chicken class I, TAP1, and TAP2 molecules interact in cells. Digitonin lysates of membranes from UG5 cells were analyzed by Western blot with mAbs to chicken class I heavy chain (F21-2), TAP1 (F1-11), or TAP2 (F1-3), either directly (lanes labeled “lysate”), or after immunoprecipitation (IP) with mAb to chicken class I heavy chain, β2m (F21-21), TAP1, and TAP2. Specific bands are indicated by arrows; exposure of film was chosen based on strength of signal (class I for 20 s, TAP1 for 20 min, TAP2 for 2 min), resulting in different levels of background bands. S, standards (apparent molecular mass, kDa).
Chicken TAPs Are Highly Polymorphic and Moderately Diverse in Sequence.
We determined exon sequences of TAP1 and TAP2 genes from 16 chicken lines containing seven MHC haplotypes, as well as cDNA sequences from nine of these lines representing all seven haplotypes (Fig. 2 and Tables S1–S5). For both TAP1 and TAP2, all alleles align from start to stop codon without insertions or deletions, except for differences at the 3′ end of TAP1 (in which the last 35 nt of B12 and B19 align poorly with the last 14 nt of the other haplotypes). The TAP1 sequences from lines with the same MHC haplotype are identical, but sequences from different haplotypes are unique, differing in 56 positions among 1,735 nt of well aligned coding sequence, each with 13 to 17 nt differences from the consensus sequence. The same is true for TAP2, with sequences from different haplotypes differing in 53 positions among 2,103 nt of coding sequence, each with 12 to 16 nt differences from the consensus sequence. Of these differences, there are 41 silent and 14 replacement changes in TAP1 (excluding the differences at the 3′ end), and 28 silent and 24 replacement changes in TAP2.
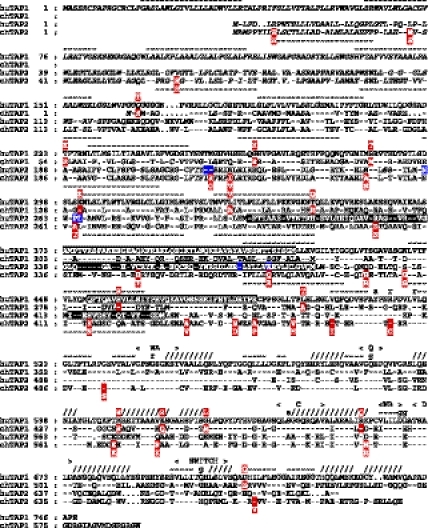
Fig. 2.
Variable residues are found throughout chicken TAP1 and TAP2 sequences, but some are in positions involved in peptide binding in mammals. Alignment of sequences and structural assignments are from ref. 26. Dashes indicate identity with human TAP1 sequence; dots indicate gaps introduced to optimize alignment. Residues shaded in red are variable in chicken TAP sequences reported in this article (allelic residues above the line for TAP1 and below the line for TAP2); those in black (positions 373–420 and 453–487 in human TAP1 and positions 301–389 and 414–433 in human TAP2) are regions that crosslink with bound peptide (27); and those in blue align with positions 217, 218, 262, 265, 266, 374, and 380 of the rat TAP2 sequence shown to be involved in determining peptide specificity (23, 24). Predicted transmembrane regions are shown as tildes above the alignment for TAP1 and below the alignment for TAP2, and residues in the tapasin-binding region are shown in italics. Nucleotide-binding domain structural features are indicated, with α-helices indicated by forward slashes and β-strands indicated by tildes above the line. WA, Walker A motif; WB, Walker B motif; C, C or signature motif; Q, Q loop; g, γ-phosphate interaction; a, adenine interaction; r, ribose interaction; s, serine in C motif.
Of the 14 positions with amino acid differences in TAP1, each allele has two or three differences from the consensus, with six between most divergent alleles. Of the 24 positions in TAP2, each allele has four to eight differences from the consensus, with 13 between the most divergent alleles. The variable positions are scattered throughout the protein chains, in the tapasin-binding region of TAP2 [chicken TAP1 lack this region (26)], the membrane-spanning domain including the proposed peptide-binding regions, and the nucleotide-binding domain. Most of the allelic differences are nonconservative replacements, four align with positions responsible for transport specificity in two rat TAP2 alleles (23, 24), five align with regions of human TAPs identified by peptide cross-linking (27), and 10 others are nearby. Thus, unlike most mammals, chicken TAPs are both highly polymorphic (with many alleles) and moderately diverse in sequence, with some variability in locations that could be functionally important.
Translocation Specificity of Cells from Different Chicken Haplotypes Differs.
We set up peptide translocation assays like those developed for mammals (20) using cell lines derived from chicken hemopoietic cells transformed with reticuloendotheliosis virus (REV). The peptides to be transported were synthesized based on sequences of self peptides isolated from chicken class I molecules, modified to include a tyrosine for iodination and an N-linked glycosylation site.
We found that chicken cell preparations support endogenous glycosylation of the iodinated peptides, indicative of being translocated into the lumen of the endoplasmic reticulum. This activity had the characteristics of TAP-mediated transport as defined for mammalian TAPs, such as dependence on ATP, intact membranes and peptide concentration, and saturability. Most importantly, cells of a particular haplotype would transport peptides known to bind class I molecules of that haplotype, but not those of a different haplotype (Fig. 3A). Thus, unlike in most mammals, peptide transport in chickens is haplotype-specific.
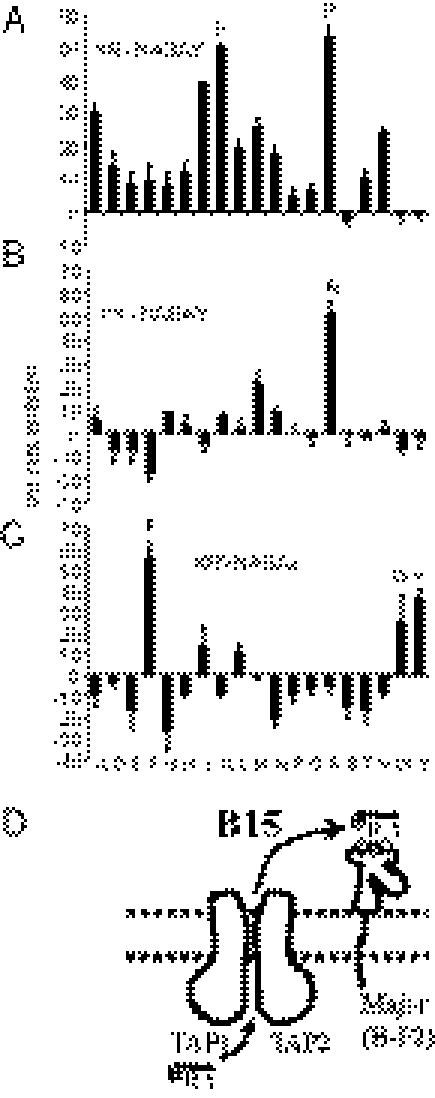
Fig. 3.
Cells from different chicken MHC haplotypes translocate different peptides, and for the B4 haplotype, translocation specificity is the same as the peptide motif of the dominantly expressed class I molecule. (A) Peptide translocation assays were performed by using iodinated indicator peptides (ADYNDSAE based on self peptide ADVEEYEE, KRYNASAY based on KRLIGKRY, GHAENYSAETL based on GHAEEYGAETL) and REV cells (B13 line UG5, B15 line TG15, B21 line TG21), with the self peptide unlabeled as a competitor, and with the amount of radioactivity bound to Con A–Sepharose after the assay indicated on the y axis. (B and C) Peptide translocation assays performed with UG5 cells, with percentage inhibition of transport of the labeled indicator peptide ADYNDSAE by cold ADVEEYEE (labeled B4) and by Ala/Ser swap and charge swap variants (as identified using position number and one-letter code) of ADVEEYEE. (D) Model of translocation of peptides with specificity of the major (BF2) but not the minor (BF1) class I molecule in the B4 haplotype. Plus sign, minus sign and empty circles indicate positive, negative, and hydrophobic residues, respectively; E, glutamic acid.
Translocation Specificity of B4 Cells Mirrors the Motif of the Dominantly Expressed Class I Molecule.
For the B4 haplotype, there are two classical class I genes, with the BF2 gene encoding the dominantly expressed (or major) class I molecule, but cDNA for the BF1 (or minor) gene barely detectable (3, 4). The peptides isolated from class I molecules on erythrocytes have a distinctive motif, mainly octamers with exclusively Asp or Glu found in both positions 2 and 5, and Glu (with small levels of hydrophobic amino acids) found in the final position. This motif is consistent with binding to the BF2 (major), which has positively charged residues in the binding site, but not the BF1 (minor) molecule, which does not (1, 3). Interestingly, compared with the other chicken TAP sequences, the B4 TAP1 sequence has positively charged residues in three positions (8), two in proposed peptide-binding regions (Fig. 2 and Tables S1–S5).
We examined inhibition of transport of an iodinated B4 indicator peptide in permeabilized UG5 cells by synthetic peptides based on the self peptide sequence but with Ala or Ser substituted in each position. Peptides with Ala at positions 2, 5, and 8 did not inhibit transport of the iodinated peptide, showing that they are not transported, whereas peptides with Ala or Ser in the other positions did inhibit transport (Fig. 3B). We then used charge-swap peptides to examine the specificity of binding for those positions, and found that only peptides with Asp or Glu at positions 2 and 5, and with Glu (but not Asp) at position 8, inhibited transport (Fig. 3C). Thus, the specificity of transport mirrors the class I peptide motif of the BF2 (major) molecule in this haplotype (Fig. 3D).
Translocation Specificity of B15 Cells Matches the Motif of the Single Class I Molecule.
In the B15 haplotypes we have examined, only the BF2 gene is functional, with the BF1 gene being disrupted (3, 4). The peptides isolated from class I molecules on B15 erythrocytes are mainly octamers and nonamers with exclusively Lys and Arg at position 1, exclusively Arg at position 2, and overwhelmingly Tyr (with very low levels of Phe and Trp) in the final position (1, 3).
We used TG15 cells to examine inhibition of transport of an iodinated B15 indicator peptide by 57 synthetic peptides based on the self peptide sequence, but each with a different amino acid (all but Cys) substituted in positions 1, 2, or 8 (Fig. 4 A–C). The only significant inhibition was by peptides with Lys or Arg (and, at a lower level, Ile and Ala) in position 1, Arg in position 2, and Tyr, Phe, Trp (and, at a lower level, Leu and Ile) in the final position. Thus, the translocation specificity mirrors the peptide motif (Fig. 4D).
Fig. 4.
For cells of the B15 haplotype, translocation specificity is the same as the peptide motif of the single class I molecule. Translocation of labeled B15 indicator peptide KRYNASAY was inhibited by individual nonradioactive peptides with amino acid substitutions (in single letter code) in positions 1 (A), 2 (B), and 8 (C). (D) Model of translocation of peptides with specificity of the single (BF2) class I molecule in the B15 haplotype. Plus sign, minus sign, and empty circles indicate positive, negative, and hydrophobic residues; R, arginine; Y, tyrosine.
Discussion
In this article, we report an extremely high level of allelic polymorphism for chicken TAP1 and TAP2 genes, with each MHC haplotype having different alleles. The translocation of peptides into the endoplasmic reticulum is also extremely specific, with each MHC haplotype transporting a different set of peptides specified in at least three peptide positions, the same as the dominantly expressed class I molecule. These properties are like those originally described for the rat (15, 16), but are far more extreme.
As hypothesized (1, 2, 8–11), coevolution of TAPs and class I genes can explain the fact of a single dominantly expressed class I molecule in the chicken. In general, the convergence of peptide-translocation specificity and the peptide-binding specificity of the dominantly expressed class I molecule means that other class I molecules will not receive peptides efficiently unless they have the same peptide-binding specificity. If a class I molecule does not receive peptides efficiently, it will not have the opportunity to present antigen or be recognized by T cells at a high level. Therefore, there will be little pressure over evolutionary time to maintain expression of such class I molecules, which will then become minor genes or pseudogenes, as is found for the BF1 locus (3, 4).
The coevolution of class I and TAP genes found in chicken likely represents the ancestral situation. Despite wide variation in the genomic structure of the MHC of nonmammalian vertebrates, many if not most share salient features with the chicken MHC, including a single classical class I gene (or more, of which only one is well expressed with high polymorphism) adjacent to one or more polymorphic TAP genes (reviewed in ref. 9; more recent reports include refs. 28–31). Indeed, the same features are found in at least one marsupial (32), so it would appear that genomic rearrangements occurred in the lineage leading to the placental mammals, most likely an inversion bringing the class III region in between the class I and class II regions, with the class I antigen processing genes (TAPs, tapasin, and inducible proteasome components) left behind in the class II region (10, 11). However, secondary evolutionary changes may also have taken place in some placental mammals, as seems clear for the rat. Similarly, changes to the ancestral configuration may have taken place in some vertebrates outside of the placental mammals, such as the Tammar wallaby, the great reed warbler, and the swallow (33–35), allowing a multigene family of class I molecules.
The results in this article highlight how genomic organization can affect gene function, and may also teach us about the origin of the MHC (10, 11). Originally, the ancestral genes for class I, TAP, and other molecules involved in antigen loading must have had different functions, and had to coevolve to work together. For instance, the ancestor of the TAP genes presumably pumped substrates for some purpose other than supplying peptides for class I molecules, and evolved to bind tapasin and to supply peptides to class I molecules. This evolutionary process would be most efficient if the genes were physically linked without high levels of recombination between them. Thus, the coevolution between alleles now demonstrated for the chicken TAPs and class I genes is conceptually like the process that must have occurred between the ancestral genes in the primordial MHC.
Materials and Methods
Animals and Cell Lines.
The chicken lines used in the present work have been described (4). The RECC-UG5 cell line (a gift of H. Hunt, Avian Disease and Oncology Laboratory, East Lansing, MI) was originally made (36) from bone marrow cells of a G-B1 bird [MHC haplotype B13, considered identical to B4 in the classical MHC (37)] infected by the highly transforming T strain of REV (REV-T). The cell lines TG15 and TG21 were made (as in ref. 38) by in vitro REV-T transformation of Concanavalin A-activated splenocytes from H.B15 and H.B21 birds at the Basel Institute for Immunology, respectively, and bear T-cell markers.
Antibodies and Biochemical Analysis.
The mAb to class I heavy chain (F21-2) and β2m (F21-21) were raised against chicken erythrocytes (39). The mAb to TAP1 and TAP2 were raised against peptides representing the C termini of chicken TAP1 (P350; DWGQQGAPGEGDRG) and TAP2 (P351; LRTRGGPYSRLLQH). Peptides glutaraldehyde-coupled to purified protein derivative and mixed with Al(OH)3 and incomplete Freund adjuvant were used to immunize bacillus Calmette–Guérin-primed BALB/c-NMRI mice four times before final boosting and fusion with SP2/0 myeloma cells.
REV-transformed cells were cultured in RPMI 1640 (supplemented with glutamine, kanamycin, and 10% FCS) at 37 °C and at 5% CO2. Cells were spun down and resuspended in 1 mL cold freeze–thaw buffer [1 mM MgCl2 in PBS solution with 0.1 mM 4-(2-amino ethyl) benzenesulfonyl fluoride (AEBSF; Pefabloc; Roche)], frozen and thawed twice, and centrifuged at 13,300 rpm at 4 °C for 1 h in a Haereus Fresco 17 microfuge (Thermo Scientific). The pellet was solubilized on ice for 30 min in digitonin lysis buffer [150 mM NaCl, 1 mM MgCl2, 10 mM TrisCl, pH 8, with 1% digitonin (Calbiochem) and 0.1 mM AEBSF] to give 108 cell equivalents per milliliter and centrifuged as described above for 10 min, and the supernatant was used immediately or frozen in aliquots.
Aliquots of lysate were incubated with mAb adhered to protein G–Sepharose (Amersham Biosciences) for 1 h to overnight on ice and washed three times with cold digitonin IP wash buffer (1 vol digitonin lysis buffer: 9 vol 150 mM NaCl, 50 mM TrisCl, pH 8). The immunoprecipitates as well as other aliquots of lysate were incubated at room temperature with SDS sample buffer (2.5% SDS, 50 mM TrisCl, pH 8, 20% glycerol, 0.1% bromophenol blue) and electrophoresed on 12% polyacrylamide gels using Laemmli buffers and the Mini-Protean Tetra Cell system (BioRad). Markers were MagicMark XP Western Standard (Invitrogen).
The gels were incubated for 5 min with rotation in 25 mM Tris, 192 mM glycine, 20% methanol, 0.0375% SDS, and then transferred in the same buffer to Hybond-C Extra (Amersham Biosciences)-supported nitrocellulose membranes for 30 min using the Trans-Blot SD semidry transfer apparatus (BioRad). Membranes were blocked overnight with 150 mM NaCl, 0.1% Tween 20, 3% milk powder, 50 mM TrisCl, pH 8, incubated with mAb for 1 h at room temperature to overnight at 4 °C, washed with 150 mM NaCl, 0.1% Tween 20, 50 mM TrisCl, pH 8, and then incubated with secondary antibody (anti-mouse IgG Fc-specific HRP-conjugated, absorbed with human, horse, and bovine serum proteins; Sigma) in the block buffer for 30 min at room temperature before washing again. The membranes were incubated with ECL reagent (Amersham Biosciences) and exposed to XAR-5 film (Konica Minolta).
TAP cDNA and Gene Sequences.
RNA was isolated from spleen and/or bone marrow of the nine Compton chicken lines using the mRNA Direct kit (Dynal), and reverse transcribed using an oligo-dT primer and SuperScript II (Invitrogen) according to manufacturer's instructions. Full-length TAP1 and TAP2 cDNA were amplified over 30 cycles (15 s at 94 °C, 30 s at 60 °C, and 2 min at 68 °C, with a final extension time of 10 min at 68 °C) from 2 μL of the cDNA preparations using 20 pmol of each primer [c1447 (ATCGGGCTCGAGATGGGGAAGATGGGCGCGG with Xho I site underlined) and c1449 (CATCGGTACCCCACCCCCTGCCCTCCCCA with Kpn I site underlined) for TAP1 from B12 and B19; c1464 (ATCGGGCTCGAGATGGGACGATGGGCGCGG with Xho I site underlined) and c1449 for TAP1 of B2, B4, B14, B15, and B21; c821 (CCCAAAGCTTAGCCATGGCGATGCCGCCCTACATTCTGC with Hind III site underlined) and c822 (ATGGGTCGACGTGCTGTAGCAGCCGGCTGTAGGGTCCG with Sal I site underlined) for TAP2 from all haplotypes] and Platinum Pfx polymerase (Invitrogen) with 1× enhancer according to manufacturer's guidelines. Products were purified by agarose gel electrophoresis and MinElute columns (Qiagen), digested with appropriate restriction enzymes, and cloned into the expression vectors pCIpac (TAP1) and pcDNA6-V5/His (TAP2). DNA sequencing was performed on both strands of multiple clones using an ABI 377 automated sequencer (Applied Biosystems).
Peptides.
Peptides were synthesized in-house using Fmoc chemistry and extensively purified by reverse-phase HPLC. For iodination, 20 nmol peptide was mixed with 50 μL PBS solution, 20 μL 2 mg/mL chloramine T (Sigma), and 1 mCi 125I at room temperature for 2.5 min, quenched with 20 μL 2 mg/mL sodium metabisulphite, and separated from free iodine by desalting on a Sephadex G10 column with peptide specific activities around 5 × 107 cpm/nmol as measured by a γ-counter.
Transport Assay.
For the transport assay (40), cells were harvested, washed twice with PBS solution, resuspended in ICT buffer (78 mM KCl, 4 mM MgCl2, 8.37 mM CaCl2, 10 mM EGTA, 1 mM DTT, 1 mg/mL BSA, 50 mM Hepes, pH 7) and divided into aliquots of 5 × 106 cells. Each sample was treated with 2 μg (1,500 hemolytic units) streptolysin O (Institute for Molecular Medicine and Hygiene, Johannes Gutenberg University) at 37 °C for 40 min to permeabilize the cells, washed twice in ice-cold ICT buffer, and resuspended in 750 μL ICT buffer. Radioactive transport peptide (1 μL, with or without competitor peptide) and ATP to 2 mM final concentration were added, and samples incubated at 41 °C for 5 min. Then cells were lysed by addition of 400 μL 1% Triton X-100, 150 mM NaCl, 50 mM TrisCl, pH 8, with 1 mM AEBSF and 1 mM iodoacetamide on ice for 10 min, and the subcellular debris was removed by centrifugation as described above for 10 min at 4 °C. The supernatant was added to 50 μL 50% Con A–Sepharose (Sigma) in 1% Triton X-100, 150 mM NaCl, 50 mM TrisCl, pH 8, and rotated at 4 °C for 2 h. The Con A–Sepharose was washed three times with 1 mL 0.1% Triton X-100, 150 mM NaCl, 50 mM TrisCl, pH 8, and counted in a γ-counter. Samples were in triplicate with necessary controls. Transport peptide without iodination was titrated into the transport assays to determine the IC50 value. This amount of peptide (generally 1–20 ng) was then used in subsequent competition assays for all peptides.
Supplementary Material
Acknowledgments
The authors thank Gillian Griffiths and Hannah Siddle for reading the manuscript. This study was supported by the UK Biotechnology and Biological Sciences Research Council and Wellcome Trust Program Grant 089305.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. JF794477–JF794490).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019496108/-/DCSupplemental.
References
- 1.Kaufman J, Völk H, Wallny H-J. A “minimal essential Mhc” and an “unrecognized Mhc”: Two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman J, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- 3.Wallny HJ, et al. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci USA. 2006;103:1434–1439. doi: 10.1073/pnas.0507386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw I, et al. Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol. 2007;178:5744–5752. doi: 10.4049/jimmunol.178.9.5744. [DOI] [PubMed] [Google Scholar]
- 5.Schat K. Immunity in Marek's disease and other tumors. In: Toivanen A, Toivanen P, editors. Avian Immunology: Basis and Practice. Vol. 2. Boca Raton, FL: CRC Press; 1987. pp. 101–128. [Google Scholar]
- 6.Plachy J, Pink JR, Hála K. Biology of the chicken MHC (B complex) Crit Rev Immunol. 1992;12:47–79. [PubMed] [Google Scholar]
- 7.Bacon L, Witter R. B-haplotype influence on Marek's disease, Rous sarcoma, and lymphoid leukosis virus-induced tumors in chickens. Avian Dis. 1993;37:53–59. doi: 10.3382/ps.0601132. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman J, et al. Gene organisation determines evolution of function in the chicken MHC. Immunol Rev. 1999;167:101–117. doi: 10.1111/j.1600-065x.1999.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman J. Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics. 1999;50:228–236. doi: 10.1007/s002510050597. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman J. The avian MHC. In: Davison T, Kaspers B, Schat K, editors. The Immunology of Birds. Amsterdam: Elsevier; 2008. pp. 159–182. [Google Scholar]
- 11.Kaufman J. The evolutionary origins of the adaptive immune system of jawed vertebrates. In: Kaufmann S, Rouse B, Sachs D, editors. The Immune Response to Infection. Washington, DC: ASM Press; 2010. pp. 41–55. [Google Scholar]
- 12.Shiina T, et al. Extended gene map reveals tripartite motif, C-type lectin and Ig-superfamily type genes within a subregion of chicken MHC-B affecting infectious disease. J Immunol. 2007;178:7162–7172. doi: 10.4049/jimmunol.178.11.7162. [DOI] [PubMed] [Google Scholar]
- 13.Koch M, et al. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Germain RN, Bentley DM, Quill H. Influence of allelic polymorphism on the assembly and surface expression of class II MHC (Ia) molecules. Cell. 1985;43:233–242. doi: 10.1016/0092-8674(85)90028-5. [DOI] [PubMed] [Google Scholar]
- 15.Powis SJ, et al. The rat cim effect: TAP allele-dependent changes in a class I MHC anchor motif and evidence against C-terminal trimming of peptides in the ER. Immunity. 1996;4:159–165. doi: 10.1016/s1074-7613(00)80680-9. [DOI] [PubMed] [Google Scholar]
- 16.Joly E, et al. Co-evolution of rat TAP transporters and MHC class I RT1-A molecules. Curr Biol. 1998;8:169–172. doi: 10.1016/s0960-9822(98)70065-x. [DOI] [PubMed] [Google Scholar]
- 17.Pamer E, Cresswell P. Mechanisms of MHC class I—restricted antigen processing. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 18.MHC Sequencing Consortium Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 19.Carrington M. Recombination within the human MHC. Immunol Rev. 1999;167:245–256. doi: 10.1111/j.1600-065x.1999.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 20.Momburg F, et al. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994;367:648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- 21.Obst R, Armandola EA, Nijenhuis M, Momburg F, Hämmerling GJ. TAP polymorphism does not influence transport of peptide variants in mice and humans. Eur J Immunol. 1995;25:2170–2176. doi: 10.1002/eji.1830250808. [DOI] [PubMed] [Google Scholar]
- 22.Walter L, Günther E. Physical mapping and evolution of the centromeric class I gene-containing region of the rat MHC. Immunogenetics. 2000;51:829–837. doi: 10.1007/s002510000219. [DOI] [PubMed] [Google Scholar]
- 23.Deverson EV, et al. Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J Immunol. 1998;160:2767–2779. [PubMed] [Google Scholar]
- 24.Momburg F, Armandola EA, Post M, Hammerling GJ. Residues in TAP2 peptide transporters controlling substrate specificity. J Immunol. 1996;156:1756–1763. [PubMed] [Google Scholar]
- 25.Wong GK, et al. International Chicken Polymorphism Map Consortium A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker BA, van Hateren A, Milne S, Beck S, Kaufman J. Chicken TAP genes differ from their human orthologues in locus organisation, size, sequence features and polymorphism. Immunogenetics. 2005;57:232–247. doi: 10.1007/s00251-005-0786-2. [DOI] [PubMed] [Google Scholar]
- 27.Nijenhuis M, Hämmerling GJ. Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J Immunol. 1996;157:5467–5477. [PubMed] [Google Scholar]
- 28.Grimholt U, et al. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- 29.Mesa CM, Thulien KJ, Moon DA, Veniamin SM, Magor KE. The dominant MHC class I gene is adjacent to the polymorphic TAP2 gene in the duck, Anas platyrhynchos. Immunogenetics. 2004;56:192–203. doi: 10.1007/s00251-004-0672-3. [DOI] [PubMed] [Google Scholar]
- 30.Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF. Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol. 2006;176:3674–3685. doi: 10.4049/jimmunol.176.6.3674. [DOI] [PubMed] [Google Scholar]
- 31.Shiina T, Hosomichi K, Hanzawa K. Comparative genomics of the poultry major histocompatibility complex. Anim Sci J. 2006;77:151–162. [Google Scholar]
- 32.Belov K, et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddle HV, et al. MHC-linked and un-linked class I genes in the wallaby. BMC Genomics. 2009;10:310. doi: 10.1186/1471-2164-10-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerdahl H, Wittzell H, von Schantz T. Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics. 1999;49:158–170. doi: 10.1007/s002510050477. [DOI] [PubMed] [Google Scholar]
- 35.Bonneaud C, et al. Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus) Immunogenetics. 2004;55:855–865. doi: 10.1007/s00251-004-0648-3. [DOI] [PubMed] [Google Scholar]
- 36.Maccubbin DL, Schierman LW. MHC-restricted cytotoxic response of chicken T cells: Expression, augmentation, and clonal characterization. J Immunol. 1986;136:12–16. [PubMed] [Google Scholar]
- 37.Miller MM, et al. 2004 Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics. 2004;56:261–279. doi: 10.1007/s00251-004-0682-1. [DOI] [PubMed] [Google Scholar]
- 38.Marmor MD, Benatar T, Ratcliffe MJ. Retroviral transformation in vitro of chicken T cells expressing either alpha/beta or gamma/delta T cell receptors by reticuloendotheliosis virus strain T. J Exp Med. 1993;177:647–656. doi: 10.1084/jem.177.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crone M, Simonsen M, Skjødt K, Linnet K, Olsson L. Mouse monoclonal antibodies to class I and class II antigens of the chicken MHC. Evidence for at least two class I products of the B complex. Immunogenetics. 1985;21:181–187. doi: 10.1007/BF00364870. [DOI] [PubMed] [Google Scholar]
- 40.Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line. 220. Immunity. 1998;8:221–231. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.