Abstract
The dynamic modification of nuclear and cytoplasmic proteins by the monosaccharide N-acetyl-glucosamine (GlcNAc) continues to emerge as an important regulator of many biological processes. Herein we describe the development of an alkynyl-modified GlcNAc analog (GlcNAlk) as a new chemical reporter of O-GlcNAc modification in living cells. This strategy is based on metabolic incorporation of reactive functionality into the GlcNAc biosynthetic pathway. When combined with the Cu(I)-catalyzed [3 + 2] azide-alkyne cycloaddition, this chemical reporter allowed for the robust in-gel fluorescent visualization of O-GlcNAc and affinity enrichment and identification of O-GlcNAc-modified proteins. Using in-gel fluorescence detection, we characterized the metabolic fates of GlcNAlk and the previously reported azido analog, GlcNAz. We confirmed previous results that GlcNAz can be metabolically interconverted to GalNAz, whereas GlcNAlk does not, thereby yielding a more specific metabolic reporter of O-GlcNAc modification. We also used GlcNAlk, in combination with a biotin affinity tag, to identify 374 proteins, 279 of which were not previously reported, and we subsequently confirmed the enrichment of three previously uncharacterized proteins. Finally we confirmed the O-GlcNAc modification of the ubiquitin ligase NEDD4-1, the first reported glycosylation of this protein.
Keywords: click chemistry, bioorthogonal labeling, proteomics
The glycosylation of serine and threonine residues by the monosaccharide GlcNAc (O-GlcNAc) is an important and abundant posttranslational modification (PTM) found in a wide range of organisms from Arabidopsis thaliana to humans (1, 2). This nuclear and cytoplasmic modification is dynamic (3), through addition and removal by O-GlcNAc transferase (OGT) and O-GlcNAase (OGA), respectively, and can compete with phosphorylation, setting it up as key regulator of signaling pathways (4, 5). A variety of proteins have been shown to be O-GlcNAc modified including regulators of transcription and translation, cytoskeletal proteins, signaling proteins, and metabolic enzymes. O-GlcNAc modification has been shown to affect protein function through changes in protein localization (6), stability (7), molecular interactions (8–10), and enzymatic activity (11); however, the biochemical consequences of many O-GlcNAc modification events are unknown.
Despite an incomplete molecular understanding of O-GlcNAc modification of many proteins, this PTM has been implicated in many biological processes. O-GlcNAc is indispensable for development in both mice and Drosophila because knockouts of OGT are lethal (12, 13). In addition, Caenorhabditis elegans mutants lacking OGT or OGA display defects in nutrient storage and Dauer formation (14, 15), and O-GlcNAc is also involved in metabolism and related diseases such as type II diabetes (16). Global O-GlcNAc modification of proteins has also been shown to be elevated when cells are subjected to a variety of cellular injuries including heat stress, oxidative stress, hypoxia, ischemia reperfusion, and trauma hemorrhage (17, 18) and additionally in several different types of cancers (19–21). Finally, several lines of evidence point toward O-GlcNAc playing a key role in neuronal function and neurodegeneration (22). For example, O-GlcNAc is enriched at synapses (23, 24), and mice with neuron-specific deletion of OGT display locomotor defects before neonatal death (25). In addition, components of proteinaceous aggregates found in neurodegenerative diseases, including the microtubule-associated protein tau and α-synuclein (26), are modified with O-GlcNAc, and reduced levels of O-GlcNAc have been observed in Alzheimer’s disease (16, 27).
Because O-GlcNAc is clearly important in a wide range of cellular processes, a handful of complementary methods have been developed for the identification and visualization of O-GlcNAcylation. Teo et al. recently generated a small panel of O-GlcNAc-specific IgG monoclonal antibodies (28). When used together, they can overcome any individual antibody’s requirement for different but overlapping peptide determinants resulting in the identification of over 200 proteins. As a complement to antibodies, two bioorthogonal reactions, Staudinger ligation and Cu(I)-catalyzed [3 + 2] azide-alkyne cycloaddition (CuAAC) (29, 30), have been used for the analysis of PTMs and small molecule–protein interactions. This approach has been applied to analyze protein glycosylation (31), acetylation (32), and lipidation (33–35), as well as covalent small molecule inhibitors (36, 37). One method that takes advantage of CuAAC is a version of the chemoenzymatic technology developed by the Hsieh–Wilson laboratory (38–43) and subsequently commercialized by Invitrogen. Specifically, an engineered β-1,4-galactosyltransferase (Y298L GalT) is used to transfer an azide-containing monosaccharide donor, UDP-GalNAz, to O-GlcNAc-modified proteins. The azide can then be reacted under CuAAC conditions with fluorescent and biotin tags for visualization and identification, respectively. Because the enzymatic addition of GalNAz is quantitative, this approach is exquisitely sensitive and has been used to identify over 200 O-GlcNAc-modified proteins, including many that overlap with those identified by antibodies.
Another approach, originally developed by Bertozzi and coworkers, relies on the promiscuity of the hexosamine biosynthetic pathway (HBP) to metabolically deliver azides into O-GlcNAc-modified proteins (44–48). Cells can be treated with the fully protected azide-bearing monosaccharide Ac4GlcNAz or Ac4GalNAz. After deprotection by cellular enzymes, GlcNAz can enter the HBP by action of GlcNAc kinase, and GalNAz can be metabolically transformed to GlcNAz, resulting in modification of proteins by GlcNAz (Fig. 1A). This approach was first used to react O-GlcNAc-modified proteins with biotin and peptide tags using the Staudinger ligation (Fig. 1B) for visualization and identification by Western blotting and mass spectrometry (44–47), resulting in the discovery of approximately 200 proteins when using GlcNAz and 18 proteins when using GalNAz (48). However, these immunoblotting methods are not ideal for analyzing quantitative changes in O-GlcNAc modification required for investigations of dynamics and regulation. In addition, several experiments have shown that CuAAC outperforms the Staudinger ligation for the analysis of cell lysates (33, 37, 49). However, the enrichment of GlcNAz-labeled proteins using CuAAC only resulted in the identification of 32 proteins (47).
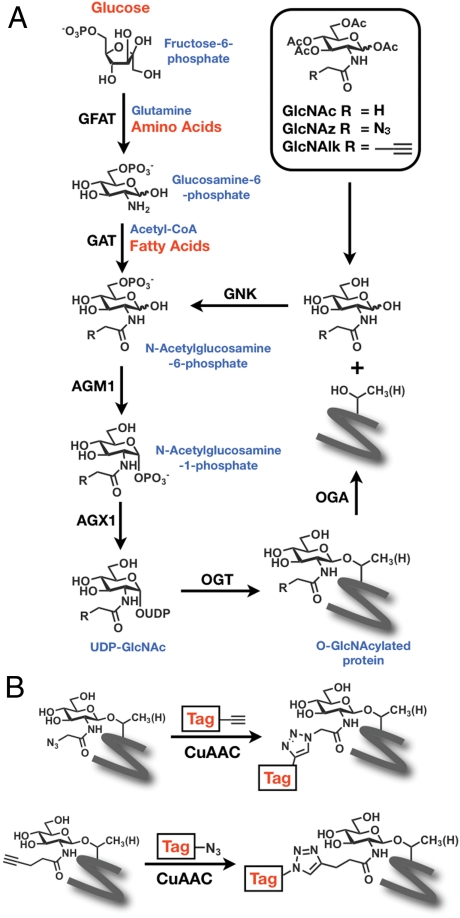
Fig. 1.
The HBP and chemical reporters of O-GlcNAc modification. (A) Glucose, glutamine, and acetyl-CoA are transformed by the metabolic enzymes of the HBP to UDP-GlcNAc, which can be used by OGT to modify protein substrates and reversed by OGA. Per-O-acetylated analogues (Ac4GlcNAz and Ac4GlcNAlk) are deacetylated by intracellular esterases, and then enter the GlcNAc salvage pathway. (B) GlcNAz and GlcNAlk bearing proteins can be covalently modified with detection tags using CuAAC.
We describe here an improvement in the use of metabolic chemical reporters of O-GlcNAc in combination with CuAAC conditions that resulted in the identification of 374 proteins, including many previously unidentified potential O-GlcNAc substrates. In addition, we observed robust fluorescent visualization of O-GlcNAz and successful metabolic incorporation and visualization of an alkyne functionalized GlcNAc analogue, termed GlcNAlk. We performed a cellular analysis of the metabolic fates of both GlcNAz and GlcNAlk, along with their corresponding galactose isomers, GalNAz and GalNAlk, demonstrating that GlcNAlk may be a more specific reporter of O-GlcNAc modification than GlcNAz. Finally, we used GlcNAlk to discover O-GlcNAcylation of the ubiquitin ligase NEDD4-1. NEDD4-1 has been shown to regulate the cell surface turnover of a handful of transmembrane receptors, participate in proliferative signaling in cancer, and facilitate the budding of viral particles including HIV (50, 51). Despite its importance in a variety of biological processes, the regulation of NEDD4-1 is not well understood. Our discovery of the O-GlcNAcylation of NEDD4-1 potentially uncovers an important regulatory mechanism.
Results
Fluorescent Detection of O-GlcNAc-Labeled Proteins.
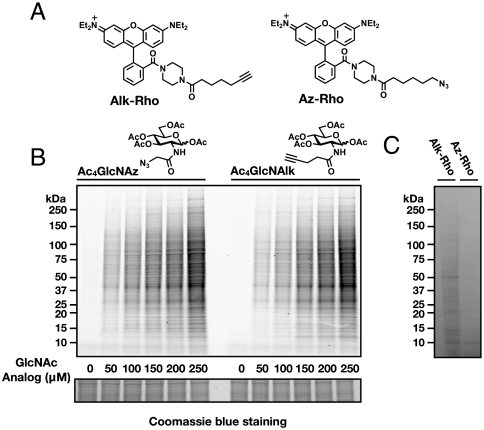
Recent studies by ourselves and others have demonstrated improved detection of metabolic chemical reporters with fluorescent tags under CuAAC reaction conditions (33, 42), suggesting that O-GlcNAz visualization could be improved. We therefore synthesized Ac4GlcNAz (Fig. 1A) and the corresponding alkyne-bearing fluorescent detection tag, alkynyl-rhodamine (Alk-Rho, Fig. 2A) to explore the detection of O-GlcNAc-modified proteins by CuAAC. HEK293 cells were metabolically labeled with various concentrations of Ac4GlcNAz for 16 h in low-glucose medium (1 g/L), thereby maximizing the uptake of our chemical reporter through the GlcNAc salvage pathway. The cells were then washed, lysed, and the soluble fraction was reacted with Alk-Rho under CuAAC conditions. In-gel fluorescence scanning revealed robust labeling of a variety of proteins at all concentrations of Ac4GlcNAz tested (Fig. 2B). Previous comparative analyses of chemical reporters have demonstrated that alkynyl-chemical reporters, in combination with the azido-detection tags, have improved sensitivity when compared to the reverse CuAAC orientation due to decreased background signal (33, 49, 52). Because of this observation, we next asked if the orientation of the CuAAC azide and alkyne partners could be reversed (Fig. 1B). Toward this end, we synthesized the alkyne-modified GlcNAc analogue, Ac4GlcNAlk (Fig. 1A), and azide-bearing fluorescent detection tag (Az-Rho, Fig. 2A). HEK293 cells were metabolically labeled under identical conditions to GlcNAz and reacted with Az-Rho. In-gel fluorescence scanning again revealed robust detection of a large number of proteins (Fig. 2B). Importantly the profile of GlcNAlk-labeled proteins was similar to GlcNAz, suggesting that the two carbohydrates are metabolically equivalent. At high contrast, the GlcNAlk orientation did indeed display lower background signal when compared to GlcNAz (Fig. 2C), which may prove critical at lower concentrations of metabolic chemical reporters and for specific O-GlcNAc substrates that are modified at substoichiometric levels.
Fig. 2.
Fluorescent detection of O-GlcNAc-modified proteins. (A) Fluorescent-tags Alk-Rho and Az-Rho for CuAAC-dependent detection of GlcNAz and GlcNAlk (B) HEK293 cells were treated with the indicated concentrations of Ac4GlcNAz or Ac4GlcNAlk for 16 h and analyzed by in-gel fluorescence scanning. Coomassie blue staining demonstrated equal protein loading. (C) Higher contrast of lanes corresponding to 0 μM in B to allow for comparison of background levels.
GlcNAz and GlcNAlk Are Metabolically Incorporated and Removed at Similar Rates.
Cellular analysis of O-GlcNAc modifications with chemical reporters requires that they are efficiently metabolized by the HBP, added to proteins by OGT, and removed by OGA (Fig. 1A). The metabolic processing of GlcNAz by the enzymes of the GlcNAc salvage pathway has been previously analyzed in vitro (44), demonstrating that GlcNAz is efficiently processed by the GlcNAc salvage pathway and used by OGT and OGA. To qualitatively determine if GlcNAlk is metabolically incorporated as efficiently as GlcNAz, HEK293 cells were treated with Ac4GlcNAz (200 μM) or Ac4GlcNAlk (200 μM) for different lengths of time. In-gel fluorescence scanning after CuAAC revealed that proteins are modified with GlcNAz and GlcNAlk at similar rates (Fig. S1A), suggesting that GlcNAlk is also an efficient substrate for the GlcNAc salvage pathway and OGT. A diverse spectrum of labeled proteins was detectable in as little as 2 h with fluorescence, representing an improvement compared to previous reports using the Staudinger ligation or CuAAC with immunotags and Western blotting, where a time course was not reported or only a subset of proteins was detected (44–47). To determine if GlcNAlk is also a reliable substrate for OGA, HEK293 cells were labeled with Ac4GlcNAz (200 μM) or Ac4GlcNAlk (200 μM) for 16 h. At this time, the cells were washed with PBS and media containing Ac4GlcNAc (200 μM) was added. Cells were then harvested at different times, lysed, and the soluble fractions reacted with the appropriate rhodamine tag. Analysis by in-gel fluorescence scanning showed that the chemical reporter dependent fluorescence signal was lost at the same rate (Fig. S1B), suggesting that O-GlcNAlk-modified proteins are efficient substrates for OGA. Importantly the global half-life of the chemical reporter modification (t1/2 = 12–24 h) corresponds well with specific O-GlcNAc-modified proteins that were analyzed by radioactive glucosamine treatment (α-crystallin t1/2 = ∼ 10 h; cytokeratin t1/2 = ∼ 55 h) (53, 54).
GlcNAz and GlcNAlk Have Multiple Metabolic Fates.
To take maximum advantage of environmental nutrients, mammalian cells are equipped with enzymes that enable crosstalk between various saccharide biosynthetic pathways including the HBP (Fig. 3A). GlcNAz and GlcNAlk could therefore have multiple metabolic fates and access multiple glycosylation pathways that could all contribute to fluorescent signal. In addition to O-GlcNAc, GlcNAc is also directly incorporated into the conserved core pentasaccharide of N-linked glycans or the branches of both N- and mucin-type O-linked oligosaccharides (blue pathway, Fig. 3A). Additionally, UDP-GlcNAc can be enzymatically converted to UDP-GalNAc and subsequently incorporated at the core of mucin-type O-linked glycans (red pathway, Fig. 3A). As stated above, GlcNAz has been incorporated into O-GlcNAc (44) and other studies have shown that GalNAz can be interconverted to GlcNAz and be similarly incorporated (48, 55). In addition, GalNAz is also incorporated into mucin-type O-linked glycans (48, 56). Despite previous experiments, these metabolic chemical reporters have not been compared directly on both a specific cell surface protein and a O-GlcNAc-modified protein, and the alkyne analogs (GlcNAlk and GalNAlk) have not been previously reported.
Fig. 3.
Characterizing GlcNAz and GlcNAlk protein labeling. (A) Monosaccharide chemical reports have several possible metabolic fates. GlcNAz and GlcNAlk can enter the glucosamine salvage pathway (blue pathway) and potentially label O-GlcNAc-modified proteins and N-linked glycans. They could also be reversibly converted to the corresponding GalNAc analogs, GalNAz and GalNAlk (red pathway), resulting in the potential labeling of mucin-type O-linked glycosylation. (B and C) COS-7 cells were transfected with a plasmid encoding GlyCAM-IgG, treated with the indicated chemical reporter, and analyzed by in-gel fluorescence scanning. PNGase-F treatment prior to CuAAC was performed to selectively remove N-linked glycans. (D and E) COS-7 cells were transfected with a plasmid encoding FoxO1A, treated with the indicated chemical reporter, and analyzed by in-gel fluorescence. For comparison experiments B and C, as well as experiments D and E, fluorescent levels were measured and normalized simultaneously.
To perform this comprehensive analysis, we first used the chimeric glycoprotein, GlyCAM-IgG (57), which contains both N-linked and mucin-type O-linked glycans. GlyCAM-IgG was expressed in COS-7 cells treated with Ac4GlcNAz (200 μM) or Ac4GlcNAlk (200 μM) in high-glucose (4.5 g/mL) media. As a control for GalNAc metabolic labeling, we also synthesized Ac4GalNAz and Ac4GalNAlk and simultaneously expressed GlyCAM-IgG in the presence of these sugars under identical conditions. Immunoprecipitation followed by CuAAC and in-gel fluorescence scanning revealed robust chemical reporter dependent labeling in the case of all sugars (Fig. 3B). This result is somewhat contradictory to previous reports where significantly less Ac4GlcNAz incorporation into cell surfaces compared to Ac4GalNAz was detected by the Staudinger ligation (58). These results could simply reflect a difference in the secondary labeling chemistry (Staudinger ligation vs. CuAAC), treatment time (3 d vs. 16 h), concentration of chemical reporter used (50 vs. 200 μM), and/or proteins analyzed (global cell surface vs. GlyCAM-IgG) (56). To determine if GlcNAz or GlcNAlk were incorporated into the N-linked glycan of the Ig domain, metabolically labeled GlyCAM-IgG was treated with Peptide N-glycosidase F (PNGase-F) prior to CuAAC (Fig. 3B). Significant loss of signal was observed for both GlcNAz and GlcNAlk, revealing that they are both incorporated into N-linked glycans, whereas much less signal was lost from GalNAz in mucin-type glycans (56). Next, we expressed GlyCAM-IgG in cells treated with our four chemical reporters (200 μM) in low-glucose (1 g/L) media. We observed significantly less metabolic labeling across all chemical reporters (Fig. 3C), suggesting that, under conditions of low glucose, both GlcNAc and GalNAc derivatives may be funneled into different metabolic pathways or there may be a general decrease in cell surface glycosylation.
We next used an insulin-insensitive mutant of FoxO1, FoxO1A (59), that has been previously shown to be constitutively O-GlcNAc modified. FLAG-tagged FoxO1A was expressed in COS-7 cells treated with one of our four chemical reporters (200 μM) in low-glucose (1 g/L) media. FoxO1A was then enriched, subjected to CuAAC, and analyzed by in-gel fluorescence scanning and Western blotting (Fig. 3D and Fig. S2A). As expected, both GlcNAz and GlcNAlk exhibit robust labeling of FoxO1A. In addition, GalNAz shows labeling of FoxO1A, consistent with interconversion from previous reports (48, 55). In contrast, GalNAlk displays very low global labeling of cellular proteins (Fig. S2A) and modification of FoxO1A. We attribute the multiple bands in our fluorescent gel and Western blotting to phosphorylation (60) and/or ubiquitination (61) of FoxO1A. We next repeated this experiment in COS-7 cells in high-glucose (4.5 g/L) media, where the biosynthesis of competitive UDP-GlcNAc should be greater, and essentially no labeling of FoxO1A was observed (Fig. 3E and Fig. S2B). In addition, global labeling of proteins by GlcNAz, GlcNAlk, and to a lesser extent GalNAz, was reduced, whereas GalNAlk labeling remained unchanged (Fig. S2B).
Taken together, these data suggest unique metabolic fates for each chemical reporter: GlcNAz and GlcNAlk are incorporated into both the IgG domain N-linked glycan (Fig. 3B) and O-GlcNAc on FoxO1A. However, because we observe very low global levels of labeling of these reporters under high-glucose conditions, which is required for robust GlyCAM-IgG labeling, this incorporation must be limited to a subset of N-linked glycans. In addition, GalNAz labels both mucin-type glycans on GlyCAM-IgG and O-GlcNAc on FoxO1A confirming previous experiments (48, 55, 56). Because this interconversion is reversible, GlcNAz could result in metabolic conversion to GalNAz and labeling of mucin-type O-linked glycans. In contrast, our second chemical reporter, GalNAlk, is distinct from the three other monosaccharides tested. GalNAlk has notably lower global levels of labeling (Fig. S2A) but labels GlyCAM-IgG strongly and not FoxO1A, suggesting UDP-GalNAlk can be generated in cell but not efficiently interconverted to GlcNAlk.
Different Cell Types Have Diverse Labeling Patterns
Given that our chemical reporters efficiently label a known O-GlcNAc-modified protein, we tested the generality of GlcNAlk against a panel of cell lines. HEK293, CHO, COS-7, HeLa, Jurkat, Mcf-7, and NIH3T3 cells were treated with Ac4GlcNAlk (200 μM) for 16 h. In-gel fluorescence scanning revealed a striking diversity in the intensity and pattern of O-GlcNAc-modified proteins (Fig. S3). Although some O-GlcNAc-modified proteins are common among the different cell types, unique patterns are readily apparent (Mcf-7 and NIH3T3 cells). In addition, the global levels of O-GlcNAc modification vary greatly, even between cell types with similar modification patterns (HEK293 and Jurkat cells). These data demonstrate the ability of our chemical reporters and fluorescent detection method to monitor specific changes in O-GlcNAc modification patterns and levels that are key to understanding specific cellular outcomes.
O-GlcNAc-Modified Proteins Can Be Identified with GlcNAlk
To identify O-GlcNAc-modified proteins, we performed a large-scale enrichment using GlcNAlk as a chemical handle. NIH3T3 cells in low-glucose media (1 g/L) were treated with Ac4GlcNAlk (200 μM), lysed under denaturing conditions (4% SDS), and reacted with an azido-biotin cleavable affinity tag (Azido-azo-biotin, Fig. S4A) using CuAAC. Labeled proteins were enriched with streptavidin beads and selectively eluted with sodium dithionite (Fig. S4B). The specificity of GlcNAlk-dependent retrieval was demonstrated by Coomassie blue staining (Fig. S4C), and the corresponding proteins were identified by gel-based proteomic mass spectrometry. Identified proteins were compiled and categorized into high- and medium-confidence lists based on the number of assigned spectra and the fold increase above control (DMSO vehicle treated) samples (Tables S1 and S2). We identified 374 proteins by GlcNAlk labeling, representing proteins with diverse cellular functions (Fig. S4D), with 142 and 232 proteins falling into the high- and medium-confidence lists respectively (Tables S1 and S2). Of these proteins, 63 (44%) of high- and 32 (14%) of medium-confidence hits have been previously reported using large-scale proteomic techniques including elimination-addition chemistry (62), metabolic chemical reporters (45–48), enzymatic extension (26, 39–42), lectin affinity chromatography (63), and O-GlcNAc-specific antibodies (28). We also identified 23 proteins that we determined could not be O-GlcNAc modified due to their localization in the secretory pathway, the cell membrane, or extracellular space (Table S3). All of these proteins have confirmed (7 proteins) or potential (16 proteins) N-linked glycosylation sites, confirming the PNGase-F sensitive GlcNAlk labeling of Glycam-IgG (Fig. 3 B and C).
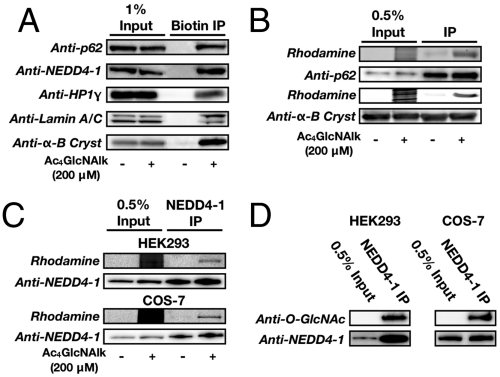
To confirm the proteins identified above, we performed Western blot analysis of the GlcNAlk enriched proteome using antibodies against the known O-GlcNAc substrate p62 (64) and three previously uncharacterized proteins (NEDD4-1, HP1, and Lamin A), confirming their specific recovery (Fig. 4A). In addition, we transfected COS-7 cells with a construct encoding α-B crystallin, which is only O-GlcNAc modified at 5–10%, and were able to use our biotin enrichment strategy selectively recover this protein (Fig. 4A). We next asked if we could visualize the incorporation of GlcNAlk using our fluorescent tags. Nuclearporin p62 was immunoprecipitated from NIH3T3 cells, and fluorescent signal was selectively detected from cells treated with Ac4GlcNAlk (Fig. 4B). We also detected fluorescent signal from α-B crystallin, from COS-7 cells, confirming our ability to detect substoichiometrically modified proteins (Fig. 4B).
Fig. 4.
Identification of O-GlcNAlk-modified proteins. (A) O-GlcNAlk-modified proteins were enriched from NIH3T3 cells treated with Ac4GlcNAlk (200 μM) using azido-azo-biotin and analyzed by Western blotting. (B) Known O-GlcNAcylated proteins p62 and α-B crystallin were immunoprecipitated from cells treated with Ac4GlcNAlk (200 μM) and analyzed by in-gel fluorescence. (C) NEDD4-1 was selectively enriched from cells treated with Ac4GlcNAlk (200 μM) and analyzed by in-gel fluorescence. (D) NEDD4-1 was immunoprecipitated from cells and analyzed by Western blotting.
We next focused on the O-GlcNAc modification of the ubiquitin ligase NEDD4-1. The family of NEDD4-like E3 ligases represent a small but important subset of enzymes capable of transferring ubiquitin to protein substrates. The founding member of this class of E3 ligases, NEDD4-1, plays an important role in neuronal development, cell metabolism, receptor endocytosis, and tumorigenesis and cell growth (50, 51). However, the regulation of NEDD4-1 remains poorly understood. To confirm O-GlcNAc modification of NEDD4-1, HA-tagged NEDD4-1 was expressed in both HEK293 and COS-7 cells. After immunoprecipitation and CuAAC with azido-rhodamine, GlcNAlk-dependent fluorescent signal was readily detected on NEDD4-1 (Fig. 4C and Fig S5A). Finally, NEDD4-1 was expressed and immunoprecipitated from HEK293 cells and O-GlcNAc modification was detected using an anti-O-GlcNAc antibody (CTD110.6) (Fig. 4D and Fig S5B), demonstrating that NEDD4-1 is a bona fide O-GlcNAc substrate.
Discussion
The modification of cytosolic and nuclear proteins by O-GlcNAc is indispensable for proper cellular function. To allow for the detection of O-GlcNAc modification, we have developed chemical reporters that can metabolically enter the GlcNAc salvage pathway, allowing for robust fluorescent detection of O-GlcNAc-modified proteins using CuAAC. Although the azide-modified GlcNAc analog, GlcNAz, has been used previously to detect O-GlcNAc-modified proteins using immunoblotting methods, our alkyne analog, GlcNAlk, in combination with an azide bearing fluorophore, may yield a more specific mode for detection of O-GlcNAc-modified proteins after CuAAC because it may not be converted to GalNAlk. Comparison of GlcNAz and GlcNAlk revealed that the analogs are metabolically incorporated at similar rates into a large collection of cellular proteins, and they are both removed from proteins at rates consistent with published radioactive O-GlcNAc probes.
We next analyzed the metabolic fates of GlcNAz and GlcNAlk. To determine if GlcNAz or GlcNAlk could label N-linked or mucin-type O-linked glycans, we used the chimeric, secreted protein GlyCAM-IgG. Under high-glucose culture conditions, both GlcNAz and GlcNAlk are incorporated into GlyCAM-IgG. Much of this labeling was PNGase-F sensitive, demonstrating that our chemical reporters are incorporated into N-linked glycans. Under the same set of conditions, treatment of cells with GlcNAz and GlcNAlk does not result in detectable labeling of a known O-GlcNAc-modified protein, FoxO1A, and very little global protein labeling occurs. In contrast, under low-glucose conditions, GlyCAM-IgG labeling by GlcNAz and GlcNAlk is greatly reduced, and the same chemical reporters robustly label FoxO1A. As controls for metabolic conversion of GlcNAz and GlcNAlk to the corresponding galactose analogs, we also synthesized GalNAz and GalNAlk. Consistent with prior experiments, under high-glucose conditions the majority of GalNAz is incorporated into mucin-type O-linked glycans (56). Under low-glucose conditions, GlyCAM-IgG labeling is reduced, and FoxO1A is labeled by GalNAz, through transformation to GlcNAz and subsequent O-GlcNAc modification (48, 55). We hypothesize that GalNAz modifies a combination of mucin-type O-linked glycans and O-GlcNAc-modified proteins and that the ratio of these modifications can be adjusted by cell culture conditions. Our second chemical reporter, GalNAlk, also labels GlyCAM-IgG under high-glucose conditions. The global levels of GalNAlk labeling are low, and it is not readily converted to GlcNAlk as FoxO1A is not labeled by GalNAlk. Further experiments will be needed to test the exact location of this monosaccharide.
Because GlcNAlk and GalNAlk appear to not be interconverted, GlcNAlk may be a more specific reporter of O-GlcNAc modification when compared to GlcNAz and GalNAz. We therefore used a cleavable biotin affinity tag (azido-azo-biotin) in combination with GlcNAlk to identify 374 proteins, including 279 proteins that had not been previously identified as potential O-GlcNAc substrates. Because GlcNAlk is also potentially incorporated into N-linked glycans, we also identified 23 N-linked glycosylated proteins that we determined could not contain O-GlcNAc modification sites due to their localization. Despite these contaminating proteins, the over 10-fold difference in number of proteins identified supports our conclusion that the majority of GlcNAlk-labeled proteins are potential O-GlcNAc substrates. Additionally we confirmed the presence of O-GlcNAc as the first glycosylation event on NEDD4-1. We are currently focusing on understanding the affects of O-GlcNAcylation on NEDD4-1 stability, localization, and interaction with substrates.
In summary, the methods described here represent an improvement in the analysis of O-GlcNAc-modified proteins with metabolic chemical reporters. The use of fluorescent reporters, combined with CuAAC, allows for the dynamic analysis of O-GlcNAc modification levels and patterns that cannot be readily determined by published immunoblotting methods. Our analysis of different monosaccharide chemical reporters reveals that, in contrast to previously reported GlcNAz (44) and GalNAz (48), GlcNAlk may not be readily interconverted with GalNAlk, making it more specific for O-GlcNAc modification. In addition, unlike methods that measure steady-state levels of modification, the metabolic chemical reporters could be used to examine the dynamic removal of O-GlcNAlk in a pulse–chase format, which we are currently exploring.
Materials and Methods
Chemical Synthesis.
Details describing the synthesis of the chemical reporters, fluorescent tags, and azido-azo-biotin can be found in SI Text.
CuAAC.
Nonidet P-40 (NP-40) cell lysate (200 μg; see SI Text for details) was diluted with cold 1% NP-40 lysis buffer to obtain a desired concentration of 1 μg/μL. Newly made click chemistry cocktail (12 μL) was added to each sample [azido- or alkynyl-rhodamine tag (100 μM, 10 mM stock solution in DMSO); tris(2-carboxyethyl)phosphine hydrochloride (1 mM, 50 mM freshly prepared stock solution in water); tris[(1-benzyl-1-H-1,2,3-triazol-4-yl)methyl]amine (100 μM, 10 mM stock solution in DMSO); CuSO4•5H2O (1 mM, 50 mM freshly prepared stock solution in water)] for a total reaction volume of 200 μL. The reaction was gently vortexed and allowed to sit at room temperature for 1 h. Upon completion, 1 mL of ice cold methanol was added to the reaction, and it was placed at -80 °C for 2 h to precipitate proteins. The reactions were then centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was removed, the pellet was allowed to air dry for 5 min, and then 50 μL 4% SDS buffer (4% SDS, 150 mM NaCl, 50 mM triethanolamine, pH 7.4) was added to each sample. The mixture was sonicated in a bath sonicator to ensure complete dissolution, and 50 μL of 2x loading buffer (20% glycerol, 0.2% bromophenol blue, 1.4% β-mercaptoethanol) was then added. The samples were boiled for 5 min at 97 °C, and 30 μg of protein was then loaded per lane for SDS-PAGE gel separation (4–20% Tris•HCl Criterion Gel, Bio-Rad).
In-Gel Fluorescence Scanning.
Following SDS-PAGE gel separation, the gel was incubated in destaining solution (50% methanol, 40% H2O, 10% glacial acetic acid) for 5 min followed by H2O for an additional 5 min prior to scanning. The gel was scanned on a Molecular Imager FX (Bio-Rad) using a 580-nm laser for excitation and a 620-nm bandpass filter for detection.
Immunoprecipitation and Protein Identification.
Details concerning the enrichment of all proteins and concerning the biotin enrichment and protein identification by mass spectrometry can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank K-.L. Guan (University of Michigan Medical School, Ann Arbor, MI) for the plasmid encoding FoxO1A, S. Rosen (University of California, San Francisco, CA) for the plasmid encoding GlyCAM-IgG, X. Jiang (Memorial Sloan-Kettering Cancer Center, New York) for the plasmid encoding NEDD4-1, C. Glembotski (San Diego State University, San Diego, CA) for the plasmid encoding α-B crystallin, and the Rockefeller University Proteomics Resource Center and Guillaume Charron for MS analysis. This work was supported by the University of Southern California and the American Cancer Society Grant IRG-58-007-51 (to M.R.P) and the Ellison Medical Foundation and the National Institutes of Health National Institute of General Medical Sciences Grant 1RO1GM087544 O1A2 (to H.C.H).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102458108/-/DCSupplemental.
References
- 1.Zachara NE, Hart GW. The emerging significance of O-GlcNAc in cellular regulation. Chem Rev. 2002;102:431–438. doi: 10.1021/cr000406u. [DOI] [PubMed] [Google Scholar]
- 2.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005;312:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 4.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: Implications for multiple signaling pathways. J Cell Sci. 2010;123:13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandy JC, Rountree AE, Bijur GN. Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1. FEBS Lett. 2006;580:3051–3058. doi: 10.1016/j.febslet.2006.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardivillé S, Hoedt E, Mariller C, Benaïssa M, Pierce A. O-GlcNAcylation/phosphorylation cycling at Ser10 controls both transcriptional activity and stability of delta-lactoferrin. J Biol Chem. 2010;285:19205–19218. doi: 10.1074/jbc.M109.080572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiromura M, et al. YY1 is regulated by O-linked N-acetylglucosaminylation (O-GlcNAcylation) J Biol Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 9.Gewinner C, et al. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 2004;279:3563–3572. doi: 10.1074/jbc.M306449200. [DOI] [PubMed] [Google Scholar]
- 10.Li X, et al. O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein beta: role during adipocyte differentiation. J Biol Chem. 2009;284:19248–19254. doi: 10.1074/jbc.M109.005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musicki B, Kramer M, Becker R, Burnett A. Inactivation of phosphorylated endothelial nitric oxide synthase (Ser-1177) by O-GlcNAc in diabetes-associated erectile dysfunction. Proc Natl Acad Sci USA. 2005;102:11870–11875. doi: 10.1073/pnas.0502488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair DAR, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanover JA, et al. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsythe ME, et al. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: Oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and Dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dias W, Hart G. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 17.Zachara N, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 18.Ngoh G, Jones S. New Insights into metabolic signaling and cell survival: The role of β-O-linkage of N-acetylglucosamine. J Pharmacol Exp Ther. 2008;327:602–609. doi: 10.1124/jpet.108.143263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J, et al. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J Biol Chem. 2009;284:34777–34784. doi: 10.1074/jbc.M109.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldwell SA, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, et al. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia. 2010;24:1588–1598. doi: 10.1038/leu.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rexach JE, Clark PM, Hsieh-Wilson LC. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole RN, Hart GW. Cytosolic O-glycosylation is abundant in nerve terminals. J Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- 24.Skorobogatko YV, et al. Human Alzheimer’s disease synaptic O-GlcNAc site mapping and iTRAQ expression proteomics with ion trap mass spectrometry. Amino Acids. 2011;40:765–779. doi: 10.1007/s00726-010-0645-9. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Enrichment and site-mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation (ETD) mass spectrometry. Mol Cell Proteomics. 2009;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Iqbal K, Grundke-Iqbal I, Hart G, Gong C. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teo CF, et al. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 30.Best MD. Click chemistry and bioorthogonal reactions: Unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 31.Agard NJ, Bertozzi CR. Chemical approaches to perturb, profile, and perceive glycans. Acc Chem Res. 2009;42:788–797. doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y-Y, Ascano JM, Hang HC. Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charron G, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 34.Charron G, Wilson J, Hang H. Chemical tools for understanding protein lipidation in eukaryotes. Curr Opin Chem Biol. 2009;13:382–391. doi: 10.1016/j.cbpa.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Martin B, Cravatt B. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cravatt B, Wright A, Kozarich J. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 37.Pratt MR, Sekedat MD, Chiang KP, Muir TW. Direct measurement of cathepsin B activity in the cytosol of apoptotic cells by an activity-based probe. Chem Biol. 2009;16:1001–1012. doi: 10.1016/j.chembiol.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khidekel N, et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 39.Tai H, Khidekel N, Ficarro S, Peters E, Hsieh-Wilson L. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J Am Chem Soc. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
- 40.Khidekel N, Ficarro S, Peters E, Hsieh-Wilson L. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khidekel N, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 42.Clark PM, et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rexach JE, et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vocadlo D, Hang H, Kim E, Hanover J, Bertozzi C. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprung R, et al. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 46.Nandi A, et al. Global identification of O-GlcNAc-modified proteins. Anal Chem. 2006;78:452–458. doi: 10.1021/ac051207j. [DOI] [PubMed] [Google Scholar]
- 47.Gurcel C, et al. Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging. Anal Bioanal Chem. 2008;390:2089–2097. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 48.Boyce M, et al. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agard N, Baskin J, Prescher J, Lo A, Bertozzi C. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 50.Harvey KF, Kumar S. Nedd4-like proteins: An emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 51.Yang B, Kumar S. Nedd4 and Nedd4-2: Closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 2010;17:68–77. doi: 10.1038/cdd.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speers A, Cravatt B. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Roquemore EP, Chevrier MR, Cotter RJ, Hart GW. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry. 1996;35:3578–3586. doi: 10.1021/bi951918j. [DOI] [PubMed] [Google Scholar]
- 54.Chou CF, Smith AJ, Omary MB. Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J Biol Chem. 1992;267:3901–3906. [PubMed] [Google Scholar]
- 55.Banerjee PS, Ostapchuk P, Hearing P, Carrico I. Chemoselective attachment of small molecule effector functionality to human adenoviruses facilitates gene delivery to cancer cells. J Am Chem Soc. 2010;132:13615–13617. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hang H, Yu C, Kato D, Bertozzi C. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bistrup A, et al. Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J Cell Biol. 1999;145:899–910. doi: 10.1083/jcb.145.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luchansky SJ, Yarema KJ, Takahashi S, Bertozzi CR. GlcNAc 2-epimerase can serve a catabolic role in sialic acid metabolism. J Biol Chem. 2003;278:8035–8042. doi: 10.1074/jbc.M212127200. [DOI] [PubMed] [Google Scholar]
- 59.Tang ED, Nuñez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 60.Tran H, Brunet A, Griffith EC, Greenberg ME. The many forks in FOXO’s road. Sci STKE. 2003;172:re5. doi: 10.1126/stke.2003.172.re5. [DOI] [PubMed] [Google Scholar]
- 61.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 62.Wells L, et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 63.Vosseller K, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Lubas WA, Smith M, Starr CM, Hanover JA. Analysis of nuclear pore protein p62 glycosylation. Biochemistry. 1995;34:1686–1694. doi: 10.1021/bi00005a025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.