Abstract
Human lysyl-tRNA synthetase is bound to the multi-tRNA synthetase complex (MSC) that maintains and regulates the aminoacylation and nuclear functions of LysRS. The p38 scaffold protein binds LysRS to the MSC and, only with the appropriate cue, mobilizes LysRS for redirection to the nucleus to interact with the microphthalmia associated transcription factor (MITF). In recent work, an (α2)2 LysRS tetramer crystallized to yield a high-resolution structure and raised the question of how LysRS is arranged (dimer or tetramer) in the MSC to interact with p38. To understand the structural organization of the LysRS-p38 complex that regulates LysRS mobilization, we investigated the complex by use of small angle X-ray scattering and hydrogen-deuterium exchange with mass spectrometry in solution. The structure revealed a surprising α2β1∶β1α2 organization in which a dimeric p38 scaffold holds two LysRS α2 dimers in a parallel configuration. Each of the N-terminal 48 residues of p38 binds one LysRS dimer and, in so doing, brings two copies of the LysRS dimer into the MSC. The results suggest that this unique geometry, which reconfigures the LysRS tetramer from α2∶α2 to α2β1∶β1α2, is designed to control both retention and mobilization of LysRS from the MSC.
Keywords: noncanonical function, p38/AIMP2, structural plasticity
Aminoacyl-tRNA synthetases (aaRSs) are essential enzymes that catalyze the attachment of amino acids onto their cognate tRNAs (1, 2). Higher eukaryotic aaRSs have evolved noncatalytic domain/sequences that are lacking in their ancestor forms (3–6). Among these, a number of sequence additions and three entire proteins (p43, p38, p18) evolved to form a high molecular mass multi-tRNA synthetase complex (MSC) that is uniquely present in higher eukaryotes (from fly to human) (7–12). Comprised of nine aaRSs and the three auxiliary proteins (or named as AIMP1, AIMP2, AIMP3, respectively), MSC is regarded as a reservoir that harbors almost half of the cellular tRNA synthetases, and that controls their flow between their canonical functions and those beyond translation (13, 14).
Lysyl-tRNA synthetase (LysRS) is a component of the MSC, where it specifically interacts with the auxiliary, scaffold protein p38/AIMP2 (15–18). Mammalian LysRS has other physiological roles in addition to aminoacylation (19). For example, full-length LysRS functions as a cytokine that binds to macrophages and peripheral blood mononuclear cells to activate their migration and TNF-α production (20). Furthermore, in stimulated mast cells, LysRS is released from the cytoplasmic MSC, and then translocates to the nucleus, where it forms a complex with the microphthalmia associated transcription factor (MITF) to drive the expression of genes that regulate the immune response (21, 22). A major question is to understand how p38 regulates the assembly and release of LysRS from the MSC.
Although an α2 homodimer is the functional unit of LysRS for aminoacylation, we were able to crystallize an α2α2 tetramer and obtain a high-resolution structure that revealed details of the dimer and tetramer interface interactions. We previously speculated that the tetramer interface of LysRS was exploited by the p38 dimer to give an α2β2 subcomplex in the MSC (23). However, some published work (24, 25) suggested that extra amounts of LysRS were incorporated into the MSC compared to other MSC-bound tRNA synthetases. For these reasons, we wanted to investigate the stoichiometry and configuration of the LysRS interaction with p38, as the basis for LysRS mobilization from the MSC. Pursuant to this objective, we first solved the structure of human LysRS in solution and confirmed the existence of a stable tetramer. In further work, subcomplexes of LysRS with MSC p38 were reconstituted in vitro, and investigated by small angle X-ray scattering. Two LysRS dimers were shown to bind to the N terminus of one p38 dimer, in an α2β1∶β1α2 geometrical configuration distinct from the α2α2 tetramer of LysRS. The model is consistent with the idea that the faces of dimer-dimer interface seen in the LysRS tetramer are used for the p38 interaction. The unique geometry of this complex provides the structural context for controlling LysRS’s release from the MSC.
Results
Human LysRS Shows a Slow Dimer-Tetramer Interconversion.
Human LysRS is the only human aaRS that crystallized in an oligomeric state that was different compared to its homologues in lower species. Gel filtration chromatography showed that a fraction of LysRS had a molecular weight 4-fold greater than that of the LysRS monomer in solution (23). In accord with this finding, our previous 2.2 Å structure revealed a tetrameric form in one crystallographic asymmetric unit as the minimal unit for crystal packing (23). Interestingly, the dimer-dimer interfaces of the tetramer (referred to as the “bottom side”) lie on the side opposite of the surface that binds tRNA (top side). This observation raised the possibility that the tetramer is fully functional and that this interface provided binding sites that evolved for interactions with other proteins, such as the p38 component of the MSC.
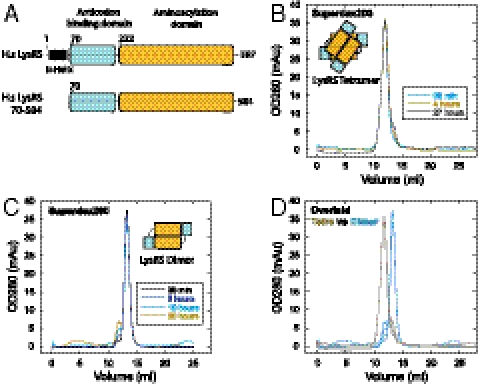
The equilibrium between the monomeric, dimeric, and tetrameric forms of human LysRS in solution suggested that the tetramer might be difficult to purify in solution. However, we observed that, for several batches of purified fractions of LysRS, the dimer and tetramer forms remained stable after long-term storage (at -80 °C). This stability suggested that the dimer-tetramer equilibrium might be slow. To assess this equilibrium, the dimer and tetramer portions of Ni-HiTrap purified human LysRS were first separated through gel filtration, then incubated at 4 °C from 30 min to up to 2 d before being reloaded on the analytical gel filtration column to determine if the dimer and tetramer could reform spontaneously. As shown in Fig. 1B, over time the dimeric form appeared and separated from the purified tetramer, indicating that a conversion from tetramer to dimer occurs. Interestingly, the dimer peak developed slowly, with < 3% of new dimers seen in the first 3 h and less than 10% forming after 27 h. The reverse equilibrium from dimer to tetramer was also kinetically slow (Fig. 1 C and D). These slow conversions of the quaternary forms suggested that conformational changes having significant activation energies occur during formation and dissociation of tetramer (26).
Fig. 1.
Slow dimer—tetramer interconversion of human lysyl-tRNA synthetase. (A) Schematic of human LysRS. The canonical fragment LysRS70-584 was used in this study because its compact structure is critical to the SAXS data analysis, and because its crystal structure has been solved. It is known that the N-terminal helix region of LysRS does not interact with p38 (17). (B–D). Sequential gel filtration analysis shows the slow conversion of dimeric LysRS to the tetramer form over 36 h, and vice versa. D is the overlay of B and C.
The Structure of Tetrameric LysRS in Solution Resembles Its Crystal Structure.
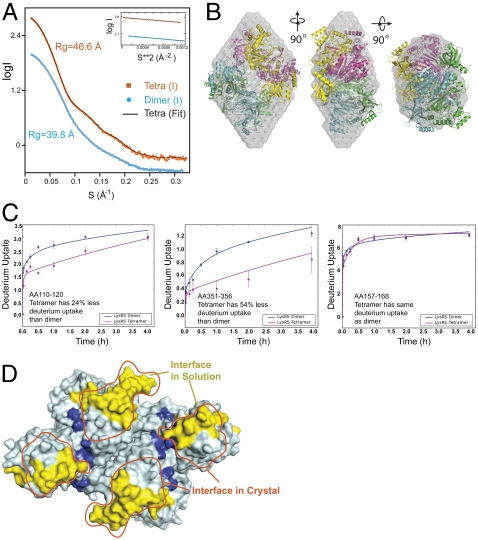
To define alterations that occur during the dimer to tetramer transitions of LysRS, tetrameric human LysRS was flash frozen immediately after being purified from dimeric LysRS. This sample was then rapidly thawed for Small Angle X-ray Scattering (SAXS) analysis, which was collected with X-ray exposures of 0.5–5 s. Based on gel filtration, minimal amount of tetrameric LysRS reequilibrated to a dimer over this time frame (2 h). As a control, SAXS data for the isolated dimer was also collected. The radius of gyration (Rg) increased from 39.8 ± 0.2 Å (with maximum diameter of 160 Å) to 46.6 ± 0.3 Å (maximum diameter of 205 Å) in the transition of the dimer to the tetramer (Fig. 2A). From the SAXS data, the apparent molecular mass, approximated by the Porod method, is 220 kDa, close to that predicted for tetrameric LysRS (60 kDa × 4,240 Da). Ab initio model building also showed a shape similar to that of the tetrameric crystal structure (Fig. 2B). Thus, tetrameric LysRS in solution has a geometric organization that, with one dimer facing the other, is similar to that of the crystalline tetrameric structure.
Fig. 2.
The structure of the LysRS tetramer in solution resembles that in the LysRS crystal structure. (A) Solution scattering data for human LysRS in its isolated dimeric and tetrameric state. Theoretical scattering profile calculated from the ab initio model with the lowest χ value is shown for the tetramer (black line). The inset shows Guinier plots (including linear fits) at the low-angle region (Smax × Rg < 1.3) indicating no aggregation in the samples. (B) SAXS-based shape reconstruction of the LysRS tetramer in solution. The crystal structure of tetrameric LysRS was docked into the envelope (gray) manually. Three orthogonal views are shown. (C) HDX-MS analysis of dimeric and tetrameric LysRS. HDX time curves for the LysRS tetramer (red) or dimer (blue) showed less solvent exposure in both the anticodon binding domain (residues 110-120) and the catalytic domain (residues 351-356). Most of the LysRS tetramer peptides showed no difference. (D) Surface representation of HDX data on the LysRS dimer structure. Less solvent-exposed peptides in LysRS tetramer are colored in yellow, with more solvent exposed peptides in dark blue. The tetramer interface found in the crystal structure is circled in red (Only the bottom view is shown).
The resolution of SAXS does not allow identification of the interface of the two LysRS dimers in solution. To do so we used hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS) (27, 28). The idea was that, for comparison of the deuterium exchange of the tetramer vs. the dimer, only those positions that were covered up in the tetramer would be less in exchange from what was observed with the dimer. For this analysis, the isolated dimer and tetramer LysRS fractions were subjected to HDX-MS analysis. The deuterium uptake was monitored over time, for each of the peptide fragments generated by proteolysis after exchange was stopped. A few short fragments in the tetramer had increased deuterium uptake that might reflect conformational changes that occur in the transition of the dimer to the tetramer (Fig. S1). Only four peptide fragments of the tetramer (residues 110-120, 133-148, 351-356, and 357-396) showed a decrease in deuterium uptake (of 8%–54%, Fig. S1, Fig. 2C). When mapped onto the crystal structure, the regions 110-120 and 351-396 are located at the bottom side of LysRS (in the N-terminal anticodon binding domain and the C-terminal catalytic domain, respectively), where they superimpose with the tetrameric interface found in the crystal structure (Fig. 2D). (Peptide 133-148 is slightly outside of but also close to the tetramer interface.) Thus, both SAXS and HDX-MS analysis are consistent with human LysRS being present in a tetrameric form in solution, similar to that seen in the LysRS crystal structure.
In Vitro Construction of the Human LysRS-p38 Complex.
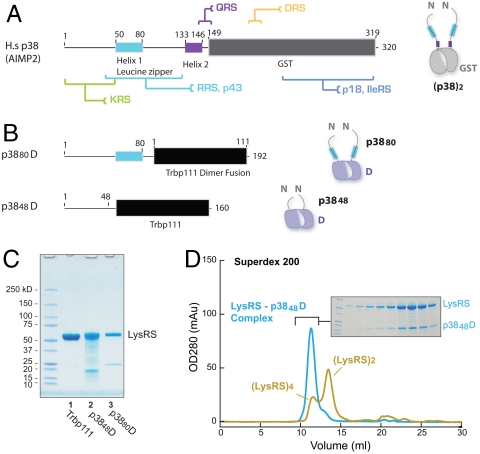
In mammalian cells, most LysRS is found within the MSC (22, 29, 30) where it directly binds to p38 (17, 31). This scaffold protein is essential for formation of the MSC and, accordingly, knockout of p38 in mice causes embryonic lethality (18). Indeed, p38 is a core component of the MSC where it interacts with LysRS, p43, ArgRS, AspRS, GlnRS, and p18 (17, 18, 31–34). Mammalian p38 contains ∼50 unstructured residues at its N terminus, followed by a coiled-coil region (50-80), another unstructured region (residue 81-132), and a short helical region (133-146) (Fig. 3A) (17, 33). The C-terminal half of p38 has a domain with structural homology to GST (35, 36), which is also found in mammalian MetRS, p18, CysRS, and ValRS, as well as in EF1β and EF1γ (35, 37, 38). All of these GST domains mediate protein-protein interactions (11, 39, 40). Particularly, several protein-binding sites have been identified in p38 (Fig. 3A) (17, 18, 32).
Fig. 3.
In vitro construction of the LysRS-p38 complex. (A) The domain organization of human MSC p38 and its binding partners. (B) p38 fusion constructs with a dimer platform. (C) SDS-PAGE analysis of the purified LysRS-p3848D and LysRS-p3880D complex after Ni-HiTrap and StrepII-tactin tandem-purification. A Ni-HiTrap purified sample of LysRS-Trbp111 is shown as a control. (D) Gel filtration analysis of purified LysRS-p3848D complex and LysRS alone. In the inset, each fraction of the LysRS-p3848D peak was loaded and stained to visualize the components.
To gain insights into how LysRS assembles into the MSC, we reconstructed the LysRS-p38 subcomplex in vitro. The possibility of forming stable complex containing only p38 and LysRS has been previously described (17). First, we attempted to generate recombinant human p38. However, trials with full-length p38 or of p38 fused in-frame with GST or MBP failed to yield soluble and stable protein, possibly due to p38’s extreme hydrophobicity. With this consideration in mind, we turned to an alternative approach to construct the subcomplex. Previous work showed that the N-terminal 42 residues of p38 can bind to LysRS, and that the N-terminal 80 residues of p38 have a binding affinity for LysRS similar to that of full-length p38 (32, 41). We predicted that fusing the LysRS-binding domain of p38 onto a soluble protein could generate a soluble and functional form. Because the GST domain of p38 interacts with itself to form homodimers and, consistently, because two p38 molecules are present in each MSC (9, 32), to more closely mimic the native state of p38, a dimeric protein is the preferred substitute for the GST domain of p38. For these reasons, we chose Escherichia coli Trbp111 (also named ygjh), a highly soluble tRNA-binding dimeric protein (42) that has both of its N termini on the same side of its dimeric structure, as seen in dimeric GST domain structures (Fig. S2 A and B). Thus, all of these features help to ensure that a complex of LysRS with the p38-Trbp111 fusion would mimic that of the LysRS-p38 complex in the MSC.
Two chimeric constructs were created by fusing the N-terminal 48 or 80 residues of human p38 with E. coli Trbp111, along with a StrepII affinity tag at C terminus (Fig. 3B). The resulting p3848D (D represents a dimer fusion) or p3880D fusions were coexpressed with human LysRS. Complexes were detected and purified by use of Ni-HiTrap for His-tagged LysRS followed by StrepII-tactin beads for the p38 fusion protein. Both p3848D and p3880D formed stable complexes with human LysRS (Fig. 3C), whereas coexpressed Trbp111 did not. Thus, the N-terminal 48 and 80 residues of p38 can form a stable and specific complex with human LysRS with Trbp111 as a platform. Further, because the N-terminal 48 residues of p38 are sufficient for binding to LysRS, we established that the LysRS binding site of p38 does not overlap with that for the previously reported p43/ArgRS binding site (residues 50-80) (33). This strategy also allowed large quantities of the stable complex to be purified, with a yield of ∼2 mg pure LysRS-p3848D complex from each liter of bacterial culture. Finally, analytic gel filtration of the purified LysRS-p3848D complex showed a single peak, indicating that the designed p3848D fusion protein forms a tight and homogenous complex with human LysRS (Fig. 3D).
Solution Structure of Human LysRS-p3848D Reveals a α4β2 Complex.
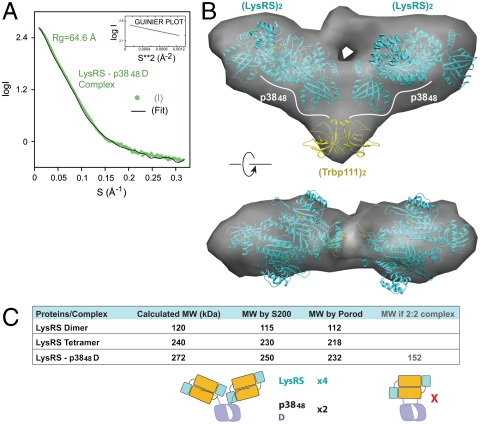
SAXS analysis was performed to visualize the structure of the LysRS-p3848D complex. The complex remained stable and did not aggregate during the short intervals of X-ray exposure (Fig. 4A). Analysis of the SAXS data established the radius of gyration of the complex as 64.6 Å, which is larger than tetrameric LysRS (Rg = 46.6 Å, see also Fig. S3). Shape reconstructions for the LysRS-p3848D complex were then generated, with a constraint of 2-fold symmetry imposed on the complex. The final envelope showed that the LysRS-p3848D complex has an extended structure with a diameter of ∼220 Å, a thickness of ∼70 Å, and a butterfly like shape (Fig. 4B). Surprisingly, based on our crystal structure of human LysRS, two LysRS dimers fit into the envelope with each dimer at one wing. The remaining space, which is located at the center of the bottom of the envelope, could only fit one Trbp111 dimer. This model thus placed LysRS and Trbp111, and showed that LysRS-p3848D forms an α4β2 complex. The bottom side of the LysRS dimer, which is the protein-protein interface in both the crystal and solution structures, is positioned to interact with the N-terminal segment of p38.
Fig. 4.
SAXS-based shape reconstruction of LysRS-p3848D complex. (A) Solution scattering data for human LysRS-p3848D complex with a Guinier plot insert. Theoretical scattering profile calculated from the ab initio model with the lowest χ value is shown (black line). (B) The overall envelope (gray) from shape reconstructions was averaged from 20 independent models. Crystal structures of the LysRS dimer and Trbp111 dimer were docked into the envelope manually. (C) Molecular mass statistics from gel filtration and SAXS data analysis. Molecular mass by Porod was estimated by mw = Vp/2, where Vp is the excluded volume (Porod volume) calculated from the SAXS data by use of PRIMUS (56).
We also calculated the molecular weight of the complex by two independent methods. An analytic gel filtration profile calibrated with standard proteins showed a molecular mass of 250 kDa for the complex. Further, Porod analysis of the SAXS data showed a similar molecular mass (232 kDa). Both molecular masses are close to the predicted molecular mass of an α4β2 (4∶2) complex (272 kDa), and are far from that of a α2β2 (2∶2) complex (of 152 kDa). Thus, our results support that an α4β2 complex is formed between LysRS and p3848D (Fig. 4C). Importantly, this architecture can form only if each of the N-terminal 48 residues of p38 binds to one LysRS dimer (Fig. 4B).
Monomeric p38 Associates with LysRS as a Half (α2β1) Complex.
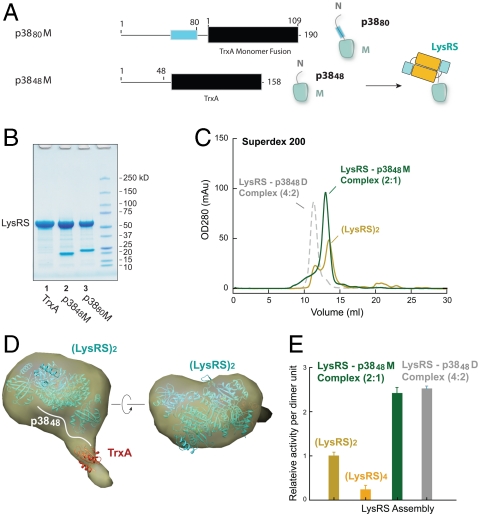
To further test the above conclusion that a single p38 N-terminal fragment binds to one LysRS dimer, we created monomeric p38 fusion proteins that can no longer dimerize. According to our model, such a separation of p38 molecules should generate a complex with LysRS that is half the size of the LysRS-p3848D complex (Fig. 5A).
Fig. 5.
A half LysRS-p38 complex is formed by monomeric p38. (A) Constructs of monomeric p38 fusions by use of a monomer platform, and a schematic diagram of the LysRS-p3848M complex. (B) SDS-PAGE analysis shows p3848M and p3880M form stable complexes with human LysRS. (C) Gel filtration analysis shows a half (2∶1) complex of LysRS-p3848M compared to LysRS-p3848D (4∶2). (D) The overall envelope (yellow) from shape reconstructions was averaged from 20 independent models. Crystal structures of the LysRS dimer and TrxA monomer were docked into the envelope manually. The weak density of TrxA suggests that it is flexible in solution. (E) Relative Lys-tRNALys aminoacylation activity of the LysRS-p38 complexes and of the LysRS dimer and tetramer forms. Each enzyme assembly (5 and 10 nM) and human tRNALys3 (8 μM) were used in reactions containing 100 μM [3H]-Lys, 2 mM ATP and 10 mM MgCl2 at 37 °C. Data are shown as mean ± SD (N = 3) and represent the activity per unit of dimer.
Two chimeric constructs were created by fusing the N-terminal 48 (or 80) residues of p38 to the E. coli TrxA (Thioredoxin) gene. Distinct from dimeric Trbp111, TrxA is present as a pure monomer in solution (Fig. S2). Tandem affinity purification for p3848M (M represents a monomer) or p3880M coexpressed with LysRS established that human LysRS formed a stable complex with both p3848M and p3880M, and that TrxA alone did not interact with LysRS (Fig. 5B). Large quantities of LysRS-p3848M were then purified and subjected to analytical gel filtration. A single homogeneous peak of the LysRS-p3848M complex eluted at a molecular mass of ∼130 kDa (Fig. 5C)—a size that is only slightly larger than that of the LysRS dimer (∼115 kDa on gel filtration), and about half the size of the LysRS-p3848D complex (at ∼250 kDa). SAXS analysis for the LysRS-p3848M complex also showed it has an estimated molecular mass of 121 kDa, and that it contains only one LysRS dimer (α2) and one p3848M fusion (β1) (Fig. 5D, Fig. S4). Thus, the LysRS dimer binds to one N-terminal segment of p38.
In this and earlier works (17, 32, 41), LysRS interacted with the N-terminal 80 residues of p38 (1-48 residues in particular) and did not interact with the C terminus of p38 (84-320). In addition, the SAXS envelopes of the LysRS-p38 complexes showed that the N terminus of p38 binds to the dimeric form of LysRS, independent of the rest of the sequences at the C terminus (M or D fusion). Because the same bottom side of the LysRS dimer is used to form the interface for the LysRS tetramer and for binding to p38, the LysRS-p38 complex and the LysRS tetramer are mutually exclusive. For these reasons, the C-terminal (80-149) region of p38 will not change the relative positions of the LysRS dimers to allow the binding of tetrameric LysRS.
To further understand the biological relevance of the various complexes we investigated, aminoacylation assays were carried out, and the results were compared with a normalization to the amount of LysRS dimer in each complex. When the tetrameric LysRS was immediately assayed after its isolation from gel filtration, the catalytic activity (apparent kcat/Km) for aminoacylation per dimer unit was decreased by 4-fold relative to the LysRS dimer (Fig. 5E). The catalytic activity per dimer unit of the LysRS-p3848M complex (which contains one LysRS dimer) was increased by 2.3-fold and that of the LysRS-p3848D complex (which contains two LysRS dimers) was increased by 2.4-fold (Fig. 5E, Fig. S5). (In terms of apparent free energies, all of these activity differences are minimal, and amount to less than 1 kcal mol-1.) These results show that, consistent with them being well folded and organized structures, the p38-fusion/LysRS complexes have robust biological activities. The activity of tetrameric LysRS is also consistent with the possibility of a biological function for the tetrameric form outside of the MSC.
Discussion
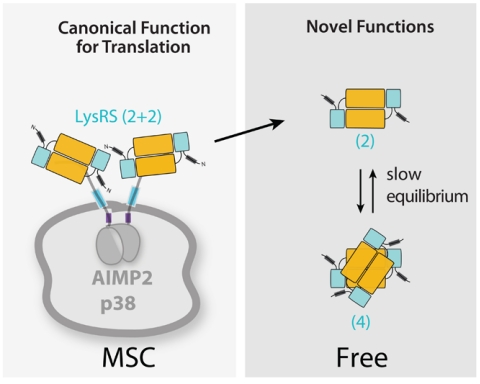
The LysRS-p3848D structure provides a model for a subcomplex of the mammalian MSC. An unexpected aspect of this structure is the one-to-one binding of the symmetric LysRS dimer to the N-terminal segment of p38. The architecture of this α2β1∶β1α2 complex is such that two juxtaposed LysRS dimers are held together by the two N-terminal segments of dimerized p38. This architecture is consistent with two LysRS dimers (i.e., four molecules of LysRS) binding to each MSC (Fig. 6) (24, 25). These findings further define the composition of MSC, in which most components were proposed to have a stoichiometry of one per MSC (31, 43–45). Close inspection of the published work on previously purified MSCs, from various species, further indicates that, in many preparations, LysRS has twice the density expected for two LysRS molecules per MSC (24, 31, 46, 47). Thus, the organization of the in vitro reconstructed complex reported here may reflect the native architectural arrangement of p38 and LysRS in the mammalian MSC.
Fig. 6.
Distinct geometries may control the mobilization of mammalian LysRS and be associated with its multifunctionality. In the MSC, each N terminus of p38 binds to one LysRS dimer, resulting in four LysRS monomers (two LysRS dimers) in a side-by-side configuration. Freed from the MSC, LysRS is either in a dimeric or a cross-like tetrameric form. Different interfaces for protein or RNA partners could form when the geometry changes.
Why extra copies of LysRS might have been selected for incorporation into the MSC is not clear. Systematic depletion of each component of the MSC by RNA interference has been reported in Hela cells (48). This work showed that the binding of LysRS to the MSC is independent of other tRNA synthetase components. In contrast, the incorporation of other components, and the formation of the entire complex, mostly depends on LysRS. The critical role of LysRS for the assembly and stability of the MSC underscores the importance of maintaining a basal level of LysRS within the MSC, even when it is specifically released in response to external or internal stimulations. Indeed, some variance in the stoichiometry of binding of LysRS to the MSC has been reported (43, 45, 49).
Although the α2β1∶β1α2 geometry of the LysRS-p38 interaction allows for the incorporation of four molecules of LysRS into the MSC, the four molecules of LysRS do not form a homotetramer. Instead, p38 holds two independent functional α2 dimers of LysRS. This design is likely to be critical for mobilizing LysRS from the MSC. The question of the form of LysRS that gets released from the MSC remains open. Whether the released form is α2β1, as seen in our LysRS-p3848M complex, or is the α2 homodimer, would depend on the relative affinity between LysRS and p38, on one hand, and the p38-p38 dimer interaction mediated by the GST domains, on the other. Phosphorylation of LysRS is involved in the release of LysRS from the MSC in immunologically activated mast cells (22). Because a phosphorylated LysRS would more likely affect the LysRS-p38 interaction than the p38-p38 dimer interaction, an α2 LysRS would be the released form. The remaining α2β1∶β1 complex may promote assembly of other components into the MSC or rearrange into a different α2β2 configuration, similar to what we have previously envisioned (23). Finally, because of the slow conversion determined here of the dimeric to the tetrameric form, we speculate that the unique functions of a mobilized LysRS most likely are carried out in the α2 dimeric form.
Methods
Expression Constructs.
Human LysRS (70-584, Fig. 1A) was constructed with a C-terminal His6x-tag in the pET20b vector (23), and into pET28a for coexpression experiments. The gene encoding human p38 was synthesized by codon optimization and cloned in vector pET20b or pBAD (Invitrogen).
Protein and Complex Preparation.
All proteins were expressed in the bacterial strain BL21(DE3) by use of the vector pET20b (Novagen). Human LysRS70-584 was with a Ni-HiTrap affinity column (GE Healthcare). E. coli Trbp111 and TrxA were purified by StrepII-tactin beads (IBA). Complexes were coexpressed and purified by the use of a Ni-HiTrap column and a StrepII-tactin column. Samples were assessed by SDS-PAGE gel and visualized with Coomassie blue stain.
Gel Filtration Chromatography.
Proteins were loaded to a Superdex 200 column (GE healthcare) in a buffer (20 mM Hepes-NaOH pH 7.5, 150 mM NaCl, 5% glycerol) as for SAXS analyses.
Small Angle X-Ray Scattering Data Collection and Analysis.
All SAXS data were collected at the SIBYLS beamline (50, 51). SAXS data were collected for three serial dilutions of each sample at maximum of 6.0 and minimum 0.5 mg/mL. Data were collected from two short exposures (0.5 s) and one long exposure (5 s) for each protein sample at room temperature (18–21 °C).
Hydrogen/Deuterium Exchange (HDX) Fourier Transform Ion Cyclotron Resonance (FT-ICR) Mass Spectrometry.
HDX samples (20 μM) was mixed with 45 μL of 20 mM Hepes, pH 7.5, 150 mM NaCl in D2O to initiate each HDX reaction, followed by simultaneous quench and proteolysis (52). Microelectrosprayed HDX samples were directed to a custom-built hybrid linear trap quadrupole 14.5-Tesla FT-ICR mass spectrometer (ThermoFisher) (53). The total data acquisition period for each sample was 6 min. Data were analyzed by an in-house analysis package (54). Time-course deuterium incorporation levels were generated by an MEM fitting method (55).
Supplementary Material
Acknowledgments.
We thank Dr. Michal Hammel, Kevin Dyer, and Jane Tanamachi for the assistance with SAXS data collection, and Dr. John Cleveland for critical review of the manuscript. We also thank Dr. Mark R. Emmett at the National High Magnetic Field Lab for the assistance with the HDX-MS experiments. This work was supported in part by grants from National Institutes of Health (NIH) (GM 15539, 23562, and 78359), National Science Foundation (NSF) (DMR 06-54118), by a fellowship from the National Foundation for Cancer Research, and by funding from The State of Florida to Scripps Florida.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100224108/-/DCSupplemental.
References
- 1.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Mirande M. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- 4.Francklyn C, Musier-Forsyth K, Martinis SA. Aminoacyl-tRNA synthetases in biology and disease: new evidence for structural and functional diversity in an ancient family of enzymes. RNA. 1997;3:954–960. [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 6.Kise Y, et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 7.Kellermann O, et al. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. I. Species specificity of the polypeptide composition. J Biol Chem. 1982;257:11041–11048. [PubMed] [Google Scholar]
- 8.Deutscher MP. The eucaryotic aminoacyl-tRNA synthetase complex: suggestions for its structure and function. J Cell Biol. 1984;99:373–377. doi: 10.1083/jcb.99.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerjan P, Cerini C, Semeriva M, Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim Biophys Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 10.Rho SB, et al. Interaction between human tRNA synthetases involves repeated sequence elements. Proc Natl Acad Sci USA. 1996;93:10128–10133. doi: 10.1073/pnas.93.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rho SB, et al. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci USA. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melamed D, et al. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 gag protein is important for virion assembly and release. J Virol. 2004;78:9675–9688. doi: 10.1128/JVI.78.18.9675-9688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Dang CV, Yang DC. Affinity chromatography of rat liver aminoacyl-tRNA synthetase complex. Biochem Biophys Res Commun. 1978;80:709–714. doi: 10.1016/0006-291x(78)91302-5. [DOI] [PubMed] [Google Scholar]
- 16.Mirande M, Cirakoglu B, Waller JP. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. III. Assignment of aminoacyl-tRNA synthetase activities to the polypeptide components of the complexes. J Biol Chem. 1982;257:11056–11063. [PubMed] [Google Scholar]
- 17.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 18.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc Natl Acad Sci USA. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nechushtan H, Kim S, Kay G, Razin E. Chapter 1: The physiological role of lysyl tRNA synthetase in the immune system. Adv Immunol. 2009;103:1–27. doi: 10.1016/S0065-2776(09)03001-6. [DOI] [PubMed] [Google Scholar]
- 20.Park SG, et al. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc Natl Acad Sci USA. 2005;102:6356–6361. doi: 10.1073/pnas.0500226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 22.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34:603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 23.Guo M, Ignatov M, Musier-Forsyth K, Schimmel P, Yang XL. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: implications for multisynthetase complex formation. Proc Natl Acad Sci USA. 2008;105:2331–2336. doi: 10.1073/pnas.0712072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirande M, Le Corre D, Waller JP. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur J Biochem. 1985;147:281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- 25.Mirande M. Aminoacyl-tRNA Synthetases Complexes. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Georgetown: Eurekah; 2005. pp. 298–308. [Google Scholar]
- 26.Benaroudj N, Triniolles F, Ladjimi MM. Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSC70. J Biol Chem. 1996;271:18471–18476. doi: 10.1074/jbc.271.31.18471. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 29.Halwani R, et al. Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J Virol. 2004;78:7553–7564. doi: 10.1128/JVI.78.14.7553-7564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29:419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminska M, et al. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J Biol Chem. 2009;284:6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- 32.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 33.Ahn HC, Kim S, Lee BJ. Solution structure and p43 binding of the p38 leucine zipper motif: coiled-coil interactions mediate the association between p38 and p43. FEBS Lett. 2003;542:119–124. doi: 10.1016/s0014-5793(03)00362-4. [DOI] [PubMed] [Google Scholar]
- 34.Guzzo CM, Yang DC. Lysyl-tRNA synthetase interacts with EF1alpha, aspartyl-tRNA synthetase and p38 in vitro. Biochem Biophys Res Commun. 2008;365:718–723. doi: 10.1016/j.bbrc.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 35.Simader H, et al. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic Acids Res. 2006;34:3968–3979. doi: 10.1093/nar/gkl560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo M, Schimmel P, Yang XL. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584:434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin EV, et al. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain—study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KJ, et al. Determination of three-dimensional structure and residues of the novel tumor suppressor AIMP3/p18 required for the interaction with ATM. J Biol Chem. 2008;283:14032–14040. doi: 10.1074/jbc.M800859200. [DOI] [PubMed] [Google Scholar]
- 39.Negrutskii BS, Shalak VF, Kerjan P, El'skaya AV, Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J Biol Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 40.Kim JE, et al. An elongation factor-associating domain is inserted into human cysteinyl-tRNA synthetase by alternative splicing. Nucleic Acids Res. 2000;28:2866–2872. doi: 10.1093/nar/28.15.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MJ, et al. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 42.Swairjo MA, Morales AJ, Wang CC, Ortiz AR, Schimmel P. Crystal structure of trbp111: a structure-specific tRNA-binding protein. EMBO J. 2000;19:6287–6298. doi: 10.1093/emboj/19.23.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norcum MT, Warrington JA. Structural analysis of the multienzyme aminoacyl-tRNA synthetase complex: a three-domain model based on reversible chemical crosslinking. Protein Sci. 1998;7:79–87. doi: 10.1002/pro.5560070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JM, Kim JY, Kim S. Molecular network and functional implications of macromolecular tRNA synthetase complex. Biochem Biophys Res Commun. 2003;303:985–993. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe CL, Warrington JA, Treadwell L, Norcum MT. A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J Biol Chem. 2005;280:38870–38878. doi: 10.1074/jbc.M502759200. [DOI] [PubMed] [Google Scholar]
- 46.Shalak V, et al. The EMAPII cytokine is released from the mammalian multisynthetase complex after cleavage of its p43/proEMAPII component. J Biol Chem. 2001;276:23769–23776. doi: 10.1074/jbc.M100489200. [DOI] [PubMed] [Google Scholar]
- 47.Kaminska M, Shalak V, Francin M, Mirande M. Viral hijacking of mitochondrial lysyl-tRNA synthetase. J Virol. 2007;81:68–73. doi: 10.1128/JVI.01267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han JM, et al. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 49.Godar DE, Garcia V, Jacobo A, Aebi U, Yang DC. Structural organization of the multienzyme complex of mammalian aminoacyl-tRNA synthetases. Biochemistry. 1988;27:6921–6928. doi: 10.1021/bi00418a038. [DOI] [PubMed] [Google Scholar]
- 50.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 51.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang HM, et al. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal Chem. 2010;82:1450–1454. doi: 10.1021/ac902550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaub TM, et al. High-performance mass spectrometry: Fourier transform ion cyclotron resonance at 14.5 Tesla. Anal Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 54.Kazazic S, et al. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectr. 2010;21:550–558. doi: 10.1016/j.jasms.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Li W, Logan TM, Li M, Marshall AG. Human recombinant [C22A] FK506-binding protein amide hydrogen exchange rates from mass spectrometry match and extend those from NMR. Protein Sci. 1997;6:2203–2217. doi: 10.1002/pro.5560061015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.