Abstract
The ubiquitin–proteasome pathway plays an important role in the pathogenesis of neurodegeneration, but mechanisms controlling expression of components in this pathway remain poorly understood. Nuclear factor E2-related factor 1 (Nrf1) transcription factor has been shown to regulate expression of antioxidant and cytoprotective genes. To determine the function of Nrf1 in the brain, mice with a late-stage deletion of Nrf1 in neuronal cells were generated. Loss of Nrf1 leads to impaired proteasome function and neurodegeneration. Gene expression profiling and RT-PCR analysis revealed a coordinate down-regulation of various proteasomal genes including PsmB6, which encodes a catalytic subunit of the proteasome. Transcriptional analysis and chromatin immunoprecipitation experiments demonstrated that PsmB6 is an Nrf1 target gene. These findings reveal Nrf1 as a key transcriptional regulator required for the expression of proteasomal genes in neurons and suggest that perturbations of Nrf1 function may contribute to the pathogenesis of neurodegenerative diseases.
Keywords: oxidative stress, conditional knockout, antioxidant response element
The ubiquitin–proteasome system (UPS) is one of the main pathways for intracellular protein degradation (1). Proteins destined for proteasomal degradation are specifically tagged with ubiquitin moieties mediated by a set of E1, E2, and E3 enzymes (2). Proteolysis of ubiquitinated substrates occurs in the 20S core complex, which is made up of α-protein and β-protein subunits encoded by different PsmA and PsmB genes (3). Proteolytic activity resides in three of the β-subunits, β-1, β-2, and β-5, which are encoded by PsmB6, -B7, and -B5 genes, respectively (4). The 20S core particle is capped at each end by a 19S complex that binds and unfolds ubiquitinated substrates, facilitating their entry into the 20S core particle. The 19S is made of ATPase and non-ATPase protein subunits encoded by the PsmC and PsmD genes, respectively. Together, the 20S and 19S complexes make up the 26S particle. Abnormal UPS function has been implicated in numerous pathological conditions (5). Malignancies can result from stabilization of oncoproteins or destabilization of tumor suppressors, and impaired UPS function has been implicated in neurodegenerative disorders (6, 7). Although a common pathological hallmark in these degenerative diseases is the accumulation of ubiquitinated protein aggregates, a direct link between aberrant UPS function and neurodegeneration has not been firmly established.
Nuclear factor erythroid-derived 2-related factor 1 (Nrf1), also known as NFE2L1/LCRF1/TCF11, is a member of the CNC subfamily of basic-leucine zipper (bZIP) transcription factors that also includes Nrf2 and Nrf3 (8). CNC factors heterodimerize with small-Maf proteins and bind DNA motifs including the antioxidant response element (ARE), which regulates expression of genes involved in oxidative stress response (9). Numerous studies indicate a pivotal role for Nrf2 in regulating ARE-driven gene expression (10). Although Nrf1 can direct ARE-mediated expression of genes involved in oxidative stress response, it has also been implicated in the control of a variety of cellular processes (11). Absence of Nrf1 in knockout mice results in lethality late in gestation that is most likely due to abnormal fetal liver erythpoiesis and anemia (12). Nrf1 is required for the survival of hepatocytes, and a deficiency in Nrf1 in hepatocytes leads to spontaneous development of steatohepatitis and hepatic neoplasia (13, 14).
Here we describe the generation and analysis of CaMK2cre-directed conditional Nrf1 knockouts to determine the function of Nrf1 in the brain, where it is highly expressed. We demonstrate that conditional knockout of Nrf1 in the brain leads to proteasome impairment and progressive degeneration in cortical neurons. Our findings establish a critical role for Nrf1 in maintaining proteasome function within the CNS and provide evidence that Nrf1 is an important transcriptional regulator of proteasome genes.
Results
Generation of Nrf1 Brain-Specific Conditional Knockout.
In situ hybridization (ISH) of Nrf1-specific probe was widespread in the mouse brain. High levels were detected in the cortex, cornu ammonis (CA) subregions and dentate gyrus of the hippocampus, and choroids plexus in adult mice brains (Fig. S1A). Nrf1 immunoreactivity was prominently detected in the mouse brain in a pattern similar to that seen by ISH, and double-labeling showed overlap between Nrf1 staining and cells positive for the neural marker NeuN (Fig. S1B). Cultured cortical neurons also showed strong immunostaining for Nrf1 (Fig. S1C). These results indicate that Nrf1 is highly expressed in neurons. To address the role of Nrf1 in the adult brain, we generated mice with deletion of Nrf1 selectively in the brain to bypass embryonic lethality in constitutive Nrf1 knockout mice. The Nrf1 flox mouse was crossed with the Calcium-calmodulin-dependent Protein Kinase Type 2–Cre (Camk2Cre) transgenic mouse to generate Camk2Cre;Nrf1−/flox animals, herein referred to as Nrf1BKO. Cre expression in Camk2Cre mice has been shown previously to occur at 1 mo of age and to be confined primarily to differentiated neurons in the forebrain (15). In accord with this, the recombined Nrf1 allele was detected in the cortex but not the cerebellum of Nrf1BKO mouse (Fig. S2A). In situ mRNA hybridization using Nrf1-specific riboprobe showed Cre/loxP-mediated Nrf1 deletion in the cortex and hippocampus of Nrf1BKO brain (Fig. S2B). Disruption of Nrf1 was further verified by immunofluorescence staining in the hippocampus (Fig. S2C).
Nrf1BKO Mice Show Age-Dependent Forebrain Atrophy.
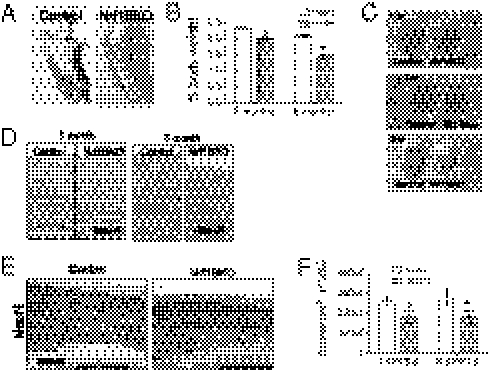
Nrf1BKO mice were born at the expected ratio without evidence of developmental defects, and they were indistinguishable from their control littermates at weaning. However, a number of behavioral abnormalities were observed in the course of maintaining these animals. At 3 to 4 mo of age, Nrf1BKO animals showed abnormal hindlimb-clasping reflexes that are often observed in mouse models of neurodegeneration (Fig. 1A) (16). To examine whether Nrf1BKO mice show age-dependent neuronal loss, we compared brain weights of Nrf1BKO and control mice. No significant differences could be observed at weaning, but Nrf1BKO brain weights were dramatically lower by 6 mo (Fig. 1B). Gross examinations showed that reduction was attributable to forebrain atrophy (Fig. 1C). Histologic evaluation indicated that the thickness of the cortex and hippocampus in Nrf1BKO mice was reduced (Fig. 1D) as a result of decreased number of NeuN-positive cells in Nrf1BKO brain (Fig. 1E). Neuronal loss was quantitatively assessed by stereological analysis. Nrf1BKO mice showed a 23% and 38% (P < 0.05, respectively) reduction in the volume of the cortex at 3 and 6 mo, respectively (Fig. 1F). These results indicate that Nrf1BKO mice suffer significant neuronal loss.
Fig. 1.
Nrf1BKO mice show age-dependent brain atrophy. (A) Nrf1BKO mouse displaying limb-clasping reflex during tail hanging. This is characterized by adduction of hind limbs close to the body with paws clasped together. (B) Normalized brain weights at 2 and 6 mo of age. Means (n = 4 per group) ± SEM. *P < 0.05. (C) Representative brains at 1, 3, and 6 mo. (D) Representative brain images of coronal sections from Nrf1BKO mice and control littermates. Sections were stained with H&E. (E) Brain sections of 6-mo-old control and Nrf1BKO mice stained for NeuN. (F) Stereological cell counts of cortical NeuN immunoreactive neurons in 3- and 6-mo-old Nrf1BKO and control littermates. Means (n = 3 samples per group) ± SEM. *P < 0.05.
Apoptotic Cell Death and Accumulation of Ubiquitin in Neurons of Nrf1BKO Mice.
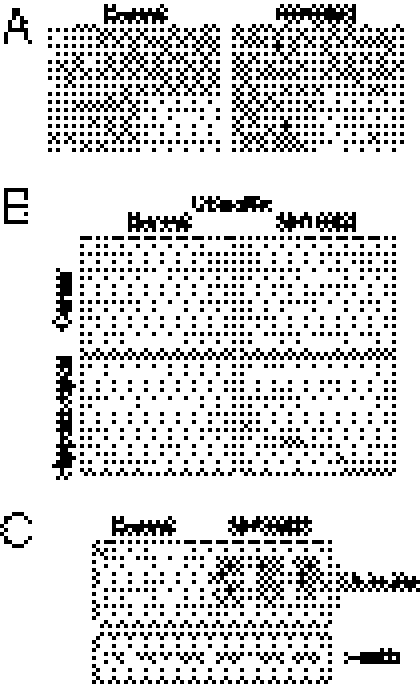
To identify degenerating neurons, Fluoro Jade B staining was done. Fluoro Jade B staining was observed in the cortex and hippocampus of Nrf1BKO brains compared with controls (Fig. S3A). Numerous apoptotic nuclei were seen in H&E-stained sections of Nrf1BKO brains compared with control brains (Fig. 2A). Consistent with this, Nrf1BKO brains showed activated caspase-3 staining (24 ± 7 positive cells per 20× field), whereas none was detected in controls (Fig. 2A, Inset). In addition, activation of caspase-3 was colocalized to neurons (Fig. S3C). Activated caspase-9, a protease involved in the upstream regulation of apoptosis, was also detected in Nrf1BKO brains (Fig. S3B). Increased GFAP immunostaining was also observed (Fig. S3D), suggesting that astrogliosis accompanies neuronal damage. Interestingly, numerous ubiquitin-positive cells were detected as early as 1 mo of age, and they were primarily in the cerebral cortex and CA subfields of the hippocampus (Fig. 2B). Double-staining experiments showed that ubiquitin-immunoreactive cells are also positive for NeuN, indicating that neurons were affected (Fig. S3E). Western blotting showed an increase in high-molecular-weight ubiquitin–protein conjugates in Nrf1BKO brains compared with controls (Fig. 2C).
Fig. 2.
Histopathology of neurodegeneration in Nrf1BKO mice. (A) Representative 6-wk-old brain sections stained with H&E. Note pyknotic cells in the Nrf1BKO brain. Insets: Activated caspase-3 immunostaining. (B) One-month-old brains immunostained for ubiquitin. Note ubiquitin-positive inclusions in the cerebral cortex and hippocampus of Nrf1BKO brains. (C) Western blot analysis of total brain lysates with ubiquitin antibody.
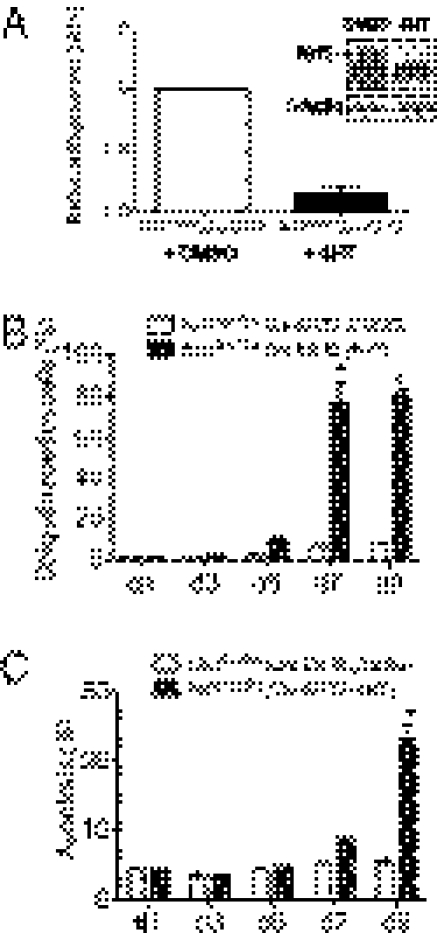
To determine whether abnormalities observed were a direct effect of Nrf1 loss of function in neurons, we analyzed neuronal cells derived from Nrf1flox/flox mice bred to mice expressing tamoxifen-inducible Cre recombinase (Cre-ERT2). This strategy allows the inactivation of Nrf1 in cells by treatment with 4-hydroxytamoxifen (4HT). Quantitative RT-PCR analysis and immunoblotting to verify the efficiency of tamoxifen-induced recombination in cultures of Nrf1flox/flox/Cre-ERT2 neuronal cells showed that Nrf1 expression was markedly reduced after 72-h treatment (Fig. 3A). After 5 d of treatment, Nrf1flox/flox/Cre-ERT2 neuronal cultures showed fourfold increase in number of ubiquitin immunoreactive cells compared with Nrf1flox/flox/Cre-ERT2 cultures treated with vehicle (Fig. 3B). By 7 d, most of the cells in 4HT-treated Nrf1flox/flox/Cre-ERT2 cultures were ubiquitin positive (Fig. 3B and Fig. S3F). TUNEL labeling did not reveal a significant difference between vehicle-treated Nrf1flox/flox/Cre-ERT2 at the different time points investigated. However, a significant increase in apoptosis was detected in Nrf1flox/flox/Cre-ERT2 cells compared with Nrf1+/+;Cre-ERT2 cells after 9 d of 4HT treatment (Fig. 3C and Fig. S3G). These results suggest that the defects in Nrf1BKO brains are directly associated with Nrf1 deficiency in neurons, and accumulation of ubiquitin is not a consequence of cell death.
Fig. 3.
Accumulation of ubiquitinated proteins and apoptosis are induced by loss of Nrf1 in neurons. (A) Nrf1 expression in Nrf1flox/flox;Cre-ERT2 neuronal cultures treated with DMSO or 4HT. Nrf1 mRNA levels were measured by quantitative RT-PCR. Data were normalized to the 18s rRNA; means ± SEM, n = 3. *P < 0.05. Inset: Western blot analysis of Nrf1 in DMSO and 4HT treated Nrf1flox/flox;Cre-ERT2 neuronal cells. (B) Quantitation of ubiquitin immunostaining in Nrf1flox/flox;Cre-ERT2 neuronal cultures at various days after treatment with DMSO or 4HT. (C) Quantitation of TUNEL labeling in Nrf1flox/flox;Cre-ERT2 neuronal cultures at various days after treatment with DMSO or 4HT.
Impaired Proteasomal Function in Nrf1BKO Brains.
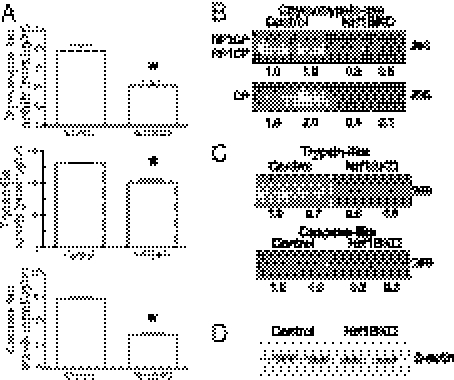
The accumulation of ubiquitinated proteins suggested that proteasome impairment could be involved in neuronal damage observed in the Nrf1BKO mice. Indeed, Nrf1BKO brains showed a 30% decrease in chymotrypsin-like activity compared with controls (Fig. 4A). Trypsin-like and caspase-like activities were also diminished by 20% and 50%, respectively in Nrf1BKO brains (Fig. 4A). To confirm these results, in-gel assay to measure proteasome activity was performed. The chymotrypsin-like activities of both the 26S and the 20S proteasomes were markedly reduced in Nrf1BKO brains compared with controls (Fig. 4B). Similarly, in-gel measurements of trypin-like and caspase-like activities of Nrf1BKO brains were also diminished compared with controls (Fig. 4C). Immunoblotting against actin showed that equal amounts of brain lysates were used for the in-gel studies (Fig. 4D).
Fig. 4.
Proteasome activity is impaired in Nrf1BKO brains. (A) Proteolytic activities of the proteasome subtypes from 1-mo-old brains. Mean values ± SEM (control n = 6, Nrf1BKO n = 6). *P ≤ 0.05. (B) Measurement of chymotrypsin-like activity by in-gel assay with fluorogenic Suc-LLVY-AMC as a substrate. Fluorescence from free AMC was visualized on a UV transilluminator. Six control and six NrfBKO samples were analyzed, and two representative samples from each genotype are shown. Upper: The 26S proteasome that appears in two forms—RP1CP, 20S CP with one bound regulatory particle; and RP2CP, 20S CP with two bound regulatory particles. Lower: The faster-migrating free 20S core particle. Densitometric quantitations for 26S and 20S levels are shown. (C) In-gel analysis of trypsin-like and caspase-like activities in control and Nrf1BKO brains. Two representative samples from each genotype are shown. Densitometric quantitations are shown. (D) Amounts of protein used for in-gel assays were evaluated by β-actin immunoblotting.
Nrf1 Deficiency in Cells Leads to Impaired Proteasome Function and Hypersensitivity to Proteasome Inhibition.
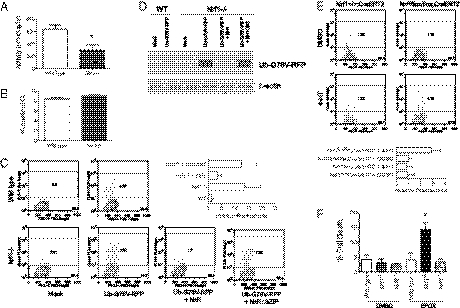
We next examined whether proteasome function is also impaired in primary mouse embryonic fibroblasts (MEFs) from Nrf1−/− animals. A significant decrease in proteasome activity was observed in Nrf1−/− MEF cells in comparison with wild-type MEF cells (Fig. 5A). In line with these data, a lentiviral shRNA-mediated system that provided efficient knockdown of endogenous Nrf1 in 293 cells resulted in 40% decrease in chymotrypsin-like activity in 293 cells (Fig. S4 A and B). To measure intracellular proteasome activity, we monitored steady-state levels of red fluorescent protein (RFP) in cells transfected with UbG76V-RFP expression plasmid. The UbG76V-RFP is a highly unstable protein targeted for degradation by the proteasome and is normally present at low levels in transfected cells unless proteasome activity is impaired (17). Although both wild-type and Nrf1−/− MEF cells transfected with UbG76V-RFP showed similar expression of RFP mRNA (Fig. 5B), RFP fluorescent levels were markedly elevated in Nrf1−/− MEF cells compared with wild-type cells (Fig. 5C). In contrast, fluorescent levels in wild-type cells were close to background levels (Fig. 5C). Confirming the instability of the UbG76V-RFP protein in wild-type cells, Nrf1−/− cells showed increased UbG76V-RFP protein levels (Fig. 5D). Transfection of wild-type Nrf1 cDNA, but not a bZIP deletion mutant of Nrf1, was able to restore UbG76V-RFP clearance in Nrf1−/− cells (Fig. 5 C and D). In addition, transfection of Nrf2 also did not restore UbG76V-RFP clearance in Nrf1−/− cells, indicating that effects are specific to Nrf1 (Fig. S4C). To further assess the potential for a direct contribution of Nrf1 deficiency to proteasome impairment, clearance of UbG76V-RFP was also monitored in MEF cells rendered Nrf1 deficient. MEF cells generated from Nrf1flox/flox;Cre-ERT2 mice were treated with DMSO or 4HT for 72 h and then transfected with UbG76V-RFP. RFP fluorescent levels were significantly higher in 4HT-treated Nrf1flox/flox;Cre-ERT2 cells compared with the same cells treated with DMSO, as well as Nrf1+/+;Cre-ERT2 control cells treated with 4HT or DMSO (Fig. 5E).
Fig. 5.
Nrf1 deficiency leads to impairment in turnover of UbG76V-RFP and sensitization to proteasome inhibitor-induced cell death. (A) Chymotrypsin-like activities in MEF cells. Mean values ± SEM (n = 3 per genotype). *P ≤ 0.05. (B) RFP mRNA levels in cells transfected with UbG76V-RFP were measured by Quantitative RT-PCR. Values are expressed as threshold cycle (Ct) corrected for 18s rRNA levels. Mean values ± SEM (n = 3 per genotype). (C) Flow cytometric analysis and (D) immunoblot analysis of cells transfected with the UbG76V-RFP along with Nrf1, or Nrf1ΔbZIP mutant expression vectors. (E) Flow cytometric analysis of RFP fluorescence in Nrf1flox/flox;Cre-ERT2 and Nrf1+/+;Cre-ERT2 MEFs transfected with UbG76V-RFP and treated with DMSO or 4HT. (F) A comparison of viability between wild-type, Nrf1−/−, and Nrf2−/− cells cultured with vehicle or 10 μM of epoxomicin (EPOX) for 24 h. Cell death was determined by propidium iodide staining and FACS analysis. Graph shows mean ± SEM (n = 3 per genotype). *P ≤ 0.05.
To assess the functional effect of Nrf1 deficiency on proteolytic stress, we tested whether Nrf1−/− MEFs and Nrf1 knockdown cells are sensitized to proteasome inhibition. Nrf1−/− MEF cells treated with epoxomicin showed a threefold increase in cell death compared with wild-type and Nrf2−/− cells (Fig. 5F). Similarly, treatment with MG132 caused increased cell death and reduced the colony-forming capability of Nrf1 knockdown cells compared with scramble cells (Fig. S4D).
Nrf1BKO Brains Do Not Show Increased Oxidative Stress.
Because the degradative capacity of the proteasome may be compromised by oxidative stress and Nrf1 has been shown to be involved in the antioxidant pathway, we examined whether proteasomal defects in Nrf1BKO brains are associated with increased oxidative stress. The ratios of reduced to oxidized glutathione (GSH/GSSG) levels in control and Nrf1BKO brain tissues were similar (Fig. S5A), and no significant differences in GSH and GSSG levels were seen in control and Nrf1BKO brain tissues (Fig. S5 A and C). As a positive control, we examined GSH and GSSG levels in Nrf2 knockouts. GSH/GSSG ratio was twofold lower in Nrf2 knockout brains (Fig. S5A). Consistent with the primary role of Nrf2 in coordinating cellular defense against oxidative stress in neurons (18), the reduction in GSH/GSSG ratio in Nrf2 knockout brains was associated with an elevation in GSSG level (Fig. S5C). We next measured intracellular reactive oxygen species (ROS) levels in neuronal cells. No increase in ROS levels was detected in Nrf1flox/flox/Cre-ERT2 neurons treated with 4HT compared with neurons treated with vehicle (Fig. S5D). Although Nrf2 knockout brains showed evidence of oxidative stress, impairment in proteasome activity was not detected (Fig. S5E). Together, these results suggest that proteasome dysfunction is not linked to oxidative stress in Nrf1BKO brains.
Proteasome Gene Expression Is Down-regulated in Nrf1-Deficient Brains and Cells.
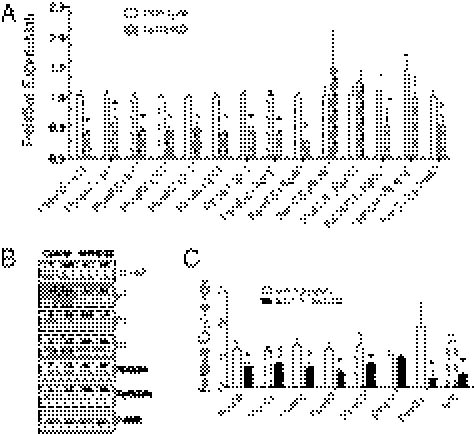
To better understand the Nrf1BKO phenotype, microarray transcriptional profiling was done. Frontal cortices of Nrf1BKO and matched controls were analyzed by oligonucleotide arrays. A total of 1,149 genes were identified by ANOVA (P < 0.05) as differentially expressed. Among these genes, 574 were underexpressed, and 575 genes were overexpressed in Nrf1BKO frontal cortex compared with control. This dataset was then analyzed using Ingenuity Pathway Analysis (IPA) software to identify processes that might be affected in the Nrf1 brain knockouts. The top canonical pathway identified was associated with proteasome function (Fig. S6). Other top-scoring networks identified also included High-affinity IgE receptor (Fc-epsilon RI) signaling, axon guidance, and inflammatory response. The increase in inflammatory response is consistent with the ongoing neurodegeneration and astrocytic response observed in the Nrf1BKO brains. However, the relevance of Fc-epsilon RI signaling and axon guidance is not clear and was not pursued further here. On the basis of proteasomal dysfunction exhibited by Nrf1BKO brains, genes representative of the 20S core and 19S regulatory complex of the proteasome were further studied. Quantitative RT-PCR showed down-regulated expression of genes encoding various subunits of the 20S core, as well the 19S regulatory complex in Nrf1BKO brains (Fig. 6A). In accord with RT-PCR results, Western blotting showed decreased levels of α-subunits and catalytic β-subunits of the 20S core, as well as RPT2 and RPT5 of the 19S complex (Fig. 6B). To determine whether expression of some of the above proteasome genes was directly linked to Nrf1, we analyzed their expression in Nrf1flox/flox/Cre-ERT2 and Nrf1+/+/Cre-ERT2 neurons after tamoxifen treatment. Consistent with results obtained from knockout brains, expression of PsmA6, -B2, -B6, -B7, and -D11 was decreased in Nrf1flox/flox/Cre-ERT2 neurons compared with tamoxifen-treated Nrf1+/+/Cre-ERT2 neurons (Fig. 6C). In addition, the knockdown of Nrf1 in 293 cells resulted in a reduction of α-subunits and catalytic β-subunits compared with cells transduced with vector or with a scrambled shRNA (Fig. S4A). Together these data suggest that proteasome dysfunction in Nrf1BKO can be explained by alterations in proteasome subunit content.
Fig. 6.
Nrf1 deficiency in mouse brains leads to coordinate down-regulation of proteasome genes. (A) Comparison of proteasome gene expression by quantitative RT-PCR analysis. Mean values ± SEM (n = 3 for each genotype). *P < 0.05. (B) Levels of proteasome subunits analyzed by Western blotting. Densitometric quantitations of band intensities are shown. (C) Quantitative RT-PCR analysis of mRNA encoding proteasomal genes in Nrf1+/+/Cre-ERT2 and Nrf1flox/flox/Cre-ERT2 neuronal cultures after tamoxifen treatment. Results are means ± SEM, n = 4 for each group. *P < 0.05.
PsmB6 Is a Direct Nrf1 Target Gene.
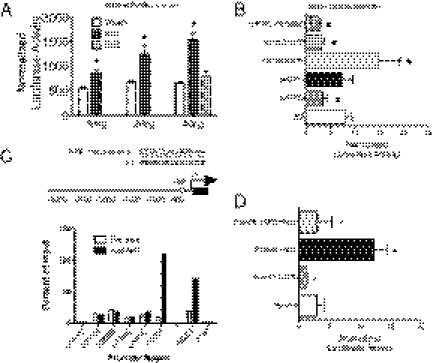
We next determined whether proteasome genes are directly under the control of Nrf1. On the basis of our expression results above, PsmB6 (encoding the catalytic β1-subunit) was chosen as a representative target gene. The promoter region of PsmB6 gene (3.0 kb) was isolated from genomic DNA by PCR amplification and cloned into the pGL3Basic luciferase reporter plasmid. Nrf1 activated the PsmB6 luciferase reporter in a dose-dependent manner (Fig. 7A). Nrf2 cotransfection did not increase reporter expression, suggesting that the activation is Nrf1 specific (Fig. 7A). To determine whether the PsmB6 luciferase reporter expression is Nrf1 dependent, we compared expression of the PsmB6 luciferase reporter plasmid in wild-type and Nrf1 knockout MEFs. Luciferase expression in Nrf1 knockout MEFs was threefold lower compared with wild-type and Nrf2 knockout MEFs (Fig. 7B), and low expression of the reporter was rescued by cotransfection of Nrf1 expression plasmid (Fig. 7B). Cotransfection of bZIP deletion-mutant Nrf1, or Nrf2, did not rescue promoter activity (Fig. 7B). To further substantiate the role of Nrf1 in PsmB6 activation, luciferase reporter activity was reduced by 50% in Nrf1 knockdown cells (Fig. S4E). Next, chromatin immunoprecipitation assays were done to examine endogenous Nrf1 protein interactions with PsmB6 genomic sequences. Chromatin isolated from MEF cells was immunoprecipitated with an anti-Nrf1 antibody or preimmune rabbit IgG as control and subjected to PCR amplification using primers spanning 3 kb of the PsmB6 gene promoter region (Fig. 7C). Enrichment of Nrf1 was detected with a primer pair spanning the region +80 to −500 nt from the transcriptional start site of PsmB6 gene (Fig. 7C). Sequence inspection revealed a perfect consensus Nrf1 binding site at −38 nt. Next, a deletion of the ARE in the −38 region was generated to verify its role in the regulation of the PsmB6 gene promoter. Both basal and activated luciferase expression by Nrf1 cotransfection was blunted when the ARE region was deleted (Fig. 7D). These data provide support that Nrf1 directly regulates PsmB6 expression in vivo.
Fig. 7.
Nrf1 regulates PsmB6 gene. (A) Transactivation of the mouse PsmB6 promoter by Nrf1. PsmB6 luciferase reporter was transfected along with vector, Nrf1, or Nrf2 cDNA into 293 cells. Activities represent the mean of at least three independent experiments ± SEM. *P < 0.05. (B) Activity of the PsmB6 luciferase reporter in wild-type, Nrf1−/−, Nrf2−/−, or Nrf1−/− fibroblasts expressing Nrf1 cDNA, Nrf2 cDNA, or Nrf1 cDNA containing a deletion in the bZIP domain. Results represent the mean of at least three independent experiments ± SEM. *P < 0.05. (C) Chromatin immunoprecipitation of PsmB6 promoter in vivo. Graph represents real-time PCR amplification of chromatin template precipitated with either anti-Nrf1 or rabbit preimmune IgG. Primers were targeted to various regions spaced 500 bp apart along the PsmB6 promoter. NQO1 and LDH promoters were used as positive and negative controls, respectively. Nonimmunoprecipitated chromatin (1%) was used as an input control. The open box indicates the location of a consensus-binding site for Nrf1 in the PsmB6 promoter. (D) Activity of wild-type PsmB6 promoter and mutant promoter containing a deletion of the −38 ARE site. Reporter constructs were transfected along with vector or Nrf1 cDNA into MEF cells. Activities represent the mean of three independent experiments ± SEM. *P < 0.05.
Discussion
Although Nrf1 has been shown to regulate antioxidant gene expression, additional roles of Nrf1 in development and cellular function were investigated. Nrf1 is highly expressed in neural tissues, but little is known about its function in the brain. Here we show that CaMK2cre-directed conditional knockout of Nrf1 (Nrf1BKO) leads to neuronal apoptosis and age-dependent brain atrophy. Our data also indicate that Nrf1 deficiency in neural cells does not induce measurable oxidative stress in brain tissues of Nrf1BKO mice. Thus, the neurodegenerative phenotype in Nrf1BKO mice is not mechanistically linked to oxidative stress. Instead, the knockout of Nrf1 in both mouse brains and cells produces defects in proteasome function and alterations in proteasome gene expression, and cell-based studies indicate that this defect is directly associated with Nrf1 deficiency and not secondary to a systemic process in the knockout animals. Because lowered proteasome function induces toxicity and apoptosis in neurons and other cells (19, 20), this suggests that proteasomal impairment is a major causative factor by which Nrf1 deficiency promotes neuronal degeneration. Our results also indicate that Nrf1 is an important effector of proteasome gene expression in the nervous system. The link between proteasome function and Nrf1 has implications for a variety of neurological disorders associated with proteasomal impairment.
The expression of proteasomal genes in yeast is controlled by Rpn4 transcription factor through a 9-bp motif known as proteasome associated control element (PACE) (21). Proteasomal genes are also regulated in a coordinate manner in mammalian cells, but little is known about the cis-active regulatory elements and transcriptional regulators that are involved (22). Transcriptional regulation of many cytoprotective genes is regulated through the ARE (23). For example, AREs control expression of both the catalytic and regulatory subunits of glutamylcysteine ligase that are involved in the biosynthesis of GSH (24, 25). The ARE also regulates phase I and phase II detoxification enzymes and multidrug resistance-associated transporters. In addition to oxidative stress-related genes, AREs have also been identified in promoters of a number of proteasomal genes (26, 27). Although Nrf2 has been shown to activate expression of proteasomal genes, proteasome activity was affected in Nrf2 knockout brains. This suggests that proteasome genes expression is not dependent on Nrf2 in neural cells. One possible explanation is that the level of Nrf2 expression may not be high enough in cortical neurons. Alternatively, the repertoire of target genes regulated by Nrf1 is uniquely different from Nrf2. This possibility would also explain the absence of oxidative stress in Nrf1BKO brains. Our data indicate that Nrf1 is a main regulator of proteasomal gene expression. On the basis of the IPA analysis of our expression profiling data, it seems that the proteasome pathway is one of the key processes affected by loss of Nrf1 in neurons. Interestingly, however, ARE-driven genes involved in oxidative stress did not appear in our microarray analysis. In line with a role for Nrf1 in proteasome gene expression, mRNA analysis and Western blotting showed a coordinate down-regulation of both α-structural and β-catalytic subunits of the 20S subunits, as well as components of the 19S regulatory subcomplex in Nrf1BKO brains. The marked accumulation of ubiquitinated protein aggregates suggests that the expression of proteasome genes is sufficiently diminished by loss of Nrf1 to affect constitutive proteasome function in Nrf1BKO brains. In further support of the requirement for Nrf1 in proteasomal gene expression, proteasome expression was down-regulated by tamoxifen-induced knockout of Nrf1 in neurons, and knockdown of Nrf1 activity by shRNA in 293 cells impaired proteasome expression and function in a parallel fashion. Our data also demonstrate that PsmB6, chosen as a representative target gene on the basis of our gene expression analysis, is directly regulated by Nrf1. Sequence inspection also revealed putative AREs in the 5′ flanking regions of various proteasomal genes, such as PsmA5, PsmB2, PsmB1, and PsmC3. However, further studies are required to examine whether Nrf1 also contributes directly to expression of these genes. Aside from its role in constitutive expression of proteasome genes, we have also demonstrated that Nrf1 is required for inhibitor-induced proteasome gene expression (28). On the basis of these findings, we propose that Nrf1 is essential for expression of mammalian 26S proteasome under basal as well as stress-induced conditions.
In summary, this study evaluated the role of Nrf1 in vivo in the adult brain. The results of our present study clearly demonstrate that Nrf1 is required for normal expression of proteasome genes in neural cells, and loss of Nrf1 in neurons causes neurodegeneration. These findings provide information on the role of Nrf1 beyond the oxidative stress response. Because neuronal degeneration is frequently observed in patients and animal models with defective proteasome function, these findings raise the possibility that Nrf1 signaling might play a more general role in neurodegenerative diseases. The Nrf1BKO mouse may provide a useful model to examine the involvement of proteasome dysfunction in the pathogenesis of neurodegenerative diseases.
Materials and Methods
See SI Materials and Methods for greater detail.
Transcriptional Profiling and Quantitative RT-PCR.
Profiling was done on Affymetrix oligonucleotide arrays. Primer sequences for PCR are listed in Table S1. Quantitative RT-PCR data was calculated as 2(Ct test gene − Ct 18s).
Transient Transfection and Luciferase Assays.
Cells were transfected using Lipofectamine and extracts were measured with Luciferase Reporter Assay Kit (Promega).
Primary Neuron Cultures.
Cultures were generated from cortices of embryonic day 14–15 embryos.
Supplementary Material
Acknowledgments
We thank Zhenrong Xu for assistance with mouse work. This study was supported in part by National Institutes of Health Grants NS065223 (to J.Y.C.), MH085801 (to M.P.V.), and NS048393 and RR024858 (to E.J.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019209108/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 4.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 5.Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9(Suppl 1):S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 7.McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat Rev Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- 8.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 10.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JY, et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, et al. Nrf1 is critical for redox balance and survival of liver cells during development. Mol Cell Biol. 2003;23:4673–4686. doi: 10.1128/MCB.23.13.4673-4686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, et al. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci USA. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B, et al. Cortical degeneration in the absence of neurotrophin signaling: Dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26:233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 16.Côté F, Collard JF, Julien JP. Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: A mouse model of amyotrophic lateral sclerosis. Cell. 1993;73:35–46. doi: 10.1016/0092-8674(93)90158-m. [DOI] [PubMed] [Google Scholar]
- 17.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 18.Lee JM, Shih AY, Murphy TH, Johnson JA. NF-E2-related factor-2 mediates neuroprotection against mitochondrial complex I inhibitors and increased concentrations of intracellular calcium in primary cortical neurons. J Biol Chem. 2003;278:37948–37956. doi: 10.1074/jbc.M305204200. [DOI] [PubMed] [Google Scholar]
- 19.Qiu JH, et al. Proteasome inhibitors induce cytochrome c-caspase-3-like protease-mediated apoptosis in cultured cortical neurons. J Neurosci. 2000;20:259–265. doi: 10.1523/JNEUROSCI.20-01-00259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuffy LP, et al. Characterization of Puma-dependent and Puma-independent neuronal cell death pathways following prolonged proteasomal inhibition. Mol Cell Biol. 2010;30:5484–5501. doi: 10.1128/MCB.00575-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc Natl Acad Sci USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiners S, et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 24.Köhle C, Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Shen G, Kong AN. Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos. 2009;30:345–355. doi: 10.1002/bdd.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takabe W, Matsukawa N, Kodama T, Tanaka K, Noguchi N. Chemical structure-dependent gene expression of proteasome subunits via regulation of the antioxidant response element. Free Radic Res. 2006;40:21–30. doi: 10.1080/10715760500354430. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan SK, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.