Abstract
Development in plants is controlled by abiotic environmental cues such as day length, light quality, temperature, drought, and salinity. These signals are sensed by a variety of systems and transmitted by different signal transduction pathways. Ultimately, these pathways are integrated to control expression of specific target genes, which encode proteins that regulate development and differentiation. The molecular mechanisms for such integration have remained elusive. We here show that a linear 130-amino-acids-long sequence in the Med25 subunit of the Arabidopsis thaliana Mediator is a common target for the drought response element binding protein 2A, zinc finger homeodomain 1, and Myb-like transcription factors which are involved in different stress response pathways. In addition, our results show that Med25 together with drought response element binding protein 2A also function in repression of PhyB-mediated light signaling and thus integrate signals from different regulatory pathways.
Keywords: transcriptional regulation, phytochrome flowering time 1, RNA polymerase II
Transcription of protein-encoding genes in eukaryotes requires RNA polymerase II (pol II) and five general transcription factors (GTFs) involved in promoter recognition, transcription bubble formation, and initiation (1). Pol II also depends on the multiprotein Mediator coactivator, which conveys signals from promoter-bound regulatory transcription factors to the pol II/GTFs (2). Mediator is well described in eukaryotes from Saccharomyces cerevisiae to human cells and there are numerous reports on regulatory pathways that target different Mediator subunits, thus resulting in proper gene expression (3–5).
Plants have the capacity to withstand enormous variations in climate, both seasonal variations and prolonged climate changes; the extremes are some trees that live for thousands of years and thus are subjected to very large environmental changes. This ability to adapt to the environment depends on several signaling pathways and transcription factors that are regulated in response to adverse conditions. These transcription factors can affect target genes directly to increase the ability to tolerate environmental stress or more indirectly by controlling developmental processes such as vegetative growth or timing of floral transition.
We recently identified Mediator in Arabidopsis thaliana and found that it comprises a core of protein subunits that are conserved in other eukaryotes and some that are specific for plants (6). One of the former is Med25, which in human cells has been identified as target for the VP16 activator. Med25 is also associated with the retinoic acid receptor and is mutated in patients with certain subtypes of autosomal-recessive axonal Charcot–Marie–Tooth neuropathies (7–10). Plant Med25 was originally identified as phytochrome and flowering time 1 (PFT1), a nuclear protein acting in a photoreceptor pathway that induces flowering in response to suboptimal light conditions (11). PFT1 is also a key regulator of the jasmonate signaling pathway and is required for resistance to leaf-infecting necrotrophic fungal pathogens (12). Pft1 showed hypocotyl-length inhibition under both red and far-red light. Unlike phytochrome B (phyB), pft1 has a late-flowering phenotype in long-day conditions. Experiments on pft1/phyB showed that deletion of PFT1 suppresses the early flowering phenotype of phyB in both long- and short-day conditions, indicating that the main role of PFT1 in phytochrome signaling is to regulate flowering time downstream of phyB in a photoperiod-independent pathway (11). It was therefore suggested that PFT1 regulates FLOWERING LOCUS T (FT) expression directly or indirectly. Our identification of PFT1 as Med25 suggested an indirect effect where PhyB operates via unidentified regulatory transcription factors that in turn interact with Med25 (6). In line with this finding, it was recently reported that Med25/PFT1 has a repressive function in light signaling, which is completely dependent on PhyB, D, and E (13). The consequence of a Med25/PFT1 deletion is therefore not a constitutive or an autonomous effect.
Here we report on the identification of three transcription factors that have been implicated in stress response pathways and interact with a conserved activator-interacting domain (ACID) in the Arabidopsis thaliana Med25. We show that Med25 is involved in stress responses and that one of the transcription factors functions in light quality pathways downstream of PhyB.
Results
Two-Hybrid Screening for Proteins That Interact with the Conserved ACID Domain in Arabidopsis thaliana Med25.
To identify plant transcriptional regulators that operate by interaction with the Med25, we used a bait composed of amino acids 551–680 of Arabidopsis Med25, the region corresponding to the ACID domain in the human Med25 (see figure 3b in ref. 6), in a two-hybrid screen with a cDNA library generated from inflorescence meristem, floral meristem, and floral buds. We identified five independent clones coding for three different transcription factors, one coding for drought response element protein B (DREB2A; At5g05410), two clones coding for zinc finger homeodomain 1 (ZFHD1; At1g69600), and two clones coding for MYB-like (At5g29000) (Fig. S1). None of these transcription factors was previously connected to light quality pathways. DREB2A belongs to a family that also includes DREB1A-C and DREB2B. They bind to the dehydration-response element/C-repeat motif, which is involved in drought and cold stress response (14). Overexpression of full-length DREB2A does not result in activation of downstream genes, but overexpression of DREB2A lacking a repressing domain (RD) results in a growth retardation phenotype and rounded, slightly darker leaves with short petioles (15). ZFHD1 belongs to a family that binds to the promoter region of the early responsive to dehydration stress 1 (ERD1) gene and causes upregulation of several stress-inducible genes and an increase in drought tolerance (16). The MYB-like protein has not been studied in detail but was identified in a transcriptome analysis as one of 454 transcripts that are specifically expressed in plants subjected to a combination of drought and heat stress (17).
Mapping of Med25 ACID Interacting Domains in the DREB2A, ZFHD1, and MYB-Like Proteins.
We mapped the region within each transcription factor required for interaction with the Med25551-680 region (Fig. 1A). Our aim was to identify the Med25 interaction region for MYB-like and to study if the Med25-interaction regions of DREB2A and ZFHD1 overlap with their previously mapped transcriptional activation domains (TADs). We localized the minimal domain of DREB2A required for interaction with Med25551-680 to amino acids 169–254 (Fig. 1B), thus indicating no correlation to the previously identified TAD of DREB2A comprising amino acids 254-335, or to the RD located between amino acids 136 and 165 (15). We notice that the region between amino acids 1 and 169, which includes the RD, has a negative effect on the interaction with Med25551-680. We found a good correlation between the previously identified TAD in ZFHD1 (amino acids 1–102) (16) and the minimal region for interaction with Med25551-680 (amino acids 1–132; Fig. 1C). Finally, we mapped the Med25551-680 interaction domain in MYB-like to the region between amino acids 103 and 309 (Fig. 1D). No TAD has previously been reported for this protein, but the MYB-like DNA-binding domain (DBD) overlaps with a homeodomain-like region located between amino acids 184 and 248. As indicated, the intact homeodomain-like region is required for interaction of MYB-like to Med25551-680.
Fig. 1.
Identification of proteins interacting with the conserved domain of the A. thaliana Med25 protein. (A) Schematic overview of A. thaliana Med25 and the bait construct used for the two-hybrid screen. The locations of the ACID domain, the Mediator-binding von Willebrand factor A domain (vWF-A) and the Gal4 DBD are indicated. (B–D) Two-hybrid interactions. The DREB2A (B), ZFHD1 (C), and MYB-like derivatives (D) used are shown. The GAL4 activation domains (G4-AD), HA epitope tag (HA), DBD of DREB2A (AP2-ERF), zinc-finger dimerization domain of ZFHD1 (ZF), and the DBDs of ZFHD1 and MYB-like (HD) are indicated. Growth on plates without tryptophane and leucine shows the presence of both the bait and prey two-hybrid plasmids. Growth on plates without adenine and histidine indicates expression of the two reporter genes (see SI Text). The panels to the right illustrate growth in the absence (Left) and presence (Right) of Med25.
In order to confirm the interactions identified in the two-hybrid assays, we recombinantly expressed Med25551-680 and different regions of DREB2A, ZFHD1, and MYB-like, which include the amino acid sequences identified as required for Med25 interaction in the two-hybrid experiments. All proteins were highly purified and interactions between each transcription factor and Med25551-680 were studied using surface plasmon resonance. Fig. S2B shows that all three factors bind to Med25551-680, but with different kinetics.
Arabidopsis thaliana Lines Lacking DREB2A, ZFHD1, MYB-Like or Med25 Are Sensitive to Salt Stress.
There are no phenotypes described for Zfhd1 or Myb-like, but the effects of overexpression of DREB2A and ZFHD1 have been described (15, 16). We obtained seeds with transfer DNA (T-DNA) insertions in the genes that encode DREB2A, ZFHD1, and MYB-like as well as the MED25/PFT1 gene from the Arabidopsis Biological Resource Center (Fig. S3). We first studied the effects on the salt stress response by measuring the percentage of germination for each mutant at different NaCl concentrations. All mutants display increased sensitivity to salt stress compared to the wild type (Fig. 2). Consistent with the interactions between the transcription factors and Med25, also med25 showed an increased sensitivity to salt stress, which is at least as pronounced as for each of the transcription factor mutants. This increased sensitivity to salt stress is a previously unidentified phenotype of med25 and strongly indicates that Med25 functions downstream of DREB2A, ZFHD1, and MYB-like. We further notice that zfhd1, myb-like, and med25 all show higher sensitivity to NaCl than dreb2a. At 175 mM, NaCl dreb2a is 5- to 24-fold less sensitive than the other mutants (Fig. 2).
Fig. 2.
Response of different Arabidopsis mutants to salt stress. Seeds of the indicated genotypes were incubated at 4 °C for 1 d on one-half MS solid medium with different concentrations of NaCl, then placed at 23 °C for 5 d, after which germination was scored. As shown, (A) med25, (B) dreb2a, (C) zfhd1, and (D) myb-like. Each genotype was treated independently. The experiments were performed using four plates of 49 seedlings for each treatment and genotype. Data represent mean ± standard deviation of at least three individual experiments.
The Function of Med25 in Salt Stress Resistance Is Conserved Among Land Plants.
All embryophytes have physiological systems to handle drought and salt stress. To find out if the role of Med25 in stress resistance is conserved during plant evolution, we used gene targeting (18) to delete PpMED25A, the single gene encoding an intact Med25 protein in the moss Physcomitrella patens. Physcomitrella med25a show an increased sensitivity to salt, resulting in a 32% reduction in the colony diameter in the presence of 0.15 M NaCl as compared to the wild type (Fig. 3). No effect was seen in the presence of an osmotic control (0.3 M mannitol). We conclude that the role of Med25 in salt stress resistance is an ancient function that was present already in an early embryophyte.
Fig. 3.
Effect of salt on colony growth in Physcomitrella wild-type and med25a strains. (A) Representative pictures of colonies from each of the three med25a knockout strains and the wild-type control after 21 d in normal light intensity (30 μmol/m2 s). The top row shows growth on BCD with 0.15 M NaCl. The middle row shows BCD with 0.30 M mannitol as osmotic control, and the bottom row shows just BCD. (B) Average colony diameter of wild-type and med25a strains under different conditions. The mean values from four colonies (WT) or 12 colonies (med25a) ± SEM are shown. The significances of the observed differences were tested using a two-tailed two-sampled t test assuming unequal variances. The asterisk denotes a difference that is significant at p = 0.0014.
Arabidopsis thaliana Lacking Med25 Is Resistant to Drought.
Overexpression of ZFHD1 or a constitutively active form of DREB2A leads to drought resistance (15, 16). We therefore examined if med25 is drought sensitive. Because med25 has a delayed flowering phenotype that could indirectly affect the sensitivity to drought (10) (Fig. S4, compare the two lines under long-day conditions), we performed the experiments under short-day conditions where flowering is inhibited (Fig. 4). Surprisingly, we found that med25 is drought resistant compared to wild-type plants (86.2% survival compared to 33.3%). Thus, it seems that Med25 has an opposite function compared to ZFHD1 and DREB2A in response to drought. Med25 showed the same phenotype also in long-day conditions (Fig. S4).
Fig. 4.
Drought resistance of the Arabidopsis med25. (A) Phenotypes of wild-type and med25 plants after drought stress in SD conditions (9 h/15 h; light/dark). Plants were grown in normal watering conditions for 4 wk, and then split in two groups. One group (D, drought) was grown without watering for 3 wk in the same light condition and then rewatered once. The other group (C, control) was grown for 4 wk in the same light under normal watering conditions. (B) Survival of plants after drought stress was assessed 7 d after rewatering. The experiment was performed using 15 plants for each genotype and treatment. Data represent mean ± standard deviation of three individual experiments. (C) The expression of drought genes (Rd29A, Rd29B, and Dreb2a) in leaves was analyzed using quantitative RT-PCR in the indicated lines during drought stress. Results were first normalized to the expression of adenine phosphoribosyltransferase 1 and eukaryotic initiation factor 4α mRNA and then to the level of expression for each mRNA at normal watering conditions. Bars represent standard deviation of the mean for three separate experiments.
To investigate if the observed drought phenotypes are caused by changes in expression of stress-induced genes, we used quantitative RT-PCR to study the levels of the drought-induced rd29a and rd29b mRNAs in wild-type cells, med25 and dreb2A (15). In line with the phenotypes, we found that both rd29a and rd29b mRNAs are strongly up-regulated as a response to drought in cells lacking Med25 (150- to 3,200-fold) and severely down-regulated in response to drought in dreb2A (Fig. 4C). We also found that the Dreb2A mRNA is strongly up-regulated at drought in med25.
The Arabidopsis thaliana DREB2A protein is involved in light quality pathways that control flowering time.
Arabidopsis Med25 was originally identified as PFT1, a downstream effector in the PhyB pathway, but it was unclear if PhyB acts directly or indirectly on PFT1 (11). Our identification of PFT1 as the Med25 subunit of Mediator suggests that the effect is transmitted indirectly through DNA-bound transcription regulatory factors, a notion which is supported by our identification of DREB2A, ZFHD1, and MYB-like as Med25 interactors. Because they had previously only been implicated in the responses to different types of stress, we wanted to see if the mutants would display a light quality-controlled flowering time phenotype, similar to med25. We therefore measured hypocotyl-length responses and counted leaf numbers for each mutant. We found that myb-like and zfhd1 are identical to the wild type, whereas dreb2a shows phenotypes opposite to med25 in both types of experiments (Figs. 4 and 5). Thus, dreb2a has an early flowering phenotype, which is comparable to that of phyB, indicating that it could function in the phyB pathway. This early flowering phenotype is a previously unidentified and unexpected function for DREB2A.
Fig. 5.
DREB2A function in repression of PhyB-mediated light signaling and has an opposite effect on flowering time compared to Med25. (A) Hypocotyl-length in dark (gray), red light (white), and the hypocothyl length inhibition (black) of 6-d-old seedlings of the indicated genotypes grown for 5 d under 10 μmol m-2 s-1 of red light or in dark. The experiment was performed using five plates of at least 20 seedlings for each treatment and genotype. (B) Flowering time of the different genotypes grown under LD conditions (16 h light/8h dark; 22 °C/16 °C). The data in A and B represent mean ± standard deviation of at least three individual experiments. (C) A representative picture of the different indicated phenotypes cultivated in LD conditions for 4 wk. (D) A model for how the DREB2A-Mediator interactions regulate flowering time in response to light quality. The DBD, RD, Med25 interaction domain (ID), and AD of DREB2A are indicated. MedX and MedY represent two as-yet unidentified Mediator subunits.
Discussion
The results described here indicate that Med25 functions as a hub that integrates signals from different environmental cues to control development. We suggest that ZFHD1 and MYB-like function as transcriptional activators by interacting with Med25 to induce target genes that encode proteins required to respond to salt stress. These two transcription factors therefore seem to function in the expected way by interaction to a Mediator subunit via their TADs. However, our results also represent a conundrum. Because dreb2a is sensitive to drought, we expected that med25 also would be drought sensitive. Surprisingly, we found that med25 is drought resistant (Fig. 4). A similar, unexpected result is the opposite effects of dreb2a and med25 on the hypocotyl-length response and flowering time (compare Fig. 5 and Fig. S5).
Our finding that the 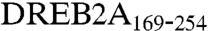 region is essential for interaction with the conserved Med25551-680 region, combined with previous mappings of DREB2A functional domains, lead us to propose a model for how Med25 integrates different signal pathways to regulate both development and expression of stress-induced target genes (Fig. 5D). DREB2A seems to function both as a transcriptional repressor and an activator depending on the circumstances. When the interaction between the RD of DREB2A and Mediator is disrupted, either by removal of the RD of DREB2A or by deletion of Med25, then DREB2A functions as an activator, both for induction of genes involved in drought (Fig. 4) and in development (14) Fig. 5). We propose that Med25 mediates the repressing effect of DREB2A by placing its RD in the vicinity of another Mediator subunit. In this model, only the shorter (30 aa) RD requires interaction with Med25, whereas the longer (81 aa) TAD can bind another Mediator subunit and thus function independently of Med25. It is possible that the Mediator protein that interacts with the RD of DREB2A is Med8 because med8 shows a more pronounced defect than med25, both in delay of flowering time and resistance to fungal pathogens (12). Moreover, DREB2A has been identified as a phosphoprotein (19) and the 30-aa-long RD in DREB2A contains no less than 11 serines and threonines, indicating that its activity might be regulated by phosphorylation.
region is essential for interaction with the conserved Med25551-680 region, combined with previous mappings of DREB2A functional domains, lead us to propose a model for how Med25 integrates different signal pathways to regulate both development and expression of stress-induced target genes (Fig. 5D). DREB2A seems to function both as a transcriptional repressor and an activator depending on the circumstances. When the interaction between the RD of DREB2A and Mediator is disrupted, either by removal of the RD of DREB2A or by deletion of Med25, then DREB2A functions as an activator, both for induction of genes involved in drought (Fig. 4) and in development (14) Fig. 5). We propose that Med25 mediates the repressing effect of DREB2A by placing its RD in the vicinity of another Mediator subunit. In this model, only the shorter (30 aa) RD requires interaction with Med25, whereas the longer (81 aa) TAD can bind another Mediator subunit and thus function independently of Med25. It is possible that the Mediator protein that interacts with the RD of DREB2A is Med8 because med8 shows a more pronounced defect than med25, both in delay of flowering time and resistance to fungal pathogens (12). Moreover, DREB2A has been identified as a phosphoprotein (19) and the 30-aa-long RD in DREB2A contains no less than 11 serines and threonines, indicating that its activity might be regulated by phosphorylation.
Our finding that Physcomitrella med25 is salt sensitive is interesting because it suggests that the function of Med25 in salt stress was present already in a common ancestor of all land plants. In this context, we note that the phytohormone abscisic acid (ABA), which functions in drought and salt stress in vascular plants, has a similar function in mosses (20). We further note that a moss cDNA that was annotated as encoding a DREB2A homolog is induced by dehydration (21). The AP2 domain transcription factors, to which DREB2A belongs, is a large protein family, and it is not easy to identify direct orthologs in mosses and angiosperms (22). Nevertheless, the fact that the moss DREB2A-related protein is induced by drought stress suggests that it could have a function analogous to that of Arabidopsis DREB2A. It is further conceivable that both Med25 and DREB2A could function as downstream components in the ABA pathway. We note that ABI3, encoding a key mediator of the ABA response (23), is among the genes that are induced by dehydration in Physcomitrella (22).
In conclusion, we suggest that previous models presented by us and others (6, 11) for how Med25 functions downstream of the PhyB photoreceptor to activate FT now can be further elaborated (Fig. 5D). Our finding that Med25 is a negative regulator of both flowering/development and drought resistance, and similar results reported by others for overexpression of DREB2A lacking its RD, suggest that DREB2A functions mainly as a repressor, and Med25 as a corepressor in these two pathways. FT cannot therefore be a direct target for repression by DREB2A acting through Med25 because disruption of the DREB2A–Med25 interaction would then cause an early flowering phenotype. Reports from other groups combined with our results presented here could be interpreted as if DREB2A and MED25 cooperate to cause down-regulation of a negative regulator of FT, such as, e.g., the FLC gene (24, 25). However, based on the model proposed by Wollenberg et al., we rather suggest that Dreb2A acts as a repressor through Med25 to down-regulate the PhyB light signaling pathway (13).
Materials and Methods
Strains and Plasmids.
A cDNA sequence encoding amino acids 551–680 of A. thaliana Med25 (At1g25540) was amplified by PCR and cloned into the Gal4 DBD vector pGBKT7. All DREB2A, ZFHD1, and MYB-like derivatives were amplified from Arabidopsis cDNA using the appropriate forward and reverse primer pairs (Table S1). Details of plasmid constructions and a list of strains are described in SI Text.
Yeast Two-Hybrid Screen.
The Yeast Two-Hybrid screen was performed according to the instructions of the Matchmaker Two-Hybrid System 3 (Clontech). Details for the two-hybrid screen are described in SI Text.
Plant Material and Growth Conditions.
All experiments were done using A. thaliana of the Columbia accession. The seed stock numbers N629555 (med25), N873547 (dreb2a), N579505 (myb-like), and N877090 (zfhd1) were obtained from the Nottingham Arabidopsis Stock Center. All mutants were identified after screening of the Salk T-DNA insertion lines (26). Homozygous plants of mutants were identified using primer sequences from http://signal.salk.edu/tdnaprimers.2.html. For flowering time and water stress, plants were grown on soil mixed with vermiculite (2∶1) under long days (LD; 16 h light) or short days (SD; 9 h light) at a temperature of 22 °C. White light sources were always from fluorescent tubes (40–70 μmol·m-2·s-1) for LD and SD conditions. For water stress, plants were grown in normal condition of watering for 3 wk (LD) or 4 wk (SD), and then split. One part was grown for 3 wk in the same light condition but without watering and then rewatering once. The other part of plants was grown in the same light and watering conditions. The survival rates of plants were assessed 7 d after rewatering in SD condition. The experiment was performed using 15 plants for each genotype and treatment.
Seeds for all experiments were sterilized using chlorine in a vapor phase. For hypocotyls measurements, seedlings were grown in one-half Murashige and Skoog basal salt mixture (MS; Sigma) agar and treated as described (27). Growth in red light conditions (10 μmol·m-2·s-1) was done in chambers of light-emitting diodes at 23 °C. After 5 d, seedlings were digitized and hypocotyls lengths were analyzed using the ImageJ program. The experiments were performed using four plates of 20–30 seedlings for each treatment and genotype.
For salt stress, seeds were grown in one-half MS-agar with different concentrations of NaCl at 4 °C for 1 d then placed at 23 °C for 5 d and scored. Each genotype was treated independently. The experiments were performed using four plates of 49 seedlings for each treatment and each genotype. Data represent mean ± standard deviation of at least three individual experiments.
Physcomitrella patens was grown as described (28). For the phenotypic analysis, small pieces of protonemal tissue from wild-type and knockout strains was inoculated on BCD (1 mM MgSO4, 1.85 mM KH2PO4, 10 mM KNO3, 45 μM FeSO4, 1 mM CaCl2, and trace elements, supplemented with 5 mM ammonium tartrate) plates as well as BCD plates containing 0.15 M NaCl or 0.3 M mannitol. Colonies from three separate med25 knockout lines and the wild-type strain were grown on the same plate in normal light intensity (30 μmol/m2 s) for 21 d before photography and scoring of growth. To estimate growth, the colony diameters on different media were measured for four colonies from each strain.
Targeted Gene Disruption in Physcomitrella.
The Physcomitrella genome contains two AtMED25-related sequences. PpMED25A (Phypa1_1:170131) encodes an intact Med25 protein. PpMED25B (Phypa1_1:92911) is an apparent pseudogene which has two frameshifts followed by stop codons in exon 7, and a deletion of 2,104 bp that starts near the end of exon 7 and ends in intron 10. This deletion removes sequences corresponding to codons 253–559 of PpMED25A and creates a third frameshift. The PpMED25A gene was PCR amplified from genomic DNA and cloned into the EcoRI site of pRS426. A selection cassette containing the hpt marker was then inserted between the two BglII sites in PpMED25A, resulting in the deletion of codons 43–838 (of 878). The targeting construct was released from the vector by SwaI digestion prior to transformation of moss protoplasts (18) and selection of stable transformants in the presence of 30 mg/L hygromycin B (Sigma H3274).
Drought Stress.
Plants (15 pots per mutant and treatment) were grown in LD conditions during 3 wk. After the second week, the pots for each mutant and treatment were randomized in different trays. After the third week, plants were not watered during 10 d for drought treatment, and watered every third day for the control. After 10 d, all leaves for each mutant and each treatment were harvested and frozen at -80 °C for RT-PCR.
Quantitative RT-PCR.
Total RNA was extracted from approximately 0.1 g leaves using the RNeasy kit (Qiagen). One microgram of total RNA was used for RT-PCR using the Superscript RT-PCR system (Invitrogen) with poly[d(T)] as primer. The resulting cDNA was used as DNA template for amplification of specific genes by RT-PCR. The details for the PCR reactions and primer sequences are found in SI Text.
Protein Expression and Purification.
Med25-ACID (amino acids 551–680) and MYB-like (amino acids 60–370) were inserted with the BamHI/NotI restriction sites into the GST containing pGEX-6P-2 vector (GE Healthcare). DREB2a (amino acids 169–355) and ZFHD1 (amino acids 1–242) was cloned into a pETM-6xhis vector (kindly provided by Günter Stier, European Molecular Biology Laboratory, Heidelberg, Germany) opened with NcoI/NotI. Protein expression was induced in BL21pLysS with 0.1 mM IPTG at 18 °C or 1 mM IPTG at 28 °C, respectively.
Med25 and MYB-like were purified with affinity chromatography using a glutathione sepharose 4B column (GE Healthcare). GST was removed using Prescission protease followed by a second run on the glutathione column. The proteins were further purified on a Mono S 5/50 GL (GE Healthcare) equilibrated with 20 mM Tris, pH 8.8, 80 mM NaCl, and 1 mM DTT.
DREB2a and ZFHD1 were purified using nickel-agarose (Qiagen) followed by a Mono S 5/50 GL equilibrated with 20 mM phosphate buffer, pH 6.2 (DREB2a) or pH 7.5 (ZFHD1), 80 mM NaCl, and 1 mM DTT. The DREB2a flow-through was applied on a Mono Q 5/50 GL and the flow-through containing ZFHD1 was run on a HiTrap Heparin 1 mL column, both equilibrated with the same buffer. Proteins were eluted in all ion-exchange experiments using a gradient from 80 mM to 1 M NaCl with an ÄKTAexplorer.
Surface Plasmon Resonance.
Sensograms were recorded at 25 °C using a Biacore 3000 instrument. Med25-ACID (0.2 μg/mL) was immobilized by amine-coupling to a CM-5 chip in 20 mM phosphate buffer pH 7.0. An empty blocked surface was used as a reference. Interaction studies were performed in a buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 10 μM ZnCl2, and 0.02% P-20. The transcription factors were analyzed at a concentration of 2 μM and at a flow rate of 40 μL/ min.
Supplementary Material
Acknowledgments.
This work was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (S.B., O.N., H.R.), the Swedish Cancer Society (S.B., H.R.), the Swedish Research Council (S.B., O.N., G.W., H.R.), the Swedish Governmental Agency for Innovation Systems (O.N., G.W., S.B.), Troëdsson Research Foundation (G.W.), and The Kempe Foundation (S.B., O.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002981108/-/DCSupplemental.
References
- 1.Conaway RC, Conaway JW. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Bourbon HM, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Björklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Yang F, DeBeaumont R, Zhou S, Näär AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittler G, et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HK, Park UH, Kim EJ, Um SJ. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 2007;26:3545–3557. doi: 10.1038/sj.emboj.7601797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard R, De Sandre-Giovannoli A, Delague V, Lévy N. Molecular genetics of autosomal-recessive axonal Charcot-Marie-Tooth neuropathies. Neuromolecular Med. 2006;8:87–106. doi: 10.1385/nmm:8:1-2:87. [DOI] [PubMed] [Google Scholar]
- 11.Cerdán PD, Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- 12.Kidd BN, et al. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell. 2009;21:2237–2252. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 2008;148:1681–1694. doi: 10.1104/pp.108.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuma Y, et al. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran L-S, et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2006;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 17.Rizhsky L, et al. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004;134:1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer DG, Zrÿd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal P, Agarwal PK, Nair S, Sopory SK, Reddy MK. Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol Genet Genomics. 2007;277:189–198. doi: 10.1007/s00438-006-0183-z. [DOI] [PubMed] [Google Scholar]
- 20.Knight CD, et al. Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell. 1995;7:499–506. doi: 10.1105/tpc.7.5.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank W, Ratnadewi D, Reski R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta. 2005;220:384–394. doi: 10.1007/s00425-004-1351-1. [DOI] [PubMed] [Google Scholar]
- 22.Richardt S, et al. Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signaling. Plant Mol Biol. 2010;72:27–45. doi: 10.1007/s11103-009-9550-6. [DOI] [PubMed] [Google Scholar]
- 23.Khandelwal A, et al. Role of ABA and ABI3 in desiccation tolerance. Science. 2010;327:546. doi: 10.1126/science.1183672. [DOI] [PubMed] [Google Scholar]
- 24.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheldon CC, et al. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso JM, Stepanova AN. T-DNA mutagenesis in Arabidopsis. Methods Mol Biol. 2003;236:177–188. doi: 10.1385/1-59259-413-1:177. [DOI] [PubMed] [Google Scholar]
- 27.Fankhauser C, Casal JJ. Phenotypic characterization of a photomorphogenic mutant. Plant J. 2004;39:747–760. doi: 10.1111/j.1365-313X.2004.02148.x. [DOI] [PubMed] [Google Scholar]
- 28.Thelander M, et al. The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol Biol. 2007;64:559–573. doi: 10.1007/s11103-007-9176-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







