Abstract
It has been widely accepted that the early spliceosome assembly begins with U1 small nuclear ribonucleoprotein (U1 snRNP) binding to the 5′ splice site (5′SS), which is assisted by the Ser/Arg (SR)-rich proteins in mammalian cells. In this process, the RS domain of SR proteins is thought to directly interact with the RS motif of U1-70K, which is subject to regulation by RS domain phosphorylation. Here we report that the early spliceosome assembly event is mediated by the RNA recognition domains (RRM) of serine/arginine-rich splicing factor 1 (SRSF1), which bridges the RRM of U1-70K to pre-mRNA by using the surface opposite to the RNA binding site. Specific mutation in the RRM of SRSF1 that disrupted the RRM–RRM interaction also inhibits the formation of spliceosomal E complex and splicing. We further demonstrate that the hypo-phosphorylated RS domain of SRSF1 interacts with its own RRM, thus competing with U1-70K binding, whereas the hyper-phosphorylated RS domain permits the formation of a ternary complex containing ESE, an SR protein, and U1 snRNP. Therefore, phosphorylation of the RS domain in SRSF1 appears to induce a key molecular switch from intra- to intermolecular interactions, suggesting a plausible mechanism for the documented requirement for the phosphorylation/dephosphorylation cycle during pre-mRNA splicing.
Keywords: RNA splicing, spliceosome complex, exonic splicing enhancer, protein phosphorylation
Pre-mRNA splicing is essential for gene expression by precise removal of intervening sequences known as introns. Because splice site sequences are often insufficient to direct faithful recognition of authentic splice sites, such a lack of sequence stringency imposes a great challenge for the splicing machinery to assemble on functional sites while avoiding numerous cryptic splice sites in the pre-mRNA (1).
Regulatory elements, such as exonic splicing enhancer (ESE) sequences, provide a key strategy to compensate for sequence variations on authentic splice sites. ESE typically consists of highly degenerate 6–8 nucleotide motifs (2, 3) that acts as positive regulators for splice site selection, and many of them are specifically recognized by SR proteins (3). ESE-bound SR proteins are involved in the recruitment of snRNPs, although the precise mechanisms of these recruitment events are only vaguely understood (4–7). This process also plays a crucial role in splicing regulation with the sequence elements, such as exonic and intronic silencer sequences (ESS and ISS, respectively) act as negative regulators by recruiting splicing repressors, such as heterogeneous nuclear RNPs (hnRNPs) (8, 9). The balance between these opposing functional elements determines the overall splicing strength in alternative splicing.
In addition to their well known activities in the regulation of both constitutive and alternative splicing, SR proteins also participate in postsplicing activities, such as mRNA nuclear export, nonsense-mediated mRNA decay, and mRNA translation (10, 11). SR proteins are characterized by having RNA recognition motifs (RRM) in the N terminus and Arg/Ser rich peptides in a C-terminal domain, referred to as the RS domain. The SR protein, SRSF1 (aka ASF/SF2), contains two RRMs and a relatively short RS domain compared to other SR proteins. The C-terminal RS domain of SRSF1 is phosphorylated by two protein kinases to generate two distinct phosphorylation states. In the cytoplasm, SRSF1 is phosphorylated at approximately 12 serines at the N-terminal portion of the RS domain by SRPK (12, 13). This partially phosphorylated or hypo-phosphorylated SRSF1 (p-SRSF1) is then imported into the nucleus where it can be further phosphorylated by the nuclear kinase CLK/STY to generate fully or hyper-phosphorylated SRSF1 (pp-SRSF1) (14). Hyper-phosphorylation is thought to facilitate the recruitment of SRSF1 to the active transcription and splicing sites (15). Dephosphorylated SRSF1 is also linked to its postsplicing functions (10, 11).
Phosphorylated SRSF1 has been shown to be crucial for U1 snRNP recruitment to the 5′SS (16, 17), which has long been thought to be mediated by the interaction between the RS domain of the SR protein and the RS-like domain present in U1-70K, a specific component of U1 snRNP (7, 18). The RS domain of SR and SR-like proteins has been shown to interact at the branchpoint sites (BPS) during different stages of spliceosome assembly (19, 20). However, the precise mechanism of SR protein-mediated spliceosome assemble is largely unknown.
In this study, we provide unique insights into biochemical mechanisms of how SRSF1 prompts early spliceosome assembly and phosphorylation regulation of this critical step. We show that SRSF1 simultaneously recognizes an ESE and U1-70K to recruit U1 snRNP to the 5′SS. However, contrary to the long-accepted model for this early spliceosome assembly process, we discovered that the interaction between SRSF1 and U1-70K is mediated by their respective RNA recognition domains (RRMs). We further show that the phosphorylation state of SRSF1 plays a modulatory role in this tripartite interaction where the fully phosphorylated RS domain allows ternary complex formation, but progressive dephosphorylation of the RS domain switches the domain to interact with its own RRM, thereby blocking the interaction of SRSF1 with U1-70K. We further show that both the RRM-mediated protein–protein interactions and the phosphorylation-induced molecular switch are linked to spliceosomal E complex formation and splicing.
Results and Discussion
Reversible Phosphorylation of the RS Domain Modulates the Stability of the ESE∶SRSF1 Complex.
To understand the role of the RS domain in ESE binding, we investigated how the RNA binding domain that includes both RRMs (RRM1/2) in the N terminus (Fig. 1A) binds ESE. We used a well-characterized ESE sequence present within the exon of Ron (receptor tyrosine kinase). SRSF1 has been shown to be involved in alternative splicing by binding to ESE (21–23). Filter binding (FB) assay revealed binding saturation at a level of only approximately 40%, suggesting weak stability of the Ron ESE∶SRSF1 (RRM1/2) complex (Fig. 1B). The mutant Ron ESE (mRon) showed negligible binding (Fig. 1C). These observations led us to conclude that SRSF1 (RRM1/2) binds to Ron ESE specifically but weakly.
Fig. 1.
Phosphorylation states of SRSF1 affect ESE binding affinity. (A) Cartoon representation of SRSF1 domain organization (upper) and coomassie stained SDS/PAGE showing unphosphorylated, hypo-phosphorylated (p-SRSF1), and hyper-phosphorylated SRSF1 (pp-SRSF1). (B and C) Filter binding assay showing the binding of SRSF1 (RRM1/2), SRSF1 (FL), p-SRSF1 (FL), and pp-SRSF1 (FL) to Ron ESE (AGGCGGAGGAAGC) and to mut Ron ESE (mRon ESE; AGGCGGUUGUUGC), respectively. The means and standard deviation (SD) of the results from three independent experiments are shown. (D) Estimated equilibrium dissociation constants (Kd) of SRSF1∶ESE complexes were measured based on FB assay (Rona) and EMSA (Ronb) (see Fig. S1 A–C). Kd was estimated as 50% of bound RNA fraction. ND denotes not determined. SD was determined from three independent experiments.
To delineate the role of the RS domain in ESE binding, we examined SRSF1 binding to Ron ESE using FB assay (Fig. 1B). We determined that unphosphorylated SRSF1 bound Ron ESE with a Kd of approximately 172 nM (Fig. 1 B and D). SRSF1 bound poorly to mRon indicating that SRSF1 binding to Ron ESE was sequence-specific (Fig. 1C). We next used p-SRSF1 and pp-SRSF1, which were generated as previously described (13) (Fig. 1A, bottom) to examine the binding affinity with the same ESE. p-SRSF1 bound Ron ESE with high affinity comparable to that of unphosphorylated SRSF1 (Fig. 1 B and D). In contrast, pp-SRSF1 showed a similar weak binding profile as in SRSF1 (RRM1/2) (Fig. 1 B and D). Electrophoretic mobility shift assay (EMSA) confirmed our conclusion from FB assay that both SRSF1 and p-SRSF1 bound Ron ESE with high affinity whereas both pp-SRSF1 and SRSF1 (RRM1/2) bound specifically but with low affinity (Fig. 2A and Fig. S1 A–C). Significantly lower ESE binding affinity of fully phosphorylated SRSF1 relative to dephosphorylated or partially phosphorylated SRSF1 indicates phosphorylation-dependent alteration of ESE recognition by SRSF1.
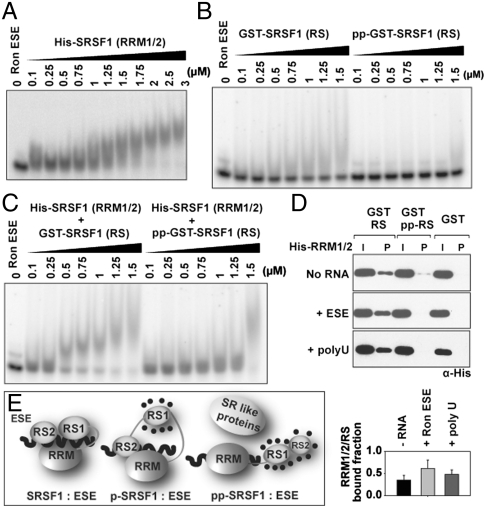
Fig. 2.
Unphosphorylated RS domain interacts with ESE∶SRSF1 (RRM1/2). (A) EMSA showing the binding of SRSF1 (RRM1/2) to Ron ESE. (B) EMSA analysis of ESE mixed with GST-RS (197–248) and GST-pp-RS (197–248). (C) EMSA showing the binding of SRSF1 (RRM1/2) to GST-RS and GST-pp-RS. (D) GST pull-down assay showing binding between His-SRSF1 (RRM1/2) (2 μg) and GST-RS/GST-pp-RS (2 μg) in the absence or presence of Ron ESE or poly U13. Input and bound proteins were detected by the Western blotting using anti-His Ab. Bound fractions was quantitated from three independent experiments (bottom). (E) The model depicting ESE binding to SRSF1 in its different phosphorylated states.
Unphosphorylated RS Domain Stabilizes the ESE∶SRSF1 Complex by Making Nonspecific Contacts with Both the RRMs and RNA.
Our study then focused on whether the high affinity of SRSF1 and p-SRSF1 with ESE is induced by their unphosphorylated RS2 domain. For this, we evaluated ESE binding to SRSF1ΔRS2 and p-SRSF1ΔRS2 (Fig. S2A). The residual RS1 domain in p-SRSF1ΔRS2 is fully phosphorylated. As shown in Fig. S2A, SRSF1ΔRS2 and p-SRSF1ΔRS2 recapitulate binding by SRSF1 and pp-SRSF1, respectively, suggesting that the RS2 domain played no specific role. It also suggests that the high affinity of the SRSF1∶ESE complex results from both RRM-mediated specific binding and charge-based binding by an unphosphorylated RS domain. Enhanced binding affinities of the ESE∶SRSF1 and ESE∶p-SRSF1 complexes mediated by the unphosphorylated RS domain might originate from different sources by its direct contact with either RNA only, RRM1/2 only, or both RNA and RRM1/2. To investigate these possibilities, we analyzed the role of free RS domain both in its unphosphorylated and phosphorylated forms. For this, two types of RS sequences, GST-RS (197–248) and RS peptides were used. Both the fusion protein and the RS peptide bound to ESE weakly in their unphosphorylated state and no binding was observed when the RS domain was phosphorylated (Fig. 2B and Fig. S2B). We next examined whether free RS domains could also stabilize the SRSF1 (RRM1/2)∶ESE complex. We found that GST-SRSF1 (RS) formed ternary complexes with SRSF1 (RRM1/2) and ESE complex (Fig. 2C). No ternary complex was observed with phosphorylated GST-SRSF1(RS) (GST-pp-RS) (Fig. S2 B and C). A 16-mer RS dipeptide repeat peptide (RS16) behaved nearly identically as the GST-RS domain. These results show that the RS domain enhances the affinity of the complex by fivefold [Kd of ESE∶RRM1/2∶RS (approximately 300 nM) and Kd of ESE∶RRM1/2 (approximately 1,500 nM)]. However, as the RS domain binds very weakly (Kd ∼ 1,250 nM), the RS-RRM1/2 interaction must contribute to the stability of the ternary complex.
To determine if the RS domain directly interacts with the RRM1/2 as well as RNA-bound RRM1/2, we performed GST pull-down assays (Fig. 2D). Our results revealed that the unphosphorylated RS domain interacts with the RRM1/2, whereas phosphorylated RS domain showed negligible binding. The RRM1/2-RS interaction was enhanced in the presence of ESE or poly-U RNA, indicating that RS binding to RNA∶RRM1/2 complex is nonspecific, electrostatic in nature. The interactions between RS and RRM1/2, and between RS and ESE suggest that RNA-dependent enhancement of RRM1/2-RS interaction is likely due to the tripartite contacts among ESE, RRM1/2, and RS domain. Phosphorylated RS domain with negative charge blocks the interactions with ESE and RRM1/2 (Fig. 2 B–D). Altogether, these observations suggest an intriguing model where the phosphorylation of the RS domain causes the dissociation of the intramolecular interaction between the RS and RRM1/2, and the “open” state of the RRM1/2 potentially interact with other proteins during spliceosome assembly (Fig. 2E).
RRMs of SRSF1 Directly Interact with U1-70K RRM: Insights into U1 snRNP Recruitment to the 5′SS.
The phosphorylation-dependent switch of the intramolecular interaction between the RRM and RS domain as described might be a critical regulatory step in the spliceosome assembly, such as the recruitment of U1 snRNP to the 5′SS and its subsequent dissociation prior to U6 snRNA binding. Although it has been previously reported that SRSF1 binds to U1-70K (7, 16, 17), we wanted to reexamine this binding in light of our model. To examine the interactions between SRSF1 and U1-70K under different states of phosphorylation, we used un-, hypo-, and hyper-phosphorylated SRSF1 (Fig. S3A). However, phosphorylated SRSF1 bound directly and indirectly to kinases (Fig. S3 B–D). To circumvent this problem, we designed different phosphomimetic versions of SRSF1. To generate unphosphorylated mimetic SRSF1 (RARA), all 18 serines in the RS domain were mutated to alanines. To mimic hypo- and hyper-phosphorylation, all 12 serines in RS1 and all 18 serines in the entire RS domain were mutated to glutamic acids to generate SRSF1 (RERA) and SRSF1 (RERE), respectively (Fig. 3A). We performed GST pull-down experiments using these GST-SRSF1 proteins and in vitro translated U1-70K (Fig. 3B). We plotted the average amount of U1-70K retained from three independent experiments (Fig. S3E).
Fig. 3.
U1-70K binding by SRSF1. (A) Schematic representation of domain maps and fragments of U1-70K (left), phosphorylation mimetic versions of SRSF1 (right), and sequences and phosphorylated sites of RS1 and RS2 domains (bottom). (B) GST pull-down assay between different GST-SRSF1 constructs (10 μg) and 5 μl of in vitro transcribed-translated [35S]-met labeled U1-70K constructs in the presence of RNase A (upper) or in the presence of cognate RNAs, Ron ESE and U1 snRNA (bottom). (C) Models depicting intra- and intermolecular binding modes within and between SRSF1 and U1-70K. (D) In vitro splicing of β-gb pre-mRNA in S100 complementation assay using WT and different phosphorylation mimics of SRSF1 (FL). Relative splicing efficiency of SRSF1 phosphomimetic mutants is shown in the bottom of the gel.
We found both WT SRSF1 (FL) and SRSF1 (RARA) had no interaction with in vitro translated U1-70K (FL) whereas both SRSF1 (RERA) and SRSF1 (RERE) showed interaction. Consistent with our model (Fig. 3C), the RS domain of wt SRSF1 and the RARA domain of SRSF1 (RARA) are involved in intramolecular interactions with its RRM1/2, thus preventing interactions with U1-70K (FL). Such intramolecular interactions are absent in SRSF1 (RRM1/2) and in SRSF1 (RERE), thereby permitting the intermolecular interactions with U1-70K (FL). Interestingly, SRSF1 (RERA) is able to interact with U1-70K (FL), demonstrating that replacement of nearly two-third serines by glutamic acids significantly weakened the intramolecular interactions, thus allowing the intermolecular interactions between p-SRSF1 and U1-70K. Using fragments of SRSF1 and U1-70K, we found that both SRSF1 (RRM1/2) and SRSF1 (RS), which do not participate in intramolecular interaction, bound to U1-70K (FL) and U1-70K (RRMs) (Fig. 3B). In addition, we observed that SRSF1 (RARA) interacts with U1-70K (RRMs) but not with U1-70K (FL). This suggests that the RRM and the RS domain of U1-70K (FL) may also participate mildly in an intramolecular interaction, but the interaction between SRSF1 (RRMs) and U1-70K (RRMs) is more predominant than U1-70K intramolecular interaction. Furthermore, the binding patterns remained the same in the presence or absence of cognate RNA elements. Our overall results by phosphomimetic mutants are consistent with the observation that U1-70K (FL) also interacts with p-SRSF1 and pp-SRSF1 (Fig. S3A) in the presence of SRPK1 (Fig. S3B). In all, these results suggest intricate and weak interactions exist within and between SRSF1 and U1-70K that are sensitive to the degree of SRSF1 phosphorylation as depicted (Fig. 3C). We next carried out in vitro splicing assay using S100 complementation to examine the impact of the intra- and intermolecular interactions in U1-70K recognition by SRSF1 (Fig. 3D). We used WT SRSF1 and its phosphomimetic mutants (RARA, RERA, and RERE). Unphosphorylated mimetic RARA mutant showed greatest splicing defect and hypo-phosphorylated mimetic RERA was partially defective. As observed previously by Cazalla et al. (24), we also found that hyper-phosphorylated mimetic RERE mutant supports splicing. These results demonstrate that disruption of the intramolecular interactions in SRSF1 by phosphorylation in the RS domain in turn permits the intermolecular interactions with U1-70K, which is essential for splicing, despite the fact that unphosphorylated SRSF1 binds to ESE with high affinity.
Disruption of the SRSF1∶U1-70K Complex Blocks Splicing.
Furthermore, to investigate how the two RRMs of SRSF1 are involved in direct interaction with U1-70K (RRM) in detail, we conducted pull-down experiments with GST-SRSF1 (RRM1/2, RRM1, and RRM2) and U1-70K (RRM) (Fig. 4A). We found that indeed the two RRMs of SRSF1 and U1-70K bound each other under stringent binding conditions (Fig. S4A). Further, the RRM1 domain, which shows high homology with SRSF2, another U1-70K binding protein, is responsible for binding to U1-70K (Fig. S4 B and C). To understand the mechanism of how the RRMs of SRSF1 recognizes U1-70K (RRM), we generated three different mutants in RRM1 domain of SRSF1 guided by the structure of SRSF1 RRM1 (PDB ID code 1X4A) (Fig. 4B). These mutants are located on distinct patches opposite to the putative RNA binding surface to determine if any of these patches is involved in U1-70K binding (shown in yellow in Fig. 4B). We mutated a pair of hydrophobic residues located on helix α1 to alanines to create m1 (I32A/V35A). In addition, we altered the charged surfaces by creating a pair of double mutants, m2 (K38A/Y39A) and m3 (D66A/D69A) (Fig. 4B). All three mutants had similar ESE binding affinities relative to WT SRSF1(RRM1/2), suggesting that these mutations had no effect on RRMs folding and were not involved in ESE binding (Fig. 4C). Two conserved RNP1 phenylalanine residues in RRM1 have been shown to be involved in ESE recognition (25) and as a control, we mutated these residues to FF-DD (F56D/F58D), which showed negligible RNA binding (Fig. 4C) as we expected.
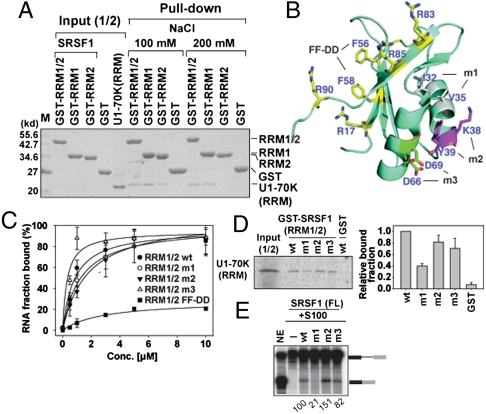
Fig. 4.
Two opposite surfaces of SRSF1 (RRMs) recruit ESE and U1-70K (RRM). (A) GST pull-down assay between GST-SRSF1 RRM1/2, RRM1, and RRM2 of 10 μg and U1-70K RRM (59–215) of 10 μg. (B) Ribbon presentation of SRSF1 (RRM1) (PDB ID code 1X4A). Putative RNA binding residues are denoted by yellow color. Residues in each mutant m1 (I32A/V35A), m2 (K38A/Y39A), and m3 (D68A/D69A) are denoted in three colors. (C) Ron ESE bindings to SRSF1 (RRM1/2) WT, m1, m2, m3, and FF-DD mutants (F56D/F58D) were measured by filter binding assay with error bars (SD) from three independent experiments. (D) Autoradiograph of GST pull-down assay between WT and mutants GST-SRSF1 (RRM1/2), and in vitro translated [35S]-met labeled U1-70K (RRM). The binding fraction of the U1-70K (RRM) to WT and mutants GST-SRSF1 (RRM1/2) were quantitated from three independent experiments. (E) In vitro splicing of β-gb pre-mRNA in S100 complementation assay using WT and mutants SRSF1 (FL) and relative splicing efficiency is quantified as shown.
Pull-down assays revealed that m1 was most defective in U1-70K (RRM) binding (Fig. 4D). Thus, our results suggest that the conserved hydrophobic patch on the SRSF1 surface, opposite to the ESE binding surface, is at least partly responsible for U1-70K recruitment. Further, to examine the effect of U1-70K recruitment from defective SRSF1 mutants in splicing, SRSF1 mutants were generated both in the context of SRSF1 (FL) and SRSF1 (RRM1/2). In vitro splicing assay with S100 complementation showed effective splicing activity in WT SRSF1 (FL), m2 (FL), and m3 (FL), whereas m1 (FL) showed defective splicing (Fig. 4E). Consistent with the splicing result of m1 (FL), m1 (RRM1/2) also showed the most pronounced splicing defect (Fig. S4D). Whereas, m2 (RRM1/2) behaved similarly to WT SRSF1 (RRM1/2) and m3 (RRM1/2) showed less splicing defect. Although the interaction between U1-70K (RRM) and m1 (RRM1/2) was only partially defective, this mutant showed dramatic splicing defect. This observation can be explained by two mutually exclusive arguments: First, we suggest that recruitment of spliceosomal component to the specific substrates is optimized through sensitive and weak interactions for splicing, and even a minor defect in binding results in a major splicing defect. Second, the same surface of SRSF1 may also be required for a second recruitment event during the spliceosome assembly such as the recruitment of U6 snRNP. Defect in multiple recruitment events due to the surface mutation would amplify the defect in overall enzymatic activity of the spliceosome. Altogether, our results confirmed that U1-70K (RRM) and SRSF1 (RRM) interact directly, and a conserved surface on RRM1 of SRSF1 bridges ESE RNA and U1-70K.
Dephosphorylated SRSF1 and U1-70K Binding Defective SRSF1 (RRM) Mutant Block Early Spliceosomal Assembly.
Interaction between U1-70K and SRSF1 is expected to facilitate U1 snRNP recruitment to the 5′SS and subsequent formation of the E complex. E complex is the first intermediate during the assembly of the mature spliceosome (C complex) that can be visualized by native gel electrophoresis. A and B complexes are two other subsequent spliceosomal intermediates assembled in the splicing process. To understand if the splicing defect of the dephosphorylated SRSF1 is affected by defective E complex formation, we tested SRSF1 phosphomimetic mutants (RARA, RERA, and RERE) for their ability to form the E complex in S100 extract. We made a new β-globin splicing template (β-gb (Ron)) by inserting the Ron ESE in exon 2 (Fig. 5A). As shown in Fig. 5B, in the absence of SRSF1, there was no E complex formation in S100 extract (Fig. 5B). SRSF1 (RARA) also failed to support the E complex formation, whereas the level of the E complex was reduced in the presence of SRSF1 (RERA). As expected, SRSF1 (RERE) mutant supported the E complex formation. We further examined the formation of later spliceosome complexes, A and B/C, in the presence of ATP (Fig. 5C) by mutating the AG dinucleotides to GG at the 3′SS junction (Fig. 5A). As expected, SRSF1 (RARA) failed to form these complexes whereas SRSF1 (RERE) supported the spliceosome complexes to similar extent as the control WT protein, SRSF1 (RSRS). Reduced levels of H complex in the presence of RARA mutant is apparently due to the formation of nonspecific aggregates, which failed to enter the gel. SRSF1 (RERA) also facilitated these later spliceosomal complexes but to a lesser extent than that of SRSF1 (RERE) or WT protein. tRNA challenge further showed that a significant fraction of the complex was the unproductive H complex (compare left and right panels in Fig. 5C. Behavior of these phosphorylated mimetic mutants in spliceosome formation is correlated with their ability to perform splicing of both WT β-gb and β-gb (Ron) substrates (Figs. 3D and 5D).
Fig. 5.
Dephosphorylated RS domain and U1-70K binding defective mutant of SRSF1 block early spliceosomal assembly steps. (A) Cartoon showing β-gb (Ron) and β-gb (Ron)ΔAG constructs. (B) Native gel analysis of the spliceosomal E complex formation by WT SRSF1 (RSRS) and different SRSF1 phosphomimetics in the presence of S100 extract and β-gb (Ron) pre-mRNA substrate. (C) Native gel analysis of the spliceosomal A and B/C complex formation of β-gb (Ron)ΔAG by SRSF1 phosphomimetics and WT (RSRS) without (left) or with 0.1 mg/mL tRNA (right) (D) In vitro splicing of the β-gb (Ron) pre-mRNA substrate by SRSF1 phosphomimetics in the presence of S100 extract. The relative splicing activities are shown at the bottom. (E) Native gel analysis of the E complex formation by WT and mutant SRSF1 (RRM1/2) in S100 extract. (F) Native gel analysis of the spliceosomal A and B/C complex formation of β-gb (Ron)ΔAG by WT and SRSF1 (RRM1/2) mutants without (left) or with 0.1 mg/mL tRNA (right) in S100 extract. (G) In vitro splicing assay of β-gb (Ron) pre-mRNA by WT and mutants SRSF1 (RRM1/2) in S100 extract. The relative splicing activities of SRSF1 (RRM1/2) mutants compared to WT as shown at bottom.
Furthermore, we investigated if the spliceosomal intermediate complex formation by WT and mutant SRSF1 (RRM1/2) also correlates with their splicing activity. EMSA showed that SRSF1 (RRM1/2) WT, m2, and m3 induced the E complex formation, whereas such complex was not observed in the case of m1 (Fig. 5E). As expected, m1 failed to support the formation of the A and B/C complexes, whereas SRSF1 (RRM1/2) WT, m2, and m3 formed the complexes (Fig. 5F). The most striking observation was the complete absence of spliceosomal complex formation by m1, which is consistent with the splicing defect observed in S100 complementation assay in both β-gb pre-mRNA substrates (Fig. 5G). Altogether, these results demonstrate that fully dephosphorylated RS domain is functionally dominant and that RRM alone can support splicing by promoting the assembly of spliceosomal E complex. The suppression of splicing by the unphosphorylated RS domain clearly implies the interplay of the RRM and de/phosphorylated RS domain in the regulation of spliceosome assembly and splicing.
Conclusion
Although numerous studies had characterized interactions between SRSF1 and U1-70K (7, 16, 17), the precise mechanism of how they interact remains unclear. Detailed molecular dissection of protein–protein and protein–RNA interactions involving SRSF1, U1-70K, and ESE with defined systems as presented in our study reveals that SRSF1 bridges pre-mRNA to U1-70K through their RRMs and that the RS domains play a modulatory role (Fig. 6). The RS domain of SRSF1 acts as a phosphorylation-dependent switch from intramolecular interaction to intermolecular interaction that then allows the binding of U1-70K. Upon phosphorylation of the RS domain, the surface of SRSF1 (RRM1/2) that binds to its own RS domain becomes exposed to U1-70K (RRM). Our study further supports the idea that phosphorylation of SR proteins are absolutely essential for splicing. It is still uncertain whether dephosphorylation is critical during splicing because the SRSF1 (RERE), which cannot undergo dephosphorylation, shows similar splicing activity as in WT. However, a large number of recent reports clearly suggest involvement of phosphatases for the second catalytic step to occur and perhaps during other assembly steps or during disassembly of the spliceosome after completion of the splicing reaction (7, 26–28). We cannot eliminate the possibility that an unphosphorylated RS domain from another SR protein or SR-related protein can complement the defect of nondephosphorylatable mutant of SRSF1 by acting in trans. Dominant function of SRSF1 (RERE) in in vitro splicing assay shown here and previously is consistent with the observation that the hyper-phosphorylated SRSF1 mimetic mutant rescued the cell death shown in SRSF1 knock-out mutant in vivo (29).
Fig. 6.
A model depicting the effect of RS phosphorylation in the E complex formation. Phosphorylation of the SRSF1 RS domain, mediated by the sequential actions of SRPK1 and CLK/STY, induces the dissociation of the RS from its RRM. Free SRSF1 (RRM) recruits U1 snRNP to the 5′SS through RRM-RRM interaction between SRSF1 and U1-70K. Released pp-RS domain interacts to the splicing factors bound to BPS/pY/3′SS to stabilize the E complex.
Splice site recognition during the E complex formation requires multiple protein–RNA and protein–protein recognition events and a single RNA–RNA recognition event (30). Many of these recognition processes are well characterized, such as RNA binding with SRSF1, SF1, and U2AF; protein–protein contacts among SF1-U2AF65, U2AF65-U2AF35, and SRSF1 (RRM1/2)-U1-70K (RRM) (derived from this study); and finally, RNA–RNA contacts between U1 snRNA and the 5′SS of the pre-mRNA (Fig. 6). Stability of the E complex is the net result of these relatively weak binary interactions. Enhancement of some binary interaction strengths can negate the contribution of weaker interactions in the formation of the E complex. This is consistent with the observation that SRSF1 (RRM1/2) alone can induce splicing in S100 complementation assay from some but not all pre-mRNA substrates (31). A strong correlation between the strength of the 3′SS and the RS domain requirement in splicing has also been reported (19, 20). We suggest that the strength of the splice sites dictates the E complex formation pathway through proper coupling of pre-mRNA and splicing factors and protein–protein interactions among splicing factors at each splice site and across the splice sites. In vitro assembly experiments with pure components using different pre-mRNA substrates are necessary to understand the underlying mechanism of how the splicing activators couples to the splice site strengths.
Materials and Methods
To analyze the E complex formation, radiolabeled β-gb (Ron) pre-mRNA was mixed with HeLa S100 extract and SR proteins as indicated under splicing conditions in the absence of ATP and MgCl2 for 40 min at room temperature. The products were resolved by 1.5% native agarose gel electrophoresis as described (32). For analysis of ATP-dependent complexes, radiolabeled β-gb (Ron)ΔAG pre-mRNA reaction mixtures (25 μl) were assembled in the presence of ATP and MgCl2 under splicing condition for 40 min at 30 °C. Then, 7 μl of the mixture were incubated with 0.5 mg/mL heparin and resolved by 2% native agarose gel for 4 h 30 min with 80 V in 0.5X TG buffer at room temperature.
For additional details on gene cloning, in vitro transcription/translation, protein purification, protein phosphorylation, EMSA, filter binding assay, GST pull-down assay, and in vitro splicing assay, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Drs. Simpson Joseph, Joseph Adams, and members of the Ghosh lab for their comments on the manuscript. We thank Dr. Joseph Adams at the University of California, San Diego, for providing the RS16 dipeptide, Dr. De-Bin Huang for Fig. 4B, and Dr. Robin Reed at Harvard Medical School for AdML splicing substrate. The work is supported by National Institutes of Health (NIH) Grant GM 084277 (to G.G.). R.S. and A.R.K. were supported by NIH Grant GM42699.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017700108/-/DCSupplemental.
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 4.Staknis D, Reed R. SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 6.Boukis LA, Liu N, Furuyama S, Bruzik JP. Ser/Arg-rich protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles. J Biol Chem. 2004;279:29647–29653. doi: 10.1074/jbc.M313209200. [DOI] [PubMed] [Google Scholar]
- 7.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Gene Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 8.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Gene Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. J Mol Cell Biol. 2003;23:1874–1884. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 11.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aubol BE, et al. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci USA. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velazquez-Dones A, et al. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 14.Ngo JC, et al. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20(1):77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 15.Misteli T, et al. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohtz JD, et al. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 17.Cao W, Garcia-Blanco MA. A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. J Biol Chem. 1998;273(32):20629–20635. doi: 10.1074/jbc.273.32.20629. [DOI] [PubMed] [Google Scholar]
- 18.Yeakley JM, et al. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 20.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Ghigna C, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 23.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 24.Cazalla D, et al. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 27.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 29.Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Wahl MC, Will CL, Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Krainer AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Gene Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.