Abstract
Genes of archaea encoding homologues of ammonia monooxygenases have been found on a widespread basis and in large amounts in almost all terrestrial and marine environments, indicating that ammonia oxidizing archaea (AOA) might play a major role in nitrification on Earth. However, only one pure isolate of this group from a marine environment has so far been obtained, demonstrating archaeal ammonia oxidation coupled with autotrophic growth similar to the bacterial counterparts. Here we describe the cultivation and isolation of an AOA from soil. It grows on ammonia or urea as an energy source and is capable of using higher ammonia concentrations than the marine isolate, Nitrosopumilus maritimus. Surprisingly, although it is able to grow chemolithoautotrophically, considerable growth rates of this strain are obtained only upon addition of low amounts of pyruvate or when grown in coculture with bacteria. Our findings expand the recognized metabolic spectrum of AOA and help explain controversial results obtained in the past on the activity and carbon assimilation of these globally distributed organisms.
Keywords: amoA, physiology, Thaumarchaeota, nitrite, NanoSIMS
Microorganisms involved in the transformation of nitrogen compounds have raised considerable interest in recent years because it is increasingly recognized that anthropogenic processes have severely perturbed the natural nitrogen cycle on Earth (1, 2). Novel microbial players and even new metabolisms with importance for the nitrogen cycle have been discovered, such as planctomycetes catalyzing anaerobic ammonia oxidation (3, 4) and archaea of the recently proposed phylum Thaumarchaeota (5, 6) capable of oxidizing ammonia to nitrite (7, 8). Ammonia oxidation represents the first step in nitrification and was long recognized to be solely performed by certain bacterial organisms of the proteobacterial phylum (9). The recent discovery of genes encoding proteins with homology to ammonia monooxygenases (amoA) in genome fragments of archaea from soil (7) and in shotgun sequences of marine environments (10), as well as the cultivation or enrichment of marine and thermophilic ammonia oxidizing archaea (AOA) (8, 11, 12) has radically changed this view, indicating that an additional, abundant, and predominant group of microorganisms (13, 14) is able to perform this process and might thus be significantly involved in global nitrogen cycling. This assumption was supported by numerous environmental studies that confirmed the presence of large numbers of archaeal amoA-like genes in various environments (reviewed in refs. 15, 16).
Environmental studies (14, 17, 18), as well as physiological investigations of the only marine cultivated isolate of AOA, Nitrosopumilus maritimus (8, 19), indicate that marine thaumarchaeota might indeed perform the bulk nitrification in the oceans. However, the situation is not so clear in terrestrial environments. Although AOA, or more precisely amoA genes of archaea, have been shown to outnumber their bacterial counterparts, sometimes even by orders of magnitudes (13, 20–22), conflicting results have been reported with respect to the role of AOA in nitrification. Upon amendment of fertilizer, growth of ammonia oxidizing bacteria (AOB) was reported to correlate with nitrification activity, whereas archaea did not seem to respond (20, 23, 24). In other soils, growth and nitrification of archaea was demonstrated (25–27). However, the active AOA group was either not investigated (25) or does not reside in significant numbers in most soil environments (26, 27). As we are aware of no study yet to conclusively correlate nitrification with autotrophic growth of group 1.1b archaea residing in soils, and because all isolates and enrichments of AOA have so far been obtained from aquatic environments (8, 11, 12), the ecological role and metabolism of soil AOA remains obscure (28). Similarly, it remains unclear whether AOA are able to assimilate organic substrates and thus have the capability to grow mixotrophically or even heterotrophically. Incorporation of labeled bicarbonate (29, 30), as well as of organic carbon (31, 32), has been observed for marine archaea, but in the latter two studies (31, 32) it was not determined whether the analyzed microbes were AOA.
Here we demonstrate the isolation of an AOA from soil. Our isolate is affiliated with group 1.1b and exhibits similarities but also specific differences to the metabolic capacity and ecophysiology of N. maritimus. In particular, it tolerates relatively high ammonia concentrations, is able to grow on urea, and needs addition of organic substrate to achieve comparable growth rates.
Results
Enrichments of AOA from Soil.
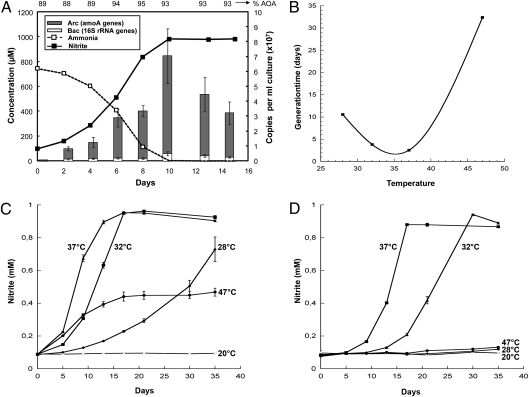
AOA were obtained in enrichment cultures with mineral media from a garden soil in Vienna. Ammonium consumption and nitrite production were followed regularly. Nitrification-positive cultures, free of bacterial ammonia oxidizers as confirmed with PCR using primers specific for AOB, were further processed. Consecutive passage of cultures over more than 2 y into medium regularly supplemented with antibiotics (streptomycin, kanamycin, ampicillin) led to two stable enrichments of AOA, termed EN76 and EN123. These cultures were analyzed by quantitative PCR (qPCR) to estimate the percentage of enrichment based on archaeal amoA and total bacterial 16S rRNA gene copy number. The percentage of archaea in the enrichments varied between 75% and 99%. Fig. 1A displays a characteristic growth curve of enrichment EN76 grown in triplicate in liquid batch culture with chemically defined mineral medium supplemented with 1 mM NH4+ and 2 mM sodium bicarbonate. Typically, after 10 d of incubation, all ammonium was consumed, which was paralleled by production of nitrite. The nearly stoichiometric oxidation of ammonium to nitrite was accompanied by growth of AOA as measured via quantification of archaeal amoA genes.
Fig. 1.
Growth of enrichments EN76 and EN123. (A) Correlation between ammonia oxidation and growth of EN76 in inorganic medium containing 1 mM NH4+, incubated at 37 °C. Near-stoichiometric conversion of ammonium (dotted lines) to nitrite (solid lines) was observed. Growth was assessed by qPCR for archaeal amoA genes and bacterial 16S rRNA genes. Numbers on top of the graph represent the percentage of archaeal enrichment at each time point. (B–D) Growth of enrichments EN76 (C) and EN123 (D) estimated by nitrite production in inorganic medium containing 1 mM NH4+ and incubated at five different temperatures. (B) Optimal temperature of enrichment EN76. Generation time was estimated from the slope of the log-transformed nitrite production measurements during exponential growth (as in C). All plotted data represent means of measurements from triplicate incubations. Error bars represent SE (in the case of ammonium and nitrite measurements, sometimes smaller than symbol size).
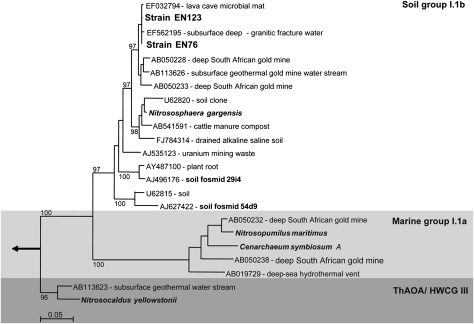
Clone libraries from the two enrichment cultures confirmed that only one unique sequence of archaeal 16S rRNA and amoA gene, respectively, could be identified in each culture. The 16S rRNA and amoA gene sequences of EN76 and EN123 differed by three and 12 point mutations, respectively, and both strains were affiliated to group 1.1b of AOA (Fig. 2). The sequence divergence from the 16S rRNA gene of Nitrososphaera gargensis, obtained in enrichments from a hot spring (11) was 3% (1,385 of 1,421 nt positions identical). A draft genome sequence of EN76 was obtained after using 454 pyrosequencing on total DNA from a highly enriched (97%) culture of EN76. From this partially assembled dataset, the full length 16S rRNA gene, a contig with the amoA, amoB, and amoC genes and a contig with genes encoding a potential urease operon were extracted. Phylogenetic analysis of the concatenated AmoAB protein sequences confirmed the affiliation of strain EN76 to the “soil” group of AOA (Fig. S1). It also confirmed the deep branching of Nitrosocaldus yellowstonii (12) at relatively good resolution, when the distantly related bacterial homologues of AMO and pMMO were included in the analysis.
Fig. 2.
Phylogenetic relationships between archaeal 16S rRNA gene sequences of strains EN76 and EN123 (N. viennensis) and all described AOA isolates or cultures, as well as relevant environmental clone sequences. Both EN76 and EN123 belong to the group 1.1b of Thaumarchaeota (formerly Crenarchaeota). The tree (1,272 nucleotide positions) was constructed by using maximum-likelihood analysis with PhyML (HKY85 with four categories). Bootstrap support was calculated 100 times. (Scale bar: 0.05 nucleotide changes per position.)
Enrichments EN76 and EN123 were tested for growth at different ammonium concentrations and at different temperatures by following nitrite production. Both enrichments grew at 1, 3, and 5 mM initial ammonium concentration (at 35 °C), but not at 20 mM. EN76 still grew at 10 mM, whereas EN123 was partially inhibited at that concentration. Differences in the ability of the two strains to grow under a range of temperatures (20, 28, 32, 37, and 47 °C) were also observed (Fig. 1 B–D). Whereas EN76 grew at all temperatures except 20 °C (Fig. 1C), EN123 showed a more narrow spectrum, growing only at 32 °C and 37 °C (Fig. 1D). These results suggest physiological differences between the two strains despite the high similarity of their 16S rRNA gene sequences. When plotting estimated generation times from the growth curves of EN76 that were obtained by measuring nitrite production, the optimal growth temperature was determined to be slightly greater than 35 °C (Fig. 1B).
Based on qPCR analysis, the EN76 culture was enriched for the archaeal AOA to approximately 97%, and contaminating bacterial species were present only at very low concentrations. According to 16S rRNA gene data, only two bacterial contaminants were detected, which were most closely affiliated with Hyphomicrobium vulgare and Mesorhizobium sp. of the α-proteobacterial order Rhizobiales.
Isolation of a Pure Culture and Physiological Characterization.
Through application of streptomycin, ampicillin, and carbenicillin, and by using a small inoculum in passages, a pure culture was obtained from the EN76 enrichment. Purity of the culture was repeatedly confirmed in successive transfers by PCR, qPCR, microscopic inspection, and plating experiments on Luria–Bertani agar plates as well as fresh water medium (FWM) agar plates. Few colonies of contaminating bacteria were only obtained from the original enrichment cultures on these agar plates, but not from the pure culture of strain EN76. Similarly, bacterial PCR products were only obtained from the enrichment culture, and cells of different morphology appeared in only the enrichment cultures, not in the pure culture.
Although the first pure subculture grew reasonably well, the strain almost ceased growing when further passaged into fresh medium. To restore growth, more than 30 different culturing conditions were applied, including addition of several organic substrates (Table S1). Only the addition of pyruvate at concentrations greater than 0.05 mM led to growth rates comparable to those obtained earlier in the coculture (Fig. 3), whereas addition of vitamins, amino acids, sugars, and other substrates had no or only a marginal effect. Addition of supernatant from the contaminated enrichment culture restored growth, but only after a long lag phase. No growth was obtained on pyruvate alone without ammonium (but with nitrate as N-source; Table S1), as checked by microscopic cell counts. No growth and no nitrite production was observed, when acetylene (0.025%) was added to the culture (containing ammonia and pyruvate), indicating that ammonia oxidation was essential for growth (Table S1). This experiment also demonstrated that acetylene is a potent inhibitor for growth of N. viennensis.
Fig. 3.
(A) Growth of the pure culture EN76 (N. viennensis) as estimated by measurements of nitrite production (solid lines) and ammonia consumption (dotted lines) when grown in duplicate either on 1 mM NH4+ (open squares) or 1 mM NH4+ supplemented with pyruvate (A+P, closed squares). Three different concentrations of added pyruvate were used (0.25, 0.5, and 1 mM) without causing significant differences in growth. Therefore, data were combined and the average from the six incubations is presented. (B) Correlation between cell density (solid bars), ammonia consumption (dotted line), and nitrite production (solid line) during growth of strain EN76 in medium with 1 mM NH4+ and supplemented with 1 mM pyruvate. Plotted data represent means of triplicate incubations. Error bars represent SEs. (C) Nitrite production of strain EN76 incubated at different initial ammonium concentrations and (D) generation times (estimated as in Fig. 1) of strain EN76 when grown on different initial pyruvate concentrations (0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1, 3, 5, and 10 mM).
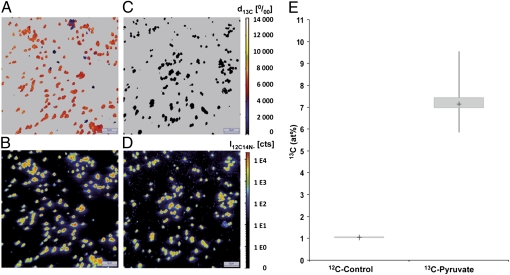
The generation time of the pure culture growing on medium with 1 mM ammonium and supplemented with pyruvate was approximately 45 h, and yielded cell densities as high as 5 × 107 cells mL−1 (Fig. 3B). Repeated transfers of this culture into media containing ammonium alone yielded growth, albeit extremely slowly (generation time of ∼23 d), demonstrating again, that the strain is, in principle, also able to grow exclusively autotrophically (Fig. 3A, open squares). To analyze whether carbon from pyruvate is incorporated into the biomass of strain EN76, incubation studies with 13C-labeled pyruvate were performed and analyzed by high-resolution nano secondary ion mass spectrometry (NanoSIMS). This technology allows highly sensitive quantitative analysis on single-cell level, i.e., it is suitable even for samples of very low biomass (as was the case for EN76 cultures). NanoSIMS analysis of EN76 cells after 10 d of incubation in the presence of 1 mM ammonium, 2 mM sodium bicarbonate, and 0.5 mM fully 13C-labeled pyruvate revealed that, after four to five generations, all cells were labeled and contained between 5.86 and 9.54 at% 13C (Fig. 4). Three-dimensional label distribution analysis within individual EN76 cells by long-term sputtering showed that all parts of the cells contained comparable 13C levels, demonstrating that the detected label resulted from incorporation into the biomass and not from absorption to the cell surface or incorporation into storage compounds. These cultures also produced 13CO2 (6.46–7.09 at%) from labeled pyruvate, as measured in the gas phase by isotope-ratio MS. Interestingly, the addition of different initial concentrations of pyruvate to the culture (0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1, 3, 5, and 10 mM) led to an increasingly positive growth effect up to concentrations of 0.1 mM (Fig. 3D). At concentrations greater than this, cultures grew with the same growth rate and to the same maximal cell density.
Fig. 4.
Incorporation of 13C-labeled carbon from pyruvate into cells of strain EN76 grown for 10 d with 0.5 mM 13C-labeled pyruvate, in mineral medium supplemented with 2 mM unlabeled sodium bicarbonate and 1 mM NH4+. Incorporation of 13C-labeled carbon was analyzed by NanoSIMS. (B and D) CN− secondary ion intensity distribution visualizing individual cells. (E) Box plot represents the relative isotopic composition [13C / (13C + 12C), given in at%] of the biomass as obtained from analysis of particular regions of interest (ROIs; n = 100). (A and C) Relative deviation of the 13C/12C isotope ratio (given in per mill) from the natural abundance level for each individual pixel of B and D. Image size is 40 × 40 μm; d, delta.
Fig. 3C shows nitrite production of the isolate when grown in medium with different initial ammonia concentrations and supplemented with pyruvate. In accordance with the enrichment culture, comparable growth rates were obtained for all ammonia concentrations tested, although an increase in the lag phase was observed at 10 and 15 mM ammonia. No growth was observed at 20 mM. For both enriched and pure culture, complete conversion of ammonia to nitrite was observed at 1 and 3 mM initial ammonium concentrations. At higher ammonia concentrations, nitrite production (and growth) ceased when approximately 3 to 3.5 mM nitrite was produced, indicating that accumulation of nitrite greater than this concentration may inhibit growth. To test this hypothesis, strain EN76 was grown in medium with 1 mM ammonium containing different initial nitrite concentrations (Fig. S2). Although retardation of growth was observed with increasing initial nitrite concentrations, complete consumption of ammonia occurred even in the presence of 10 mM nitrite, indicating that not only nitrite, but an additional intermediate or side product may inhibit growth, when cultures are grown in the laboratory at relatively high (>3.5 mM) ammonium concentrations.
The finding of an urease gene cluster in the genome of strain EN76 (Fig. S3A) indicated that this archaeon might be able to use urea instead of ammonia as sole energy source. This was confirmed by growth experiments with the enrichment culture (Fig. S3B) as well as with the isolated strain (Table S1). As expected, approximately 2 mM nitrite was generated when an initial concentration of 1 mM urea was applied to the enrichment culture. The urease gene cluster consisted of the urease encoding subunits ureA, ureB, and ureC, as well as the urease accessory proteins (ureE, ureF, ureG, and ureD; Fig. S3A). It has also been found in the genome of Cenarchaeum symbiosum (33) and in environmental datasets (34), indicating that some marine ammonia oxidizers can use this substrate as well.
The optimal pH for growth was determined to be slightly higher than 7.5 (Fig. S4). The pH was controlled with Hepes buffer in all cultures at 7.5 and did not change during incubations. Although we mostly grew the cultures without shaking, growth of strain EN76 was slightly positively affected by shaking the flasks. In particular, this shortened the lag phase of the cultures.
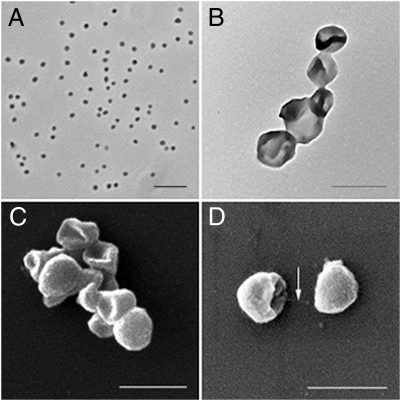
In phase-contrast microscopy (Fig. 5A) and EM (Fig. 5 B–D), the cells of strain EN76 appeared as small, slightly irregular cocci or spherically shaped. They are considerably smaller (with a diameter of approximately 0.5–0.8 μm), but with their characteristic coccoid shape almost indistinguishable from other distantly related archaea of the phylum Euryarchaeota (e.g., Haloferax volcanii) and Crenarchaeota (e.g., Sulfolobus spp.).
Fig. 5.
Microscopy pictures of strain EN76 (N. viennensis) in pure culture. (A) Phase contrast-light micrograph. (Scale bar: 5 μm.) (B) Transmission EM image of negative stained cells. (Scale bar: 1 μm.) (C and D) Scanning EM images of cells. (Scale bar: 1 μm.) The arrow in D points to a cell appendage.
Discussion
With this study, we demonstrate that AOA from soil that are affiliated with the group 1.1b of Thaumarchaeota can be enriched and maintained in pure culture, but under different conditions than the earlier isolated marine organism N. maritimus of group 1.1a. So far, ammonia oxidation in group 1.1b has only been demonstrated for N. gargensis, which has been analyzed in enrichments from a warm water spring (11). However, no study had conclusively demonstrated ammonia oxidation performed by members of this group residing in soil, although they are found in high abundance in many different terrestrial habitats (13, 22, 35, 36). Thus, the physiological capacity of these organisms has been debated (20, 24, 25). Strain EN76 represents the first isolated archaeon from group 1.1b that converts ammonia aerobically to nitrite, thus confirming that AOAs from soil have the capacity of ammonia oxidation. Strain EN76 grows well in media containing ammonium concentrations as high as 15 mM, but its growth is inhibited at a concentration of 20 mM ammonium. This is considerably higher than the inhibitory concentration of 2 to 3 mM reported for the aquatic strains N. maritimus (19) and N. gargensis (11). The inhibitory concentration of strain EN76 is rather similar to that of the most oligotrophic ammonia oxidizing bacterium reported so far, strain JL21 (21.4 mM), which was isolated from an activated sludge (37) and is affiliated to the Nitrosomonas oligotropha cluster (9), but it is still low compared with 50 to 1,000 mM maximum ammonium tolerance reported for a range of described AOB species (38). Thus, AOA and bacteria could, in principle, compete for the same resources under similar environmental conditions in soils with limiting amounts of ammonia. Of course, it is also likely that factors other than the adaptation to different ammonia concentrations contribute to the ecological fitness and niche adaptation of each group. In this context, it is interesting to note that the two enrichments reported here exhibit different ecophysiological adaptations (to temperature), although the two AOA strains are closely related. This suggests that a wide range of ecotypes can be expected to occur in the group of AOA from soil.
Unexpectedly, growth of strain EN76 in pure culture was strongly enhanced by the addition of as much as 0.1 mM pyruvate (Fig. 3D). Only upon addition of this compound, growth rates comparable to those of previous enrichment cultures and of the chemolithoautotroph N. maritimus were obtained, whereas, under purely autotrophic conditions, growth was approximately 12 times slower, indicating that this organism might be adapted to mixotrophic growth. Consistently, incorporation of cellular carbon from pyruvate in the presence of bicarbonate and ammonium was demonstrated for strain EN76 on the single-cell level by NanoSIMS (Fig. 4). However, less than 10% of the cellular carbon stemmed from pyruvate, showing that carbon assimilation in this strain is predominantly driven by fixation of bicarbonate under the tested growth conditions. A potential ability to assimilate inorganic carbon is also suggested by the identification of genes in the draft genome of N. viennensis encoding key enzymes of the potential carbon fixation pathway of AOA (the 3-hydroxypropionate/4-hydroxybutyrate cycle) that are present in all thaumarchaeotal genomes (33, 39), as well as in metagenomes (40). These include acetyl-CoA carboxylase, methylmalonyl-CoA mutase, and 4-hydroxybutyryl-CoA dehydratase. However, we would like to note that a homologous pathway is also found in strictly heterotrophic crenarchaeota (41), indicating that the mere presence of those genes does not exclude a mixotrophic/heterotrophic lifestyle. When comparing the gene content of the draft genome of N. viennensis with the genome of N. maritimus, we did not find any indications for the pyruvate dependence of N. viennensis. Further attempts to improve growth conditions and to scale up the laboratory cultures are currently under way. These steps will be essential for future detailed physiological tests and labeling studies to fully explain the growth mode and carbon flow in this organism.
More generally, it will be interesting to elucidate whether the dependence on certain organic substrates is a general feature of AOA in soil, and whether obligate heterotrophically growing Thaumarchaeota exist. Current results based on isotope labeling studies point to a range of capacities: although stable isotope probing in an agricultural soil did not demonstrate incorporation of bicarbonate into cells of archaea of group 1.1b (20), incorporation of 14C-bicarbonate by cells of N. gargensis (group 1.1b) has been revealed with FISH/microautoradiography (11). In this context, it will also be interesting to see whether N. maritimus and related strains in the ocean will show enhanced growth rates with specific organic substrates, a feature well known for nitrifying bacteria. Hommes et al. (42) have shown increased growth and incorporation of organic carbon into cellular biomass in Nitrosomonas europaea when grown with fructose and pyruvate. Similarly, uptake of pyruvate by bacterial nitrite oxidizers, generally assumed to be autotrophs, has been demonstrated (43). However, the positive growth effect of pyruvate for strain EN76 is considerably stronger than that observed for bacterial nitrifiers (Fig. 3A).
Our study highlights the importance of obtaining pure cultures to get closer insights into the growth requirements of AOA. The original cultures highly enriched for AOA did not yet give indications for a growth mode that is dependent on pyruvate. Such a dependency on an organic substrate could also explain why bacteria could not yet be eliminated from other enrichment cultures of ammonia-oxidizing archaea, like Candidatus N. gargensis (11) or Candidatus N. yellowstonii (12).
The pure isolate EN76 opens new avenues for the detailed characterization of the energy metabolism and carbon assimilation in AOA, microorganisms that range among the most abundant (44)—but still poorly understood—organisms on this planet, whose role in global carbon and nitrogen cycling still needs to be clarified.
Strain EN76 is an ammonia oxidizing archaeon isolated from soil, and we propose the following candidate status:
Nitrososphaerales order, nov.;
Nitrososphaeraceae fam. nov.; and
Nitrososphaera viennensis gen. et sp. nov.
Etymology: L. adj. nitrosus, “full of natron,” here intended to mean nitrous (nitrite producer); L. fem. n. sphaera, spherically shaped; L. viennensis, isolated from Vienna.
Locality: garden soil from Vienna.
Diagnosis: an ammonia oxidizer growing optimally upon addition of pyruvate (≤ 0.05 mM), but also to limited extent chemolithoautotrophically, spherically shaped with a diameter of approximately 0.5 to 0.8 μm, optimum pH 7.5, optimum temperature 35 °C. N. viennensis is affiliated with group 1.1b of the recently proposed phylum Thaumarchaeota (5, 6) of the domain Archaea and is closely related to Candidatus N. gargensis (97% sequence divergence in its 16S rRNA gene), an AOA that has not yet been obtained in pure culture, but has been shown to incorporate bicarbonate as carbon source when grown with ammonia in a nitrifying culture (11). Whereas N. gargensis stems from a hot spring, N. viennensis has been isolated from soil. Different from the recently isolated marine strain N. maritimus, N. viennensis is able to grow on urea and under higher ammonia concentrations approaching those tolerated by oligotrophic bacterial ammonia oxidizers.
Materials and Methods
All materials and methods are described in detail in the supplementary information.
Availability of Sequences and Strain.
The sequences of N. viennensis described in this manuscript (16S rRNA and amo genes) have been deposited in GenBank under accession numbers FR773157, FR773158, FR773159, and FR773160. Archaeal sequences from the 454 pyrosequencing have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information (accession no. SRA030754). The strain will be available upon request from the corresponding author.
Supplementary Material
Acknowledgments
We thank Birgit Wild, Margarete Watzka, Natalia Khilkevitch, and Daniela Gruber for technical assistance. This work was supported by the University of Vienna, Faculty of Life Sciences, and Fonds zur Förderung der wissenschaftlichen Forschung (Austrian Science Fund) Grant P23000 (to C.S.). A.S. was supported by a DOC-fForte fellowship of the Austrian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Sequence Read Archive (SRA) database, www.ncbi.nlm.nih.gov/sra (accession no. SRA030754). The sequences of N. viennensis described in this manuscript (16S rRNA and amo genes) have been deposited in GenBank under accession numbers FR773157, FR773158, FR773159, and FR773160.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013488108/-/DCSupplemental.
References
- 1.Rockström J, et al. A safe operating space for humanity. Nature. 2009;461:472–475. doi: 10.1038/461472a. [DOI] [PubMed] [Google Scholar]
- 2.Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 3.Jetten MSM, et al. The anaerobic oxidation of ammonium. FEMS Microbiol Rev. 1998;22:421–437. doi: 10.1111/j.1574-6976.1998.tb00379.x. [DOI] [PubMed] [Google Scholar]
- 4.Strous M, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 5.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 6.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Treusch AH, et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 8.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 9.Purkhold U, et al. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 11.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 13.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 14.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicol GW, Schleper C. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 2006;14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 17.Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- 18.Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 20.Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen XP, Zhu YG, Xia Y, Shen JP, He JZ. Ammonia-oxidizing archaea: Important players in paddy rhizosphere soil? Environ Microbiol. 2008;10:1978–1987. doi: 10.1111/j.1462-2920.2008.01613.x. [DOI] [PubMed] [Google Scholar]
- 22.He JZ, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 23.Di HJ, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- 24.Di HJ, et al. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- 25.Schauss K, et al. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- 26.Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- 27.Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 28.Schleper C. Ammonia oxidation: Different niches for bacteria and archaea? ISME J. 2010;4:1092–1094. doi: 10.1038/ismej.2010.111. [DOI] [PubMed] [Google Scholar]
- 29.Wuchter C, Schouten S, Boschker HTS, Sinninghe Damsté JS. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 30.Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- 31.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallam SJ, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinidis KT, DeLong EF. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- 35.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 36.Bates ST, et al. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2010;2010:18. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 1994;28:1523–1532. [Google Scholar]
- 38.Koops HP, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. New York: Springer-Verlag; 2003. [Google Scholar]
- 39.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 41.She Q, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hommes NG, Sayavedra-Soto LA, Arp DJ. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J Bacteriol. 2003;185:6809–6814. doi: 10.1128/JB.185.23.6809-6814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.