Abstract
Questions of if and when protein structures change within cells pervade biology and include questions of how the cytoskeleton sustains stresses on cells—particularly in mutant versus normal cells. Cysteine shotgun labeling with fluorophores is analyzed here with mass spectrometry of the spectrin–actin membrane skeleton in sheared red blood cell ghosts from normal and diseased mice. Sheared samples are compared to static samples at 37 °C in terms of cell membrane intensity in fluorescence microscopy, separated protein fluorescence, and tryptic peptide modification in liquid chromatography–tandem mass spectrometry (LC-MS/MS). Spectrin labeling proves to be the most sensitive to shear, whereas binding partners ankyrin and actin exhibit shear thresholds in labeling and both the ankyrin-binding membrane protein band 3 and the spectrin–actin stabilizer 4.1R show minimal differential labeling. Cells from 4.1R-null mice differ significantly from normal in the shear-dependent labeling of spectrin, ankyrin, and band 3: Decreased labeling of spectrin reveals less stress on the mutant network as spectrin dissociates from actin. Mapping the stress-dependent labeling kinetics of α- and β-spectrin by LC-MS/MS identifies Cys in these antiparallel chains that are either force-enhanced or force-independent in labeling, with structural analyses indicating the force-enhanced sites are sequestered either in spectrin’s triple-helical domains or in interactions with actin or ankyrin. Shear-sensitive sites identified comprehensively here in both spectrin and ankyrin appear consistent with stress relief through forced unfolding followed by cytoskeletal disruption.
Keywords: cysteine labeling, repeat domain, conformation, oxidation state, mechanotransduction
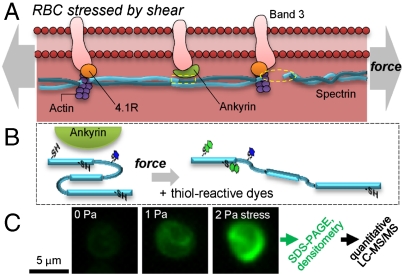
The folding and association states of the many proteins within a cell might never be assessed by high-resolution methods of diffraction or NMR applied to the cell, but for structural proteins of the cytoskeleton such questions are likely central to understanding stress responses and probably signal transduction. The simplest mammalian cell, the RBC, possesses a well-elaborated membrane skeleton (1) assembled from α- and β-spectrin plus F actin with proteins such as 4.1R (2) helping to stabilize the network and ankyrin (3) helping to attach the network to the membrane (Fig. 1A). However, it is unclear whether these proteins dissociate and/or unfold as the cytoskeleton is stressed in blood flow. Deeper insight is needed to explain, for example, the strong effects of point mutations that are sometimes distal to protein interaction sites (4). To begin to address such issues we have developed a method to map dynamically exposed cysteines in proteins of intact cells that are stressed mechanically, thermally, or by drugs (5). Cys is relatively hydrophobic, which means that it is frequently buried—wholly or partially—in protein folds as well as in protein–protein interfaces, but the thiol in cysteine is also highly reactive. Fluorescent dyes allow “shotgun” labeled cells to be imaged and the labeled proteins to be both quantified and site-by-site analyzed using Cys shotgun–mass spectrometry (CS-MS). Controlled stressing of human RBC membranes by fluid shear stresses at levels similar to in vivo blood flow stresses (up to 1–2 Pa) has thus revealed stress-enhanced Cys labeling at 37 °C, with forced unfolding of some of the dozens of three-helix repeat domains (Fig. 1B) (5). Whether other membrane skeleton proteins contribute and show stress-enhanced labeling was not previously addressed.
Fig. 1.
Shear stress response of red cell membrane cytoskeleton as detected by Cys-shotgun labeling. (A) Shearing of the membrane tends to stretch the spectrin–actin network, perhaps from a slack or curved state, with possible stressing of attachment proteins protein 4.1R and ankyrin as well as integral membrane proteins such as band 3. (B) Forces can either unfold spectrin repeats or dissociate spectrin from ankyrin (dashed box) and/or actin (dashed oval). Fluorescent thiol probes loaded into cells react with solvent exposed (blue dye) and newly exposed cysteines (green dye), revealing changes in tertiary and quaternary structure. (C) Stress-dependent labeling of the membrane can be visualized and measured in intact RBC ghosts that were labeled for the same duration at 37 °C but exposed to various physiological levels of shear stress (units of Pascal: Pa).
Denaturation studies of purified spectrin decades ago showed spectrin repeats unfold collectively and cooperatively only for T≫37 °C (6), but in more recent studies in which almost every spectrin repeat was recombinantly expressed, more than a third of the domains unfolded at or below 37 °C (7). Single molecule experiments have shown that spectrin domains—independent of melting temperature—unfold at relatively low force compared to other proteins such as titin (8), and the forced unfolding can cooperatively propagate through multiple domains (9, 10) with significant perturbations in some mutant spectrins (11). In addition, RBC ghosts with spectrin interacting fragments entrapped inside show shear-enhanced incorporation into the cytoskeleton (12). This raises the question of how much dissociation occurs under stress relative to unfolding of spectrin’s many repeat domains—especially in normal versus mutant cells, such as from 4.1R-null mice, which have mechanically fragile RBCs (13).
Results and Discussion
Stress Enhances Cys Labeling of Spectrin in Mouse Cell Membranes, but Labeling Is Low in 4.1R-Null Mutants.
Fluorescence microscopy shows that RBC membranes from normal mice have a higher intensity when labeled at higher shear stress (Fig. 1C). RBCs were lysed to remove hemoglobin and its highly reactive cysteines (14), and then Cys reactive dyes were entrapped. Blue dye [N-(Iodoacetaminoethyl)-1-naphthylamine-5-sulfonic acid (IAEDANS)] was added first to label Cys exposed on the surfaces of proteins, thus resulting in a “background” signal. Green dye [N-(4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene-2-yl) iodoacetamide] (BODIPY) was then added with the aim of separately labeling stress-exposed sites, and the resealed ghosts were then quickly split and either held “static” at 37 °C or else sheared at 37 °C at physiological stresses (< 2 Pa) in a standard stress-controlled fluid-shearing device for up to 60 min. Labeling differences with the second dye (e.g., BODIPY) are most apparent at 30–60 min, consistent with previous studies (5).
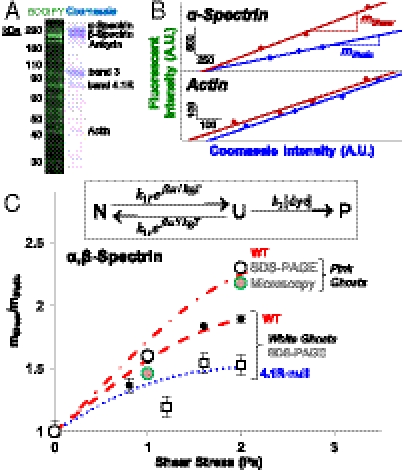
To identify membrane proteins labeled at a specific time and stress and to quantify the labeling per mass, SDS-PAGE gels were run with various loads of solubilized membranes and then imaged in both fluorescence and after Coomassie staining for protein mass (Fig. 2A). Cysteines are underutilized in many cytoskeletal proteins, occurring in spectrin, for example, at only one to two per approximately 100 amino acids (versus random usage of approximately 5%), but the proteins are nonetheless labeled robustly by the fluorescent dyes when compared to Coomassie stain. Densitometry of both signals at the various lysate concentrations (Fig. 2B) provides a specific labeling level as a slope m for each stress condition, i.e., mshear or mstatic. A ratio (mshear/mstatic) > 1 for spectrin was found for all shear stresses tested and indicates force-enhanced labeling, whereas (mshear/mstatic) = 1 was found for actin at low stress (see below). Note that static labeling is nonzero and depends on the particular dye, but such effects are common to both static and shear samples. Equal labeling of α-spectrin and β-spectrin (within 5%) is consistent with an antiparallel arrangement of molecular “springs” being equally stressed.
Fig. 2.
Spectin labeling depends on stress and on 4.1R. (A) Images of an SDS-PAGE-separated RBC ghost membrane lysate in fluorescence with BODIPY labeling or after staining with Coomassie dye, showing considerable labeling of many cytoskeletal proteins. (B) Fluorescence and Coomassie intensity of individual bands was quantified by densitometry for different amounts of the same lysate and then fit to a line (R2 > 0.95). The ratio of slopes, (mshear/mstatic), for a given shear stress and time (60 min), shows that α-spectrin labeling is shear-enhanced, whereas actin labeling is shear-independent. (C) The labeling ratio for α,β-spectrin increases with shear stress, and the results fit to fLL reaction kinetics for a force-accelerated transition from native to unfolded (Inset; see SI Text) shown as dashed lines. Densitometry of spectrin from WT pink ghosts (open circles: WT), shows the same stress-enhanced fluorescence labeling as that measured by microscopy of intact pink ghosts (pink circles). With white ghosts, labeling of spectrin from normal cells, black dots, exceeds the labeling of spectrin from 4.1R-null cells.
With increasing shear stress (for 60 min at 37 °C), spectrin’s labeling ratio increases (Fig. 2C). As with white ghosts, pink ghosts with approximately 20% of physiological hemoglobin show increased BODIPY labeling with shear stress, both for spectrin separated by SDS-PAGE and in fluorescence imaging of the sheared ghosts (after a final hemolysis). Ghost membranes and spectrin bands were labeled about twice as much after being stressed at 2 Pa versus 1 Pa, with relatively tight error bars. Additional labeling of pink ghosts perhaps reflects the enhanced viscosity and stress in such ghosts.
The 4.1R-null cells mutant show decreased spectrin labeling relative to the various wild-type samples, and the labeling appears nonlinear and almost step-like. The maximum labeling at high stress is also only 50% higher than static cell labeling. 4.1R therefore has an important effect on stress-dependent network dynamics.
Shear-enhanced labeling of spectrin was fit to a force-dependent Linderstrom–Lang (fLL) reaction (Fig. 2C, Inset), which models a stress-dependent shift in equilibrium from native folded (or associated) domains to unfolded (or dissociated) domains followed by irreversible labeling of exposed cysteines. The fLL merely assumes one overall process with respect to Cys labeling, although some or all of the molecules could start with some slack, compressed, or bent. Spectrin labeling in normal mouse RBCs fits very well to the fLL equation with just two adjustable parameters (R2 = 0.98; see Table S1) plus parameters already used for fits of normal human RBCs under stress (5) and based in part on single molecule AFM studies of forced unfolding (9–11). Although step-like data for 4.1R-null cells do not fit very well to the fLL equation (R2 = 0.76), which suggests additional processes, a lower unfolding rate for these cells also suggests less stress sustained by spectrin. Forced and rapid dissociation of spectrin from actin seems plausible because the spectrin–actin association in the absence of 4.1R is known to be very weak at Kd ∼ 10 μM (15, 16). If 4.1R normally keeps spectrin bound to actin when the membrane is stressed, then spectrin can be stretched and unfolded; but if 4.1R is missing, then spectrin will detach from actin when the membrane is stressed and spectrin stretching and unfolding will be less likely. 4.1R-null cells also have less spectrin, actin, and other membrane proteins (13, 17), and so the fewer cross-links in these cells would tend to be more rapidly stressed and dissociated, thereby relaxing spectrin before much labeling occurs. It is nonetheless conceivable that much smaller shear stress than used here might cause more unfolding and labeling of spectrin in 4.1R cells than in normal cells, but smaller stresses would also give less labeling per protein and thus be much more difficult to measure.
Stress-Enhanced Labeling of Actin and Ankyrin Exhibits a Threshold.
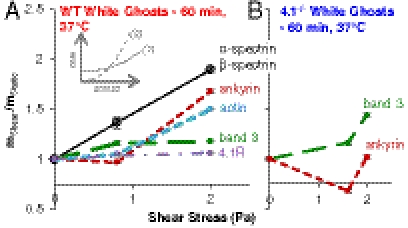
Simultaneous exposure of all proteins to thiol-reactive dye allows a comparison of any stress-dependent changes of other cytoskeletal proteins to spectrin. Although stress-dependent labeling of spectrin is essentially linear, stress-enhanced labeling of actin and ankyrin (Fig. 3A) occurs only at stress > 0.8 Pa, suggesting protein dissociation at high force. Reaction rates for a concentration n are generally accelerated by stress σ as rate = n ko exp(σ/σo), with both the unstressed rate ko and the stress scale σo being characteristic of each reaction (unfolding or dissociation). For competing reactions, if reaction-(1) has a high ko but large σo while reaction-(2) has a low ko and small σo, then there is an understandable transition from reaction-(1) dominated to reaction-(2) dominated at high stress (Fig. 3A, Inset). Here, reaction-(1) represents up to n1 = 2 × 37 = 74 unfolding reactions for spectrin tetramer domains per ankyrin dissociation reaction-(2) (i.e., n2 = 1), and if one assumes for simplicity that all of the ko’s are the same, then the net rates of unfolding versus dissociation are equal when
This occurs at σ ∼ 2 Pa (Fig. 3A), giving σo2 ≈ σo1/(1 + 2.1σo1). The stress-enhanced rate of labeling any one domain in Fig. 3A is also estimated as exp(2 Pa/σo1) ≈ 2, and so σo1 ≈ 2 Pa and σo2 ≈ 0.4 Pa. Ankyrin dissociation from spectrin might thus be more stress-sensitive (low σo) but many spectrin domains unfold first. Entropy thus dominates.
Fig. 3.
Ankyrin and actin labeling exhibit a shear threshold that depends on 4.1R. (A) With normal ghosts, a low shear stress of 0.8 Pa primarily causes force-enhanced labeling of spectrin, but at a higher shear stress of 2.0 Pa, ankyrin and actin also show considerable stress-enhanced labeling, whereas band 3 and 4.1R show little to no stress-enhanced labeling. (B) With 4.1R-null ghosts, band 3 labeling is stress-enhanced, whereas ankyrin labeling is not. Actin was not be analyzed because actin levels in 4.1R-null cells are lower than in wild type.
In contrast to large labeling changes in spectrin, actin, and ankyrin, band 3 shows only slight shear-enhanced labeling at low and high stress (+15%) whereas 4.1R shows no significant force-dependent labeling. Band 3’s large cytoplasmic tail interacts with the spectrin network through ankyrin and possesses Cys that might be stress-exposed (17, 18), whereas 4.1R is a globular stabilizer of spectrin–actin and a connector of F-actin protofilaments to the membrane. Spectrin thus seems stretched and unfolded at low stress, whereas high stress dissociates a fraction of spectrin–actin and also extends ankyrin with dissociation of a fraction of ankyrin from spectrin and/or band 3.
Loss of 4.1R limits force-enhanced labeling of ankyrin to (mshear/mstatic) = 1.0 at the highest shear stress of 2 Pa (Fig. 3B). Without 4.1R, spectrin dissociation from actin at low stress would certainly reduce the tension on spectrin and not only minimize force-enhanced unfolding and labeling of spectrin as observed (Fig. 2C) but would also minimize stress on ankyrin. Remarkably, a somewhat lower shear stress of 1.6 Pa leads to less labeling of ankyrin than in static 4.1R-null cells: (mshear/mstatic) < 1.0. Cys labeling in static cells is dominated by sites on the protein surface and also perhaps involves buried sites that are exposed at a basal rate dictated by a basal cytoskeletal stress in the RBC membrane (19). With 4.1R-null RBCs, the shear-enhanced spectrin dissociation from actin—which is decreased in abundance (17)—could therefore reduce the net tension on the ankyrin–spectrin interface to levels below static levels. At the same time, band 3 in the 4.1R-null cells at the highest shear exhibits significantly more stress-enhanced labeling than wild-type cells; previous work with 4.1R-null cells had indeed suggested that band 3 undergoes a conformational change in the absence of 4.1R (17). The exact nature of this change and its shear sensitivity needs further study, especially at small stresses, but all of the molecular responses here are distinct from normal cells.
Cys Shotgun Mass Spectrometry Kinetic Maps: Half the Sites Exhibit Force-Dependent Labeling.
Cys adducts of IAEDANS and monobromobimane (mBBr) have both been shown to survive the ionization and fragmentation processes of tandem mass spectrometry (5) (BODIPY does not remain intact), and so we used both dyes to identify and quantify tryptic peptides of spectrin that are labeled in sheared mouse cell membranes. Roughly similar in size to a tryptophan side chain, the dyes label buried Cys without greatly perturbing the refolding of proteins (5). Labeling under static conditions was compared to labeling at moderately high shear stress of 1.5 Pa for t = 30 and 60 min, and unreacted Cys in all samples (no reaction went to completion) were capped with iodoacetamide (IAM), providing a means to normalize dye results. IAEDANS was entrapped in ghosts first in an attempt to differentially label fast-reacting Cys on the protein surfaces even though the efficiency of labeling and/or MS detection of this dye is low. Lysates were separated by SDS-PAGE and spectrin bands were excised, trypsinized, and run through liquid chromatography–tandem mass spectrometry (LC-MS/MS). Sequence coverage was about 76% for α-spectrin and 68% for β-spectrin; 17 cysteines were detected, with 13 located in repeat domains, and 3 of these were predicted to be surface exposed (Table S2).
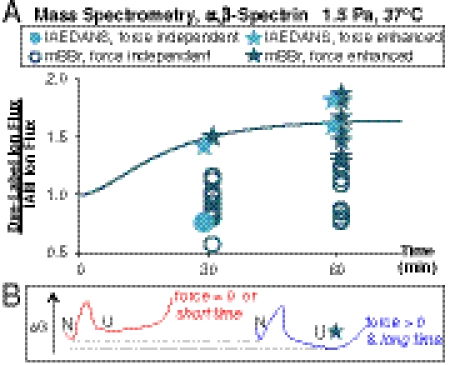
Force-enhanced labeling for individual tryptic peptides was determined from normalized ion fluxes in the first MS as the ratio for IAEDANS, Φtime = [(Cys-IAEDANS)/(Cys-IAM)], or else as φtime for mBBr, with (Φ,φ)≥1.25 appearing to be a sound criterion for force-enhanced labeling. Even at moderately high shear stress, only two Cys appear shear sensitive at 30 min in LC-MS/MS analyses, whereas nearly half of the Cys show force-enhanced labeling at 60 min (Fig. 4A). The average (Φ, φ)-values for force-enhanced Cys labeling were fitted to the fLL equation (Fig. 4A) using the same parameters as for spectrin in normal cells (Fig. 1C). This indicates that the stress-dependent average labeling measured by densitometry of SDS-PAGE can also be quantified by LC-MS/MS at the level of individual domains.
Fig. 4.
Cys shotgun–mass spectrometry labeling kinetics identify force-enhanced and force-independent sites. (A) Ion fluxes in MS of dye-labeled spectrin peptides were normalized to IAM-modified peptides in the same sample and represented by Φ and φ for IAEDANS and mBBr modified peptides, respectively. The fLL curve was generated using the same parameters as in the WT-white ghost analysis (Fig. 2C and Table S1) and used to define the cutoff for force-enhanced labeling, Φ,φ≥0.25. This cutoff was chosen such that average Φ, φ for force-enhanced cysteines fit to the fLL curve (R2 = 0.97). Starred points represent cysteines with force-enhanced labeling. After 30 min of moderate shear, only two of the detected cysteines showed significant force-enhanced labeling, whereas at 60 min, about half of the cysteines showed force-enhanced labeling. (B) Representative schematic of the fLL-based free energy landscape for force-sensitive domains. At zero stress or short times, unfolding and labeling of such domains is unlikely, whereas force and time both enhance labeling.
Tabulation of all detected labeled peptides with (Φ, φ)-values in Table S2 shows that domain locations as well as predicted accessibilities are highly variable. Solvent accessibility of Cys located within repeat domains was predicted by heptad position within the helices (20) (Fig. S1), with five sites showing force-enhanced labeling at 60 min. These domains thus seem likely to have unfolded during shear, but two Cys that were predicted to be solution-exposed also show force-enhanced labeling at 60 min. Overall accuracy of the shear/static ratio measurements by LC-MS/MS is evident in the fact that there are essentially no ratios much less than 1.0, and also only some ratios increase with time.
Labeling Kinetics at Junctional Cys Reveals Different Binding Strengths in Spectrin Associations.
Kinetic mapping of labeled peptides by LC-MS/MS shows that two Cys that are force-exposed at 30 min under high shear are located at or near sites of protein–protein interaction (Fig. 5A). Cys β1883 is directly adjacent to the spectrin–ankyrin-binding site, whereas Cys α473 is located across from the spectrin tetramer interface, which is a couple of repeats distal to the spectrin–ankyrin-binding site. Force exposure of both cysteines correlates with the increased labeling of ankyrin at high shear (Fig. 3A).
Fig. 5.
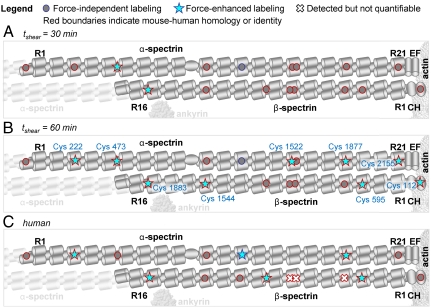
Kinetic maps of labeled Cys detected by mass spectrometry. Ghosts were sheared at a moderate shear stress (1.5 Pa), below the threshold of ankyrin and actin dissociation but high enough to expect significant spectrin unfolding (Fig 2C). The force-enhanced labeling of individual cysteines was assessed via the ion-flux ratio of modified/unmodified peptides (Tables S2 and S3). In this schematic, repeat domains are depicted as three-cylinder bundles. All cysteines detected by mass spectrometry are represented by gray circles (force-independent labeling), blue stars (force-enhanced labeling), or white x’s (unquantifiable). White x’s represent cases where only modified cysteines were detected, and the modified/unmodified ratio could not be calculated. Red outlines mark cysteines that are homologous with cysteines in human spectrin. (A) After 30 min of moderately high shear stress (1.5 Pa), only Cys located near the spectrin–ankyrin junction show force-enhanced labeling (stars). (B) After 60 min, Cys near the spectrin–actin junction as well as Cys buried within repeat domains show force-enhanced labeling. At both time points, there are also many Cys for which measurements show labeling is force-independent. (C) Many of the cysteines detected here in mouse spectrins are homologous to the Cys detected in previous studies of human RBCs (6), with some disparities likely due to sequence and/or structure differences. Amino acids and repeat assignments were made according to Uniprot files P08032 (mouse α-spectrin), P15508 (mouse β-spectrin), P02539 (human α-spectrin), and P11277 (human β-spectrin).
Cysteines near the spectrin–actin interaction—in either the CH1 domain (β112) or in repeat 21 of α-spectrin (α2155)—show shear-enhanced labeling only at 60 min in these normal cell membranes (Fig. 5B). According to densitometry results, both ankyrin and actin show force-enhanced labeling at high stress (60 min), but spectrin binding to the actin–4.1R complex is about 50-fold stronger than the cited binary interactions (15, 16) and therefore stronger than spectrin–ankyrin binding (Kd = 10-7 M) (21), which makes it likely that ankyrin dissociates first from the spectrin network. Cys α2155 was predicted to be buried within repeat 21, but its labeling seems indicative of high stress at the spectrin–actin junction.
Cysteine homology between mouse and human spectrins is striking, with past studies revealing Cys exposure in several sites seen also in mouse, including α222, α1877, and β595 (Fig. 5C). Cys in human repeat β15 also shows increased labeling in shear, which implicates dissociation from ankyrin, and shear-enhanced labeling of human ankyrin was indeed significant if small in previous studies of human membranes (5). The same cited studies did not reveal a significant shear dependence to actin labeling, consistent with cysteines in human α-spectrin repeat 21 and β-spectrin’s CH1 domain showing no shear enhancement. Therefore, in human RBCs and likely in mouse RBCs, spectrin dissociation from actin is uncoupled from forced dissociation of spectrin from ankyrin.
Several additional differences exist in the labeling patterns of human and mouse spectrins, which may argue that the transmission of force is somewhat species dependent. Cys α473 and β1544 show shear-enhanced labeling, whereas the human homologues do not. Cys α1248 in repeat α12 is not homologous to the human cysteine in that repeat, but although both are predicted to be buried, shear-enhanced labeling is seen only in human, which suggests that repeat α12 unfolds in human RBCs and perhaps not in mouse. Cys β1159 is located in the linker between repeats β8-9, based on a crystal structure (22), and labeling of this site appears independent of stress in mouse but strongly stress-dependent in humans (5). Sequence differences in the linker region could have distinct effects on linker flexibility and Cys exposure (Fig. S2A).
Cys Labeling in Relation to Atomic Models of Interaction Domains.
Homology models of mouse domains were made based on existing crystal structures, suggesting that Cys β112, β1883, and α473 are all partially exposed to solution so that some labeling is expected even in static samples. Cys β112 is within helix E of the CH1 domain and is very close to the spectrin–actin interface (Fig. 6A and Fig. S2B). Dissociation from actin is expected to greatly increase the labeling efficiency at least because of decreased steric hindrance. Although it is not yet known whether 4.1R possesses any cryptic Cys that are sequestered within folds or binding interfaces, this protein shows no stress-enhanced labeling but is certainly labeled under all conditions.
Fig. 6.
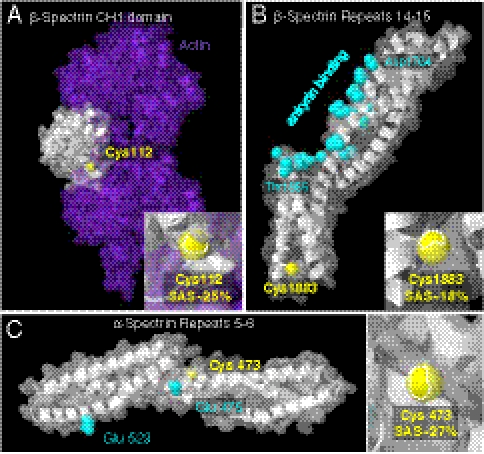
Force-enhanced labeling around spectrin binding sites. Homology models of mouse spectrin domains were made based on existing crystal structures (see Materials and Methods) and used to estimate the spatial relationship between detected Cys and previously studied, functionally important residues. Secondary structure is represented in ribbon form, and the translucent surface represents the solvent accessible surface (SAS). Thiol groups of cysteines of interest are shown yellow in spacefill with %-SAS indicated; 0% would be a buried Cys. (A) The CH1 domain in β-spectrin (white) is bound to F-actin (purple) with Cys 112 located just outside the spectrin–actin contact. (B) β-spectrin repeats 14–15 constitute the ankyrin binding site (cyan side chains), and Cys 1883 is located on the same repeat but not within the ankryin binding site. (C) α-spectrin repeat 5, located adjacent to the tetramerization site, laterally associates with β-spectrin. Side chains of amino acids in contact are cyan. Cys 473 is on the opposite face of this interacting helix.
The thiol in Cys β1883 is directed toward the hydrophobic core of this spectrin repeat and is located at the end of a domain involved in spectrin–ankyrin binding (23) (Fig. 6B and Fig. S2C). Site-directed mutagenesis in the large and unusual kink between β-repeats 14–15 has suggested a mechanosensitive, conformationally plastic binding site for ankyrin with possible relations to domain linker mutations in hemolytic anemias (24). Increased labeling of Cys β1883 in stressed cells here might thus reflect enhanced accessibility with conformational changes at moderate shear stress (1.5 Pa) or perhaps it indicates forced dissociation from ankyrin, consistent with the 5-fold higher stress sensitivity (see σo estimates above). At about the same stress, the 4.1R null cells showed reduced labeling of ankyrin (Fig. 3B), which might indicate that spectrin binding to (or shielding by) ankyrin is enhanced by relaxation of cytoskeletal tension in these cells containing decreased amounts of ankyrin and band 3 (17). Alternatively, small structural changes in the complex during shear could allow thiol probes greater access to a given cysteine without the dissociation or outright domain unfolding.
Cys α473 is located on a helix that is part of the proximal tetramerization interface between α- and β-spectrin (Fig. 6C). It is on the opposite side of the helix involved in the interaction (25), and so labeling of this residue does not necessarily imply dissociation of the spectrin tetramer. However, force-enhanced labeling of this site could indicate extra stress exerted at this junction when cells are sheared.
Ankyrin Labeling Is Shear-Enhanced in Ankyrin Repeats and Near the Spectrin Binding Site.
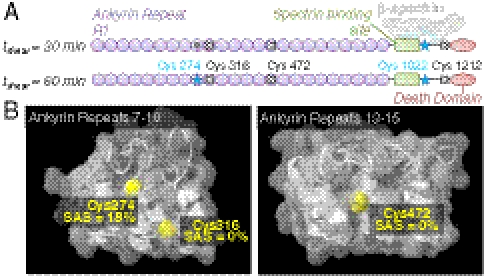
Sequence coverage of ankyrin ranged from 38% to 52%, with five detected cysteines spread along the protein (Fig. 7A and Table S4). Three detected Cys are in ankyrin repeats, two of which, Cys 316 and Cys 472, are predicted to be completely buried (Fig. 7B). Cys 274 in ankyrin repeat 8 shows force-enhanced labeling only at 60 min and contributes to the stress-enhanced labeling measured in densitometry of SDS-PAGE gels (Fig. 3A). (Φ)-values could not be calculated for Cys 316 and 472 in ankyrin repeats 9 and 14, respectively, or Cys 1212 because only peptides with fluorescently modified cysteines were detected, suggesting that these are highly reactive surface sites that are well-labeled under static conditions. Such sites would not contribute to stress-enhanced labeling. Ankyrin repeats 7–12, containing Cys 274 and 316, interact with one dimer of the band 3 tetramer, and repeats 13–24, containing Cys 472, interact with the other. The ankyrin groove surfaces directly associate with band 3 dimers (26). Cys 274 is located at the end of a helix, faces directly into the ankyrin groove, and is probably shielded by band 3 binding. Force-enhanced labeling could result from fluctuations in ankyrin–band 3 association after 60 min of shear. Cys 316 faces the interface of four repeat helices and is located on a helix in the outer row. The ankyrin superhelix is sufficiently elastic to stretch before unfolding occurs (27). Stress on the membrane propagating through band 3 could also stretch the ankyrin groove. Helices in the outer row are less tightly packed, increasing chances for solvent exposure of Cys 316 during shear. Cys 472 also faces the ankyrin groove but is not solvent accessible and does not likely participate directly in interactions with band 3. A high degree of labeling suggests that the ankyrin repeats experience a high degree of structural strain, even at short times.
Fig. 7.
Ankyrin Cys-labeling detected by mass spectrometry. Ankyrin repeat domains, structurally defined as two alpha helices separated by β-turns, are also reported to respond to mechanical stress, providing the molecular basis for overall protein flexibility. (A) The schematic respectively shows ankyrin repeats, the spectrin binding domain, and the death domain as purple circles, a green rectangle, and a red oval. Labeling of cysteines detected by mass spectrometry is represented in Fig. 5, where blue stars indicate force-enhanced labeling, gray circles represent force-independent labeling, and white x’s indicate cysteines where only the modified peptide was detected. Force-enhanced labeling of Cys 1022 occurs at both 30 and 60 min of moderate shear (1.5 Pa). Cys 316 and Cys 472 are predicted to be buried, whereas Cys 274 is partially exposed to solution. (B) Homology models based on crystal structures for human Ankyrin R illustrate spatial relationships of detected Cys to known ankyrin binding interfaces. For both structures, the ankyrin groove faces out of the page.
Cys 1022 shows force-enhanced labeling at 30 and 60 min and is within only a dozen residues of the β-spectrin–ankyrin interface. The extent and kinetics of stress-enhanced labeling of this site φ30′,60′ = (1.61,1.76) are similar to that of β-spectrin Cys 1883 φ30′,60′ = (1.52,1.68). The comparison suggests a similar mechanism of perturbation, which is detected only at high stress (Fig. 3A) and therefore suggests forced dissociation of spectrin–ankyrin, which increases Cys 1022 exposure to solvent.
Conclusions
Both spectrin unfolding and spectrin dissociation from the network appear to contribute to the RBC’s response to mechanical stress. At low stress, repeat unfolding resulted in enhanced spectrin labeling that was equal for both antiparallel chains, whereas other cytoskeletal proteins were labeled independent of stress. Simultaneous enhanced labeling of spectin, actin, and ankyrin at high stress points to coordinated events, confirmed after examination of individual domain labeling with mass spectrometry. Fast labeling of Cys at high stress proximal to the spectrin–ankyrin junction verified the weaker ankyrin–spectrin affinity, as compared to the spectrin–actin-4.1R interaction. Stress-enhanced labeling of solution-exposed Cys in the region of spectrin–actin interaction signaled quaternary structure changes in the cytoskeletal network. Loss of coordinated, stress-enhanced labeling in the absence of 4.1R illustrates this protein’s crucial role in maintaining network integrity, opposing spectrin–actin dissociation up to intermediate shear stress.
Mutant cells have the potential for abnormal molecular responses, and preliminary results on human RBCs with spectrin mutations that lead to an elliptocyte morphology and mechanical fragility similar to 4.1R deficiency (28) also indicate stress-accelerated spectrin labeling (Fig. S3). Although more study is needed to understand differences from normal cells, the shear-enhanced labeling at 60 min (1 Pa) appears similar to that of 4.1R-null cells here and below that of normal RBC (Fig. 2C). Nonetheless, the many complex mechanisms of molecular failure under stress motivate continued study of a type illustrated in Fig. S4 where we show (i) strong Cys labeling of membrane fragments generated at very high shear (4 Pa) consistent with rupture-enhanced labeling; (ii) treatment of ghosts with 1.5 M urea increases the amount of Cys labeling consistent with partial unfolding and/or dissociation; (iii) solubilized spectrin is labeled more than membrane-assembled spectrin, which seems consistent with Cys shielding by 4.1R, ankyrin, and/or actin in the membrane.
CS-MS reveals several types of structural changes, and labeling kinetics provide valuable insight into molecular processes within the network, with some tentative distinctions between weak and strong interactions. All of the spectrin Cys that were concluded here to be significantly stress-enhanced in labeling are in domains with melting temperatures Tm > 37 °C based on studies of individual domains (Fig. S5) (8) or purified intact protein (7). In other words, these domains should be mostly folded at the temperature used in these studies, but force is seen to unfold the domains in intact membranes. Tm’s otherwise correlate poorly with the extent of force-enhanced Cys labeling, which illustrates the limited physiological insight obtained from traditional melting studies. Membrane curvature on a length scale of microns likewise shows no detectable correlation with labeling (Fig. S6): Indeed, labeling appears homogeneous on the membrane within approximately 10%. The present studies thus suggest a temperature threshold to the stress-dependent, homogeneous dynamics of protein interactions and domain folding. It is also now clear that the cytoskeleton responds in a graded fashion to force, with the nature of structural change depending on the magnitude and duration of stress.
Materials and Methods
Information about reagents, the preparation of ghosts, cysteine labeling under shear, SDS-PAGE analysis, mass spectrometry analysis, and structure predictions can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Sara Saad (Universidade Estadual de Campinas, São Paulo, Brazil) for providing spectrin-mutant cells for initial study. We are grateful for support for this work from National Institutes of Health Grants R01HL062352 (to D.E.D.), P01DK032094 (to X.A., M.N., and D.E.D.), R01HL038794 (to D.W.S.), Interdisciplinary Cardiovascular T32 (C.C.K.), Penn’s National Science Foundation–Nano Science Engineering Center (D.E.D. and C.C.K.), and the Human Frontiers Sciences Program (D.E.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018887108/-/DCSupplemental.
References
- 1.Mohandas N, Gallagher PG. Red cell membrane: Past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takakuwa Y, Tchernia G, Rossi M, Benabadji M, Mohandas N. Restoration of normal membrane stability to unstable protein 4.1-deficient erythrocyte membranes by incorporation of purified protein 4.1. J Clin Invest. 1986;78:80–85. doi: 10.1172/JCI112577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett V, Healy J. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb Perspect Biol. 2009;1:a003012. doi: 10.1101/cshperspect.a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giorgi M, Cianci CD, Gallagher PG, Morrow JS. Spectrin oligomerization is cooperatively coupled to membrane assembly: A linkage targeted by many hereditary hemolytic anemias. Exp Mol Pathol. 2001;70:215–230. doi: 10.1006/exmp.2001.2377. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandts JF, Erickson L, Lysko K, Schwartz AT, Taverna RD. Calorimetric studies of the structural transitions of the human erythrocyte membrane. The involvement of spectrin in the A transition. Biochemistry. 1977;16:3450–3454. doi: 10.1021/bi00634a024. [DOI] [PubMed] [Google Scholar]
- 7.An X, et al. Conformational stabilities of the structural repeats of erythroid spectrin and their functional implications. J Biol Chem. 2006;281:10527–10532. doi: 10.1074/jbc.M513725200. [DOI] [PubMed] [Google Scholar]
- 8.Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: Low unfolding forces in helix bundles. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 9.Law R, et al. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortiz V, Nielsen SO, Klein ML, Discher DE. Unfolding a linker between helical repeats. J Mol Biol. 2005;349:638–647. doi: 10.1016/j.jmb.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 11.Johnson CP, et al. Pathogenic proline mutation in the linker between spectrin repeats: Disease caused by spectrin unfolding. Blood. 2007;109:3538–3543. doi: 10.1182/blood-2006-07-038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An X, Lecomte MC, Chasis JA, Mohandas N, Gratzer W. Shear-response of the spectrin dimer-tetramer equilibrium in the red blood cell membrane. J Biol Chem. 2002;277:31796–31800. doi: 10.1074/jbc.M204567200. [DOI] [PubMed] [Google Scholar]
- 13.Tchernia G, Mohandas N, Shohet SB. Deficiency of skeletal membrane protein band 4.1 in homozygous hereditary elliptocytosis. Implications for erythrocyte membrane stability. J Clin Invest. 1981;68:454–460. doi: 10.1172/JCI110275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiancone E, Currell DL, Vecchini P, Antonini E, Wyman J. Kinetics of the reaction of the “masked” and “free” sulfhydryl groups of human hemoglobin with p-mercuribenzoate. J Biol Chem. 1970;245:4105–4111. [PubMed] [Google Scholar]
- 15.Ohanian V, et al. Analysis of the ternary interaction of the red cell membrane skeletal proteins spectrin, actin, and 4.1. Biochemistry. 1984;23:4416–4420. doi: 10.1021/bi00314a027. [DOI] [PubMed] [Google Scholar]
- 16.Discher DE, et al. Mechanochemistry of protein 4.1’s spectrin-actin binding domain: Ternary complex interactions, membrane binding, network integration, structural strengthening. J Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomao M, et al. Protein 4.1R-dependent multiprotein complex: New insights into the structural organization of the red blood cell membrane. Proc Natl Acad Sci USA. 2008;105:8026–8031. doi: 10.1073/pnas.0803225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Kiyatkin A, Bolin JT, Low PS. Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood. 2000;96:2925–2933. [PubMed] [Google Scholar]
- 19.Discher DE, Boal DH, Boey SK. Simulations of the erythrocyte cytoskeleton at large deformation. II. Micropipette aspiration. Biophys J. 1998;75:1584–1597. doi: 10.1016/S0006-3495(98)74076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry DA, Dixon TW, Cohen C. Analysis of the three-alpha-helix motif in the spectrin superfamily of proteins. Biophys J. 1992;61:858–867. doi: 10.1016/S0006-3495(92)81893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podgorski A, Alster P, Elbaum D. Interactions among red cell membrane proteins. Biochemistry. 1988;27:609–614. doi: 10.1021/bi00402a017. [DOI] [PubMed] [Google Scholar]
- 22.Kusunoki H, MacDonald RI, Mondragón A. Structural insights into the stability and flexibility of unusual erythroid spectrin repeats. Structure. 2004;12:645–656. doi: 10.1016/j.str.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 23.Ipsaro JJ, Mondragón A. Structural basis for spectrin recognition by ankyrin. Blood. 2010;115:4093–4101. doi: 10.1182/blood-2009-11-255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stabach PR, et al. The structure of the ankyrin-binding site of β-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties. Blood. 2009;113:5377–5384. doi: 10.1182/blood-2008-10-184291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, et al. A comprehensive model of the spectrin divalent tetramer binding region deduced using homology modeling and chemical cross-linking of a mini-spectrin. J Biol Chem. 2010;285:29535–29545. doi: 10.1074/jbc.M110.145573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaely P, Tomchick DR, Machius M, Anderson RG. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee G, et al. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 28.Gallagher PG. Hereditary elliptocytosis: spectrin and protein 4.1R. Semin Hematol. 2004;41:142–164. doi: 10.1053/j.seminhematol.2004.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.