Abstract
A whole-genome phylogeny of the Escherichia coli/Shigella group was constructed by using the feature frequency profile (FFP) method. This alignment-free approach uses the frequencies of l-mer features of whole genomes to infer phylogenic distances. We present two phylogenies that accentuate different aspects of E. coli/Shigella genomic evolution: (i) one based on the compositions of all possible features of length l = 24 (∼8.4 million features), which are likely to reveal the phenetic grouping and relationship among the organisms and (ii) the other based on the compositions of core features with low frequency and low variability (∼0.56 million features), which account for ∼69% of all commonly shared features among 38 taxa examined and are likely to have genome-wide lineal evolutionary signal. Shigella appears as a single clade when all possible features are used without filtering of noncore features. However, results using core features show that Shigella consists of at least two distantly related subclades, implying that the subclades evolved into a single clade because of a high degree of convergence influenced by mobile genetic elements and niche adaptation. In both FFP trees, the basal group of the E. coli/Shigella phylogeny is the B2 phylogroup, which contains primarily uropathogenic strains, suggesting that the E. coli/Shigella ancestor was likely a facultative or opportunistic pathogen. The extant commensal strains diverged relatively late and appear to be the result of reductive evolution of genomes. We also identify clade distinguishing features and their associated genomic regions within each phylogroup. Such features may provide useful information for understanding evolution of the groups and for quick diagnostic identification of each phylogroup.
Keywords: prokaryotic phylogeny, commensal minimalism, Jensen-Shannon Divergence, phylotyping
The bacterium Escherichia coli was once considered only an innocuous commensal microbe, and Shigella was maintained as a distinct pathogenic genus because of its clinical significance. By 1982, with the identification of new enterohemorrhagic (EHEC) strains of E. coli, namely the infamous O157:H7 pathogen, a clearer picture had emerged, suggesting that the E. coli/Shigella group is an extremely diverse single enteric species (1) with a wide array of strains exploiting niches ranging from beneficial intestinal denizens to obligate extraintestinal pathogens. The pathogenic strains are a world-wide health issue: 125 million Shigellosis/dysentery infections occur annually in Asia alone (2). Consequently, the Shigella and Escherichia genera are two of the most extensively sampled among the prokaryotic genome-sequencing projects. With the availability of broadly sampled whole-genome sequences, we revisit the issues of phylogrouping and their evolutionary relationship by using an alignment-free feature frequency profile (FFP) method (3) on whole genomes. We compare our results with recent alignment-based methods that use a selected gene set.
The progenitor strains of today's commensal and pathogenic variants were likely present in the primate gut preceding the divergence of the great apes, perhaps greater than 30 MYa (4). Thus, we address several issues that relate to the evolutionary development of this clade. Which phylogroup of E. coli/Shigella may be the extant organism most closely related to the progenote of this group? Was this earliest progenote pathogenic or commensal? Which genomic features distinguish one strain from another and which are common among E. coli and Shigella? We summarize below current differences in interpretation on three relevant issues depending on analysis methods used.
Phylogroups and Divergence Order.
Early evidence using a small number of gene sequences (5) from E. coli isolates indicated that there were some clonal (monophyletic) groups within E. coli. Eventually, core gene sequence alignments established several stable phylogroups. The current classification (based on seven housekeeping genes), defined by Wirth et al. (6), divides the species into five phylogroups: A, B1, B2, D, and E. There is only a loose correlation between phylogroup and pathogenicity class, as apparent from Table 1. Shigella remains as a contentiously defined polyphyletic genus with its members clustering with several E. coli phylogroups. Regarding the order of earliest divergence, phylogenetic trees of housekeeping genes indicated that group D diverged first and that A and B1 are sister groups that separated later (7). Later analysis indicated that perhaps B2 rather than D is ancestral (8). Recent work by Touchon et al. (9) asserts that the D group (followed by the B2 group) diverges first, whereas Ogura et al. (10) showed the divergence orders of their two trees, one based on core protein-coding sequences and the other on gene content, are quite different. Although there is a natural bias toward the sequencing of pathogenic strains, it is worth noting that, generally, pathogenic strains occupy the basal positions and the commensal strains (in particular phylogroup A) appear to have more derived character in that they are deeply nested within most recent phylogenies.
Table 1.
E. coli/Shigella subgroups
| Strain | Class | Phylo- group | Serotype |
| K12-W3110 | Commensal | A | O16 |
| K12-MG1655 | Commensal | A | O16 |
| HS | Commensal | A | O9:H4 |
| K12-BW2952 | Commensal | A | O16 |
| K12-DH10B | Commensal | A | O16 |
| REL606 | Commensal | A | ? |
| BL21(DE3) | Commensal | A | O7? |
| ATCC8739 | Commensal | A | O146 |
| SE11 | Commensal | B1 | O152:H28 |
| 55989 | Enteroaggregative | B1 | O128:H2 |
| IAI1 | Commensal | B1 | O8 |
| E24377A | Enterotoxigenic | B1 | O139:H28 |
| 11128 | EHEC | B1 | O111:H- |
| 11368 | EHEC | B1 | O26:H11 |
| 12009 | EHEC | B1 | O103:H2 |
| S88 | Extracellular pathogenic E. coli | B2 | O45:K1 |
| E2348/69 | Enteropathogenic E. coli | B2 | O127:H6 |
| ED1a | Commensal | B2 | O81 |
| APEC01 | APEC | B2 | O1:K12:H7 |
| 536 | UPEC | B2 | O6:K15:H31 |
| UTI89 | UPEC | B2 | O18:K1:H7 |
| CFT083 | UPEC | B2 | O6:K2:H1 |
| UMN026 | Extracellular pathogenic E. coli | D | O17:K52:H18 |
| IAI39 | Extracellular pathogenic E. coli | D | O7:K1 |
| SMS-3-5 | Extracellular pathogenic E. coli | D | O19:H34 |
| EDL933 | EHEC | E | O157:H7 |
| TW14359 | EHEC | E | O157:H7 |
| Sakai | EHEC | E | O157:H7 |
| EC4115 | EHEC | E | O157:H7 |
| B4 Sb227 | Shigella | S | OB4 |
| B18 BS512 | Shigella | S | OB18 |
| SS Ss046 | Shigella | S | OSonnei |
| F2a 2457T | Shigella | S | OF2a |
| F2a 301 | Shigella | S | OF2a |
| F5b 8401 | Shigella | S | OF5b |
| D1 Sd197 | Shigella | S | OD1 |
Serotypes: O, somatic lipopolysaccharide; K, capsular polysaccharide; H, flagellar antigens.
Evolution of Shigella.
A particularly quarrelsome issue in E. coli systematics is the validity of the Shigella genus or at least its validity as a qualified “subclade.” The conventional view is that Shigella are E. coli strains that have acquired a specific set of genes that contribute to the Shigella pathotype. This collection of genes is observed both in a genomically incorporated form as well as in “virulence plasmid” isolates, emphasizing the mobile nature of the genes (11). Of particular interest is how Shigella developed virulence and its phylogenetic relationship with enteroinvasive E. coli. Two conflicting theories have been proposed on the origin of Shigella: either multiple independent origins (12–15) or single clonal origin (8). Pupo et al. (12) observed (with a handful of housekeeping genes) that Shigella strains form three clusters within E. coli, suggesting that three separate ancestral E. coli strains independently acquired virulence. According to this theory, the virulence plasmid would have been independently acquired by distantly related strains through lateral transfer of a pathogenicity island (PAI). Escobar-Páramo et al. (8), on the other hand, suggested that there is a single ancestral virulence plasmid that accounts for the emergence of Shigella. More recent analyses by Touchon et al. (9) and Ogura et al. (10), using two different core gene sets, indicate that some but not all Shigella species are clonally related.
Lateral Gene Transfer and Lineal Phylogeny.
As the above conflict illustrates, the lateral transfer of genes among prokaryote (archaea/bacteria) species can severely confound species boundaries and necessarily makes lineal phylogenic inference more difficult, especially when the inference is based on the alignment of a set of selected genes, one of which may be laterally transferred. E. coli is especially problematic in this regard because the species has quite a high level of recombination (16). E. coli genomes show evidence of widespread acquisition of functions by lateral gene transfer, accompanied by an equivalent level of gene deletion (17). When comparing any two E. coli strains, one may observe as little as 3% nucleotide divergence among conserved genes but, at the same time, may also observe differences in gene content up to 50% (18). Consequently, one must consider whether such frequent recombination could overwhelm the true lineal phylogenetic signal. It is hypothesized that a “core” or “backbone” set of genes exists in E. coli upon which a mosaic set of genes can be supplemented to provide the necessary genetic variation to adapt to specific environmental niches. The main contributors to this great genetic diversity among E. coli are mobile genetic elements that are responsible for widespread lateral transfers and genomic rearrangements. Phages, plasmids, transposons, and insertion sequences [collectively referred to as the “mobilome” (19)] are all possible candidate sites of recombination. In general, mobile elements are well known for genetic exchange among themselves and within the host genome, and therefore they can be composed of both conserved genes and relic genes from previous hosts. Paradoxically, recombination can be both a divergent and a convergent force. In some cases, homologous recombination, in the form of gene conversion, can actually restore sequence similarity. Touchon et al. (9) hypothesized that high levels of gene conversion occur in E. coli strains. They observed that gene conversion events are more likely than point mutations. Thus, in E. coli, contributions from recombination may far outweigh site-level mutation as an evolutionary mechanism.
Thus, the above differences highlight the fact that phylogrouping and divergence order of the groups derived based on gene-alignment methods depend highly on the choice of genes selected to align. We present here the use of an alignment-free method applied to whole genomes to address these issues.
Results
To avoid possible bias in tree building as a consequence of subjective gene selection, we used a method of alignment-free genome comparison that uses FFPs to enable an efficient comparison of whole-genome sequences (3) (Materials and Methods). The FFP method is a viable alternative to gene-based alignments, especially when substantial differences in gene order and gene content are suspected, and can be applied to genome sequence comparison even if very few common genes with high sequence identity are distributed among the genomes.
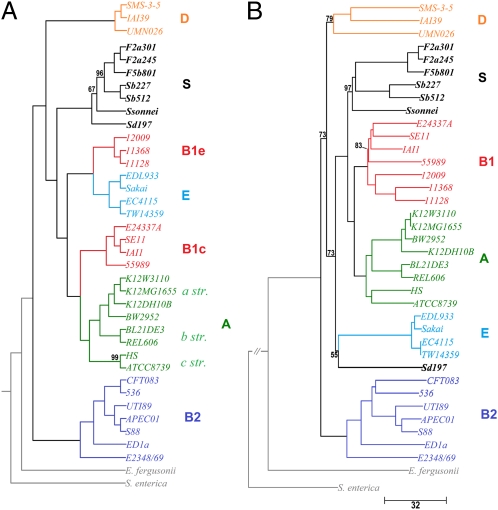
We present the phylogeny of the Escherichia/Shigella group derived by using the FFP method in two contrasts. In the first form (Fig. 1A), the compositional similarity of features of length l = 24 (∼8.4 million features) is used to build a tree without any filtering. Jensen–Shannon divergences (20) between FFPs are used as distances. Because this approach emphasizes whole-genome features, the resulting tree is likely to reflect phenetic relationships (phenetic phylogeny) among the extant organisms in general and especially if they experienced extensive convergent or divergent evolution. In the second form (Fig. 1B), we have built a phylogeny based on genomic features, not genes, commonly shared among all phylogroups, which is thus likely to reflect the lineal evolutionary history among the phylogroups (evolutionary phylogeny). This tree is based on the composition of core features (∼0.56 million features), with low frequency and low variability among genomes, which is likely to reveal lineal evolutionary signal. Because it is difficult to distinguish features corresponding to evolutionary signal from those representing lateral transfer signal, we first removed features likely to be associated with mobile or repetitive DNA by filtering out features with high frequencies. Only features ranging in frequency between 1 and 3 were used. These features are the core features of the genome—features which are likely conserved, observed in common among all genomes compared, and change rarely in frequency. We then used an unordered character state model (21) to reduce the effect of lateral transfer signal. In this case, the frequencies are treated as character states, where states are different if frequencies of a feature in two genomes are nonidentical. Distances are expressed as the sum of differences of the unordered states.
Fig. 1.
Phenetic and evolutionary alignment-free whole-genome phylogenies of E. coli/Shigella. Two method were used, both constructed from FFPs of length l = 24. The compositional phylogeny in A uses all features of length l = 24 and the Jensen–Shannon divergence (the tree is drawn unscaled). The evolutionary phylogeny in B depicts the likely evolutionary history of the phylogroups. The tree was constructed from features present in all 38 genomes and a distance derived from a multistate unordered characters model of feature frequency. Distances represent the number of character feature changes. The differences between the two trees reflect lateral transfer of features or genes within Shigella and between B1 [B1 is phenetically separated into B1e (EHEC) and B1c (commensal) subgroups] and E. The A phylogroup can be divided into a, b, and c strains. Numerical values placed at internal branches represent 10% jackknife confidence values. No value represents 100% agreement among pseudoreplicates.
In both cases, a feature length of 24 is used (Materials and Methods), and the neighbor-joining method is used to construct trees. A principle advantage of the FFP method is that the method does not require gene selection or gene boundary identification. Our trees are compared with those of Touchon et al. (9) and Ogura et al. (10), which are currently the most comprehensive treatments of the subject by gene-alignment–based methods. The former is based on the comparison of concatenated alignable regions of 1,878 core genes (∼10% of all genes) from E. coli/Shigella, and the latter is based on comparison of alignable regions of 345 orthologous protein-coding sequences and also on a gene repertoire consisting of 12,940 protein-coding sequences.
Phylogroups of E. coli/Shigella.
The phylogrouping of Touchon et al. (9) and Ogura et al. (10) are in general agreement with phylogrouping in our phylogenies based on alignment-free whole-genome FFPs. However, two of the notable differences with the tree of Touchon et al. are that they show Shigella is divided into three clades, one clustering with subgroup E, another with subgroup B1 of E. coli, and the third independent, and subgroup D is split into two separate clades. Our result in Fig. 1A shows that Shigella forms a monophyletic grouping. Additionally, the B1 group is split into two subgroups: one group clusters with subgroup E, and the other clusters with subgroup A. The E-associated group contains the non-O157 EHEC strains (Table 1), whereas the A-associated group contains the commensal strains. We define them as EHEC B1e and commensal B1c. The general conclusion is that this topology may be more associated with phenotype, gene complement, and pathotype. In the evolutionary phylogeny (Fig. 1B), on the other hand, Shigella dysenteriae is segregated from the main Shigella clade, clustering with phylogroup E (serotype O:H157; Table 1). Subgroup B1 remains monophyletic. One of the notable differences with the trees of Ogura et al. is that their gene-content tree intermixes phylogroups B1 and A.
Divergence Order.
The divergence order of the phylogroups in Touchon et al. (9) and Ogura et al. (10) are considerably different from our evolutionary phylogeny tree. The divergence order can be interpreted as the relative order by which different phylogroup lineages evolved. The divergence order in Fig. 1B (relative to the outgroup Escherichia fergusonii) is B2, D, S. dysenteriae, E, Shigella, B1, and A. The most basal phylogroup is subgroup B2, a subgroup primarily dominated by uropathogenic (UPEC)/avian pathogenic (APEC) strains (Table 1). The UPEC strains are opportunistic pathogens. One of the most derived (or recently evolved) phylogroups in our tree is subgroup A, which contains exclusively commensal strains. The A group genomes are generally smaller and encode fewer genes relative to other phylogroups, perhaps an indication of extensive reductive convergent evolution. The commensal subgroup has the smallest genomes, reflecting the absence of the wide range of virulence factors found in pathogenic strains. For Shigella, the most basal member is S. dysenteriae.
The divergence order does not strictly follow the order of pathogenicity to commensality. Such lack or correlation may reflect the fact that some extant pathovars only exhibit opportunistic pathogenic behavior in response to environmental triggers but usually coexist peaceably in the intestine with other commensals. When transferred to another environment, a radically different behavior caused by differential gene expression can be observed (22). The E. coli uropathogen colonization of the bladder and kidneys is a classic example of this triggered behavior.
Phenetic Phylogeny vs. Evolutionary Phylogeny.
We interpret the tree of Fig. 1A as representing the phylogeny of whole-genome similarities, thus similarities of extant organisms (phenetic phylogeny). The genome of an organism in a given environment is likely to be the result of the lineal evolution modulated by convergent and divergent chromosomal changes to adapt to each niche, e.g., converging via mobile element–assisted and other processes to suit specific niche environments. The tree of Fig. 1B, on the other hand, is interpreted as representing lineal evolution of the organisms (evolutionary phylogeny) under the model of evolution that evolutionary footprints can be traced among the common core features. For the practical utility of grouping extant organisms, phenetic phylogeny may be useful, but to trace back to their ancestors, an evolutionary phylogeny may be more revealing. In both FFP trees, the phylogroups are largely consistent, except for the Shigella and B1 groups. Shigella forms a single clade according to whole-genome features in Fig. 1A, but S. dysenteriae separates out from the rest in evolutionary tree of Fig. 1B, suggesting that it evolved from a different lineage (related to subgroup E) then converged to appear phenetically similar to the rest of Shigella. On the other hand, subgroup B1 is monophyletic in Fig. 1B but paraphyletic in Fig. 1A, suggesting that it diverged into two separate phenetic subgroups, EHEC B1e and commensal B1c subgroups.
Distinguishing Features (DFs) and Characteristic Genes of Phylogroups.
Table 2 shows the number of DFs for each phylogroup as well as those that are common in all phylogroups; they are shown in Venn diagram form in Fig. S1. The regions of the diagram represent features that are present in all members of each indicated set of phylogroups. For example, there are 2,889 features that distinguish phylogroup A, i.e., they are present in all members of phylogroup A but they do not appear in any other phylogroup, and the intersection of all phylogroup sets contains 1,680,010 features that occur at least once in all 36 genomes of the E. coli/Shigella group. The numbers of DFs vary from about a few thousand (B1, Shigella, and A phylogroups) to tens of thousands (D, B2, and E phylogroups) but may change as the whole-genome sequences of more members are determined. Orthogonal sampling of a subset of DFs from each phylogroup can be used as a marker probe for identification of groups.
Table 2.
DF counts
| Phylogroup set symbol | Feature count |
| U = Universal set | 8,390,656 |
| V = A ∪ B1 ∪ B2 ∪ D ∪ E ∪ A | 8,224,172 |
| V′ | 166,484 |
| A | 2,727,759 |
| B1 | 2,848,252 |
| B2 | 2,691,012 |
| D | 2,742,919 |
| E | 3,505,791 |
| S | 2,153,098 |
The set notation corresponds to the features in each phylogroup. ∪ is the union, and V′ represents the complement of V, i.e., those features not present in V but part of U.
For genomes of each phylogroup, the distribution of the DFs within coding sequences was also analyzed: Whenever a DF falls within a coding sequence boundary, this gene receives a “hit.” The coding regions with high hits are referred to as “characteristic genes.” Below we discuss a selection of the characteristic genes and their relevance to the known phenotypic characteristics of Shigella, groups B1 and D, where the phylogrouping is most different between our work and those of Touchon et al. (9) and Ogura et al. (10). A full list is provided as Dataset S1. Note that the genes identified through this process may not be specific to a particular phylogroup—we only indicate that they have distinguishing “featural” components. In fact, some genes are conserved in the whole subgroup, but a particular sequence can have a specific pattern of transversion mutations making it phylogenically distinct. General trends to note are that most groups contain characteristic fimbrial proteins or fimbrial ushers, some groups share a common PAI, and some groups share specific variants of common housekeeping genes.
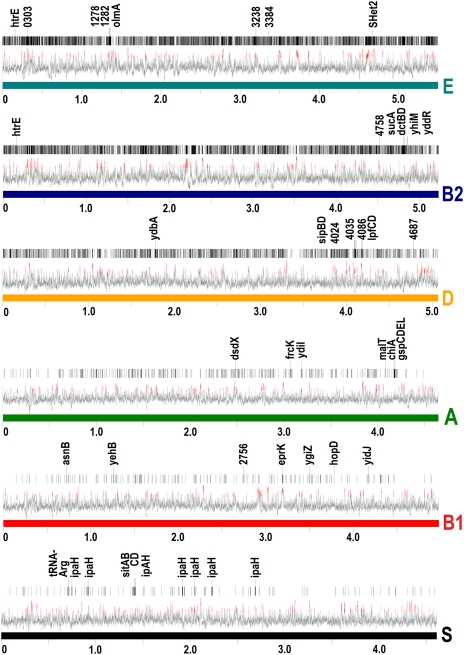
The DFs of each phylogroup were tabulated and mapped to regions of the genomic sequences of a representative member of each phylogroup (Fig. 2). Table S2 summarizes the results for three phylogroups, where the top 8–10 genes are listed sorted by the number of feature hits within the coding sequence boundaries of all of the genomes in that particular phylogroup. The percentage of group-specific feature hits located within an annotated gene coding region (by phylogroup) are: A,72%; B1, 69%; B2, 75%; S, 65%; D, 78%; and E, 73%. The remaining percentage of feature hits are located in intergenic (noncoding) regions. The numbers of DFs and shared features are depicted as a six-set Edwards’ Venn diagram (23) in Fig. S1. Table S1 describes feature counts in each region of the set diagram. A complete tabular listing of DFs by phylogroup, along with mapped genomic locations, is provided as Dataset S2.
Fig. 2.
Genomic distribution of DFs in representatives of each phylogroup. Vertical bars represent the locations of DFs within each genome. Below this is a moving average line graph of GC content; deviations greater than 2 SDs from the mean are colored in red. Characteristic genes are indicated above the features, where labels are either gene abbreviations or loci numbers. Representative strains are: K12W3110 (A), E24377A (B1), CFT083 (B2), SMS-3-5 (D), Sakai (E), and 2Fa301 (S). Genome locations are indicated in millions of base pairs.
Discussion
Divergence Order and Commensal Minimalism by Reductive Genome Evolution.
If we assume that the ancestral E. coli progenote that first infected primates closely resembles the most basal group in our evolutionary phylogeny, a likely conclusion is that this microbe was not a harmless commensal strain. Also, like the UPEC strains, it could have been an opportunistic pathogen, i.e., able to effectively switch pathogenesis on and off in response to environmental conditions. Interesting speculation has been made about the avian (APEC) origin of UPEC strains themselves: Ron (24) has proposed that UPEC strains are derived from the ingestion of APEC-contaminated poultry. APEC01 is a member of the basal B2 clade in our trees. (The reason that the APEC01 isolate does not occupy the most basal position within B2 may be because of the contribution of virulence factors.) Perhaps the connection is more significant and reflects some clues about the avian dietary habits of early anthropoid primates.
A more recently evolved phylogroup is A, which contains, among others, the commensal K12 laboratory workhorse strains. Fig. 2 shows that phylogroup A also has relatively few DFs because this group has few accessory genes, and the common core genes are shared by all other phylogroups. It appears that the K12 strains are examples of “commensal minimalism,” which occurs through the shedding of unnecessary pathogenic and related genes as the organism adapts to the host. This reductive strategy contrasts with another form of genome reduction referred to as “pathogenic minimalism” (25), which characterizes the loss of unnecessary metabolic genes in pathogenic strains. A general trend can be observed: the later evolving phylogroups have progressively smaller genomes. The simplest explanation for this observation is that the A archetype is a result of reductive evolutionary processes. The obligate commensal and obligate pathogens may be a new adaptation. Perhaps commensal strains have lost the capacity to “turn on” pathogenesis and obligate pathogens have lost the ability to turn it “off.”
Shigella Phylogroup.
The Shigella group has been shown in earlier alignment-based studies to be polyphyletic. This finding agrees, in part, with our results using FFPs derived from core features (i.e., universally common features with low frequency and variability) in that the S. dysenteriae genome appears as an outlier strain. Historically, S. dysenteriae was discovered soon after E. coli and, because of its human pathogenesis, put in a separate genus. S. dysenteriae more closely resembles the strains of group E, and, indeed, this group has many Shigella-like characters. Like Shigella, they can be lactose negative, nonmotile, indole producing at low level, and/or may not produce gas during fermentation; additionally, their invasive properties may be attributable to the presence of a plasmid similar to those of Shigella. On the other hand, in our study using phenetic phylogeny, all Shigella form a monophyletic subgroup, and the convergence signal is especially strong; therefore, the clading using this method is quite robust. This monophyly is also observed by Ogura et al. (10) when gene composition was used to build their tree, suggesting that it is caused by convergent gene loss, i.e., microbial parasitic minimalism. As Shigella-like species adapted to infect large intestinal epithelial mucosa, unnecessary accessory metabolic genes, e.g., lactose metabolism, may have generally been discarded.
In Shigella, the top characteristic genes ranked by DF hits are associated with either known PAIs, such as the invasion plasmid antigen (e.g., ipaH), or phage components, especially tail components, insertion sequences, and iron acquisition proteins associated with the sit gene cluster. Shigella are enteroinvasive, therefore the sit siderophore genes are necessary for colonization of iron-poor tissues of the host. Generally, bacteria require cytoplasmic iron concentrations of 10−6 M. However, the iron concentrations in mammalian hosts are very low (for example 10−25 M in the blood) (26). Consequently, pathogenic bacteria must scavenge for free iron by secreting siderophores that have a high affinity for insoluble ferric iron and then retrieving the complexes via specialized transport systems. Pupo et al. (12) proposed that all of the above Shigella genes have been horizontally acquired, perhaps via a single common virulence plasmid, and subsequent incorporation into the genome has occurred. DNA probes designed to match regions of ipaH can be used to characterize Shigella infections (27). Another possible probe target is a specific variant of tRNA-Arg, which is encoded within a prophage region of all strains.
Our results indicate that Shigella flexneri and Shigella boydii share a common ancestor, whereas S. dysenteriae is more distantly related and has a closer association with phylogroup E. The commonalities (in terms of DFs) in the Shigella genus are overwhelmingly dominated by features from elements that are known or strongly suspected of horizontal transfer.
B1 Phylogroup.
The B1 phylogroup is monophyletic in the evolutionary phylogeny but splits into a pathogenic group (B1e) and a commensal group (B1c) in the phenetic phylogeny. The B1e group of strains has been referred to as non O1:H157 EHEC because the pathogenesis is similar to phylogroup E. This association is likely a result of group E PAI transfers. This split has also been observed when gene composition was used for phylogrouping (10). All members of the B1 group share a near-identical variant of the plasmid maintenance proteins mvpA and mvpT, which are absent in all other phylogroups. These sequence are plasmid derived and likely mobile. Other variants of mvpA and mvpT are observed in Shigella and phylogroup E plasmids, but there is little similarity at the nucleotide level.
There are several genes that are highly conserved in this group even at the nucleotide level, although homologs (at the amino acid level) exist in other phylogroups, suggesting that these genes are acquired by the B1 group recently. A type III secretion system protein lipoprotein, eprK, is highly conserved (99–100% identical at the nucleotide level) in this group. This gene has been characterized as part of the ETT2 PAI by Prager et al. (28). This PAI is widely distributed in E. coli, in various stages of degradation. The gene gspO (hopD) is a proposed methylase/prepilin leader peptidase that is implicated in the secretion of pili structures via the general secretion system. Homologs exist in Shigella, B1, and E. Asparagine synthetase B (asnB) is extremely well conserved at the nucleotide level; however, homologs exist in all phylogroups with the closest in phylogroup A. Ornithine decarboxylase/putrescine transporter speF, as well as other arginine synthesis/metabolism–related genes, could be implicated in substrate-level ATP synthesis (glycolysis) under anaerobic conditions. Related arginine synthesis genes, in UPEC (B2) strains, are suspected to be involved in glycolysis as well as pH homoeostasis (29). Close homologs of speF exists in all phylogroups (the closest in S. sonnei); however, changes on the nucleotide level are very specific to the B1 group. Three other proteins of unknown function are specific to B1: transcriptional regulation protein (EC55989_2758); yidJ, a putative sulfatase; and ygiZ, a conserved inner membrane protein. A putative fimbrial usher protein gene, yehB, is specific to this group.
D Phylogroup.
Group D is the least-sampled phylogroup, containing only three representatives. In the phylogeny from Touchon et al. (9), the two group D genomes do not form a clade; however, our results indicate a monophyletic clade. This phylogroup shares more than 10,000 DFs; some are likely associated with PAIs. However, the features are distributed fairly evenly across the entire genome length (Fig. 2), which is highly indicative of common ancestry. The most prominent characteristic gene region is a putative invasin/intimin homolog. In EHEC species of E. coli, these proteins are involved in mechanisms for attaching and effacing host cells. The invasion variant present in this group differs substantially from other phylogroups primarily in the C-terminal region of the protein. Intimin, SipB, SipD, and ydbA are all involved in the type III secretion system, whereby the EHEC species attach and secrete proteins necessary for infection directly into the host cell via a needle-like appendage (sipD). Invasin/intimin and the various type III secretion system proteins are known to be encoded in other phylogroups on the LEE PAI, which inserts itself into pheU-tRNA (30). Two variants of the fimbrial usher protein are specific to this group.
Conclusion
Using the FFP method, we are able to examine two aspects of genomic evolution that are highly revealing: the evolution both of core features, which we suggest infers ancestral history of organisms (evolutionary phylogeny), and of the composition of all features, which is likely to reflect phenetic grouping of extant organisms (phenetic phylogeny). We can summarize our findings as follows:
i) Our phenetic phylogeny corresponded well with observed pathogenesis classes, except B1 class.
ii) Comparison of the two phylogenies suggests that Shigella originated from two distinct ancestors to become one phenetically similar group, and B1 group originated from a single ancestor but split into two phenetic groups.
iii) The basal group of E. coli/Shigella is B2 phylogroup, suggesting that the progenote of the group may have been facultative/opportunistic pathogenic organisms. Under certain environmental conditions, these strains were harmless, in others, they were pathogenic.
iv) Commensal phylogroup A is a more recently evolved branch, probably by reductive evolution of their genomes.
v) The FFP method identifies short distinguishing oligonucleotide features (24 nt, in this case) for each phylogroup. Selection of a set of orthogonal features for each phylogroup can be used to design a probe set for diagnostic characterization of subgroups of E. coli/Shigella.
Materials and Methods
Sequence Data.
The genome sequences were obtained from the National Center for Biotechnology Information ftp site. We used only the main chromosome and did not include any plasmids. Each of the genomes was translated into an RY (purine/pyrimidine)–coded form, which has been demonstrated in several applications (e.g., ref. 31) to improve nucleotide sequence comparison by reducing base composition bias, as well as to reduce the overall feature space and thus reduce computing time. Note that when we count frequencies of features in RY code, features coded in the forward direction are equivalent to those coded in the reverse complement direction.
FFP Method.
In this method, a whole genome is represented by a profile of the frequencies of all features (oligonucleotides) of an optimal length. The FFPs can be used to derive pair-wise similarity/distance information and, in turn, used to derive phylogenies. The details of the FFP method have been published (3, 32). In brief, we count the number of features of a particular length l that occur in a particular genome and construct an FFP for each genome. The FFP of each species is then compared using the Jensen–Shannon divergence measure. The divergences are assembled into a distance matrix, and then a tree is inferred by using the neighbor-joining method (Fig. 1A) as implemented in the Phylip package (33).
Tree Construction: Phenetic and Evolutionary Phylogeny.
Two methods of tree construction were used: (i) Phenetic phylogeny, which is based on FFPs of the composition of all possible features, thus reflecting phenetic aspect of phylogeny, and (ii) evolutionary phylogeny, which is based on FFPs of core feature counts as multistate unordered characters (see below), thus reflecting ancestor-to-descendant phylogenic relationships.
Phenetic phylogeny.
In this application of the FFP method, we used features (l-mers) of length l = 24, and we used the composition of all features without any filtering. The proper feature length is established by using the criterion of topological convergence. As l is increased, tree topologies converge to a single stable topology; at l=23 and l=22, the tree topologies are the same as at l = 24. A length of l = 24 yields 8,224,172 (out of a maximum of 8,390,656) features, all of which appear in at least 1 of the 38 taxa. The Jensen–Shannon divergences (20) were used for distance calculation between a pair of FFPs (Fig. 1A). To determine the robustness of the tree topologies, we sampled the feature space via a 10% jackknife procedure (i.e., sampling without replacement where the probability of individual feature sampling is 10%). One hundred pseudoreplicates were sampled, and a majority consensus tree was constructed with the Phylip CONSENSE utility.
Evolutionary phylogeny.
Because it is well known that mobile elements can affect the tree topologies, we used a modification of the FFP method designed to extract the core features, which are less prone to lateral transfer. We used a set of features composed of l-mers of length 24 and extracted only those features that were present in all taxa, which reduced the set of features to 819,528. The distribution of low-valued feature frequencies (among all 38 taxa, for the full feature set) follows a Gumbel-type extreme-value distribution, which is characterized by two parameters: μ = 0.023 and β = 0.981. Using the extreme-value cumulative-distribution function, 95% of all feature frequencies (P[X<=3] = 0.95) should occur three times or less; however, a number of features occur with very high frequencies. We removed all features that occur more than three times in any of the taxa from the FFP matrix, leaving 564,403 features. Despite this reduction, these features cover between 74% (E. coli 11368 strain) and 88% (Salmonella enterica) of the genomes. In this treatment, we view the numerical count in the FFP as a feature state (or character state). Each of these feature states is unordered, meaning that there is no arithmetic relationship between feature counts (i.e., state “1” is no closer to “2” than it is to “3”). See Wilkinson (21) for a review of these kinds of characters. FFPs for different species are compared by calculating a simple cumulative distance among all features states. Different states add 1 to the distance, whereas identical states add nothing. Therefore, genomic distance in this case represents the total number of feature state differences, not feature frequency differences. The resulting distance matrix was then used as input to the neighbor-joining tree algorithm (Fig. 1B).
Phylogroup DFs.
Phylogroup DFs were identified from the unfiltered l = 24 FFP matrix. DFs are those features that appear in all members of one phylogroup/clade but no other E. coli/Shigella phylogroups (note the outgroup taxa Sa. enterica and E. fergusonii are excluded in all cases). These are tabulated in Table 2 and correspond to the labeled regions in Fig. S1. We identified the DFs for all six phylogroups of the E. coli/Shigella subgroup. The distributions of DFs are represented in featural density diagrams in Fig. 2 for representative examples of three phylogroups. Next, we tabulated the number of times a DF is located within a specific annotated genomic region. When a feature lies within a particular genomic region, this is considered a hit. The genomic regions were ranked by total feature hits and are displayed in Table S2. The locations of these characteristic gene regions are also annotated in Fig. 2.
Supplementary Material
Acknowledgments
We thank Dr. Se-Ran Jun for discussion on her similar studies using proteome sequences, and Seong-Yong You for his help with data classification. This work was supported by World Class University Project Grant R31-2008-000-10086-0 from the Korean Ministry of Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105168108/-/DCSupplemental.
References
- 1.Brenner DJ. In: Bergey's Manual of Systematic Bacteriology. Krieg NR, Holt JG, editors. Vol. 1. Baltimore: Williams and Wilkins; 1984. pp. 408–420. [Google Scholar]
- 2.Bardhan P, Faruque ASG, Naheed A, Sack DA. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg Infect Dis. 2010;16:1718–1723. doi: 10.3201/eid1611.090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sims GE, Jun SR, Wu GA, Kim SH. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proc Natl Acad Sci USA. 2009a;106:2677–2682. doi: 10.1073/pnas.0813249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 5.Milkman R, Bridges MM. Molecular evolution of the Escherichia coli chromosome. IV. Sequence comparisons. Genetics. 1993;133:455–468. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang FS, Whittam TS, Selander RK. Evolutionary genetics of the isocitrate dehydrogenase gene (icd) in Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:6551–6559. doi: 10.1128/jb.179.21.6551-6559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escobar-Páramo P, Giudicelli C, Parsot C, Denamur E. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J Mol Evol. 2003;57:140–148. doi: 10.1007/s00239-003-2460-3. [DOI] [PubMed] [Google Scholar]
- 9.Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogura Y, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan MM, et al. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun. 2001;69:3271–3285. doi: 10.1128/IAI.69.5.3271-3285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, et al. Revisiting the molecular evolutionary history of Shigella spp. J Mol Evol. 2007;64:71–79. doi: 10.1007/s00239-006-0052-8. [DOI] [PubMed] [Google Scholar]
- 14.Haggerty LS, Martin FJ, Fitzpatrick DA, McInerney JO. Gene and genome trees conflict at many levels. Philos Trans R Soc Lond B Biol Sci. 2009;364:2209–2219. doi: 10.1098/rstb.2009.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolland K, Lambert-Zechovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology. 1998;144:2667–2672. doi: 10.1099/00221287-144-9-2667. [DOI] [PubMed] [Google Scholar]
- 16.Guttman DS, Dykhuizen DE. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 20.Lin J. Divergence measures based on the Shannon entropy. IEEE Trans Inf Theory. 1991;37:145–151. [Google Scholar]
- 21.Wilkinson M. Ordered versus unordered characters. Cladistics. 1992;8:375–385. doi: 10.1111/j.1096-0031.1992.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 22.Manning SD, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci USA. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards AWF. Cogwheels of the Mind: The Story of Venn diagrams. Baltimore, London: Johns Hopkins University Press; 2004. [Google Scholar]
- 24.Ron EZ. Host specificity of septicemic Escherichia coli: Human and avian pathogens. Curr Opin Microbiol. 2006;9:28–32. doi: 10.1016/j.mib.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 26.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 27.Venkatesan M, Buysee JM, Kopecko DJ. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli. J Clin Microbiol. 1989;27:2687–2691. doi: 10.1128/jcm.27.12.2687-2691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prager R, et al. Prevalence and deletion types of the pathogenicity island ETT2 among Escherichia coli strains from oedema disease and colibacillosis in pigs. Vet Microbiol. 2004;99:287–294. doi: 10.1016/j.vetmic.2004.01.011. 287–294. [DOI] [PubMed] [Google Scholar]
- 29.Brzuszkiewicz E, et al. How to become a uropathogen: Comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tauschek M, Strugnell RA, Robins-Browne RM. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol Microbiol. 2002;44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- 31.Prasad AB, Allard MW, Green ED. NISC Comparative Sequencing Program Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sims GE, Jun SR, Wu GA, Kim SH. Whole-genome phylogeny of mammals: Evolutionary information in genic and nongenic regions. Proc Natl Acad Sci USA. 2009b;106:17077–17082. doi: 10.1073/pnas.0909377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.