A common signaling mechanism of cell–cell and cell–environment communications in both animals and plants is mediated by receptor-like kinases (RLKs), which evolved independently in the two kingdoms but share a similar domain organization with a ligand-binding extracellular domain (ECD) connected via a single transmembrane helix to an intracellular kinase domain (KD) (1). The plant RLKs form a huge monophyletic protein superfamily with ∼440 and 790 members in Arabidopsis and rice, respectively (2). Based on sequence motifs in their ECDs, plant RLKs can be grouped into ∼20 families, with the largest family containing 1–32 tandem copies of leucine-rich repeat (LRR) (2), a widespread structural motif of ∼24 amino acids rich in leucine. These LRR-RLKs can be functionally classified into two major groups: the first group controls plant growth/development, such as BRI1, which perceives the plant steroid hormone brassinosteroids (BRs) (3), and the second group is involved in plant defense, including FLS2 and EFR, which recognize bacterial flagellin and translational elongation factor EF-Tu, respectively (4, 5). It is well known that ligand-induced homodimerization is a common mechanism to activate receptor kinases in animals (6). By contrast, plant LRR-RLKs, which likely exist as constitutive homodimers, are thought to be activated by ligand-induced heteromerization between a ligand-bound LRR-RLK and a non–ligand-binding LRR-RLK (7). Little is known about how ligands trigger the formation of such receptor–coreceptor complexes, however. In PNAS, Jaillais et al. (8) at the Salk Institute report a significant discovery that provides a key advance in our understanding of the ligand-induced formation of receptor–coreceptor complexes.
Among hundreds of plant LRR-RLKs, BAK1, having only five extracellular LRRs (eLRRs), is one of the most studied LRR-RLKs because of its multifunctionality in regulating both plant growth and defense (9). BAK1 was initially discovered as a coreceptor for the BR receptor BRI1 carrying 25 eLRRs (10, 11) and was later recognized as a coreceptor for the flagellin receptor FLS2 with 28 eLRRs (12, 13). Further studies revealed that BAK1 is also required for plant defense responses to other microbe-derived ligands [better known as microbe-associated molecular patterns (MAMPs)], including bacterial peptidoglycan and EF-Tu (12–14). BAK1 does not directly participate in ligand binding or signal transduction but is rapidly recruited to ligand-bound BRI1, FLS2, and possibly other MAMP-recognition LRR-RLKs to activate their kinase/signaling activities fully via transphosphorylation (15, 16). The BRI1/FLS2-BAK1 pairs have become paradigms for understanding the activation/signaling mechanisms of plant LRR-RLKs; however, little is known about what determines the binding specificity of the BRI1/FLS2–BAK1 complexes.
The study by Jaillais et al. (8) took advantage of a previously described gain-of-function allele of BAK1 (bak1elg) (17) to reveal a crucial role of eLRRs in determining the binding specificity and driving the receptor-coreceptor interaction. The Arabidopsis elg (elongated) mutant was originally isolated as a suppressor of a dwarf mutant deficient in the plant growth hormone gibberellins (18) and was later found to carry an Asp(D)122-Asn(N) mutation in the third LRR of BAK1 responsible for a BR-hypersensitive phenotype (17). D122 is highly conserved between BAK1 and its homologs and is predicted to be a solvent-exposed residue on the concave surface of a curved solenoid LRR structure (19), which provides a surface for binding ligand/protein (20). Using a transgenic approach, Jaillais et al. (8) confirmed the stimulatory effect of bak1elg on BR signaling and made a surprising discovery that the D122N mutation selectively affected several BAK1-dependent immune responses, with bak1elg blocking the plant immunity to peptidoglycan and flg22 (an active flagellin-derived peptide) but having no effect on the EF-Tu–triggered plant defense. Such differential behaviors of bak1elg on BR signaling/plant immunity were not caused by changes in the protein abundance or subcellular localization of BAK1, BRI1 or FLS2 but rather by altered affinity of bak1elg to bind different LRR-RLKs. A coimmunoprecipitation experiment showed that bak1elg failed to bind FLS2 in response to flg22 but interacted well with BRI1 even when the endogenous BR contents were below the level needed to maintain a detectable BRI1 binding to wild-type BAK1 (8).
That a single amino acid change in an LRR motif prevented the recruitment of BAK1 to the flg22-bound FLS2 but enhanced the BRI1-BAK1 binding was a very significant discovery because it convincingly demonstrated a crucial role of eLRRs in selecting binding partners and in driving the formation of ligand-induced LRR–RLK complexes in plants. Previous studies suggested that BR-induced BRI1-BAK1 interaction was largely mediated by their KDs, which were known to interact in vitro and in yeast (10, 11, 15), whereas a recent model suggesting the ECD involvement in the FLS2-BAK1 binding lacked any experimental support (19). The selective binding of BAK1 to different LRR-RLKs is consistent with a recent yeast two-hybrid study showing that BAK1 interacted with the LRR-containing ECD of a tomato LRR receptor-like protein, LeEix1, but failed to bind that of its closest homolog, LeEix2 (21). It is quite possible that ligand binding to an LRR-containing ECD alters its conformation, exposing a binding surface for the BAK1 LRR domain to drive the formation of a receptor–coreceptor complex. Such an induced exposure of an extracellular dimerization interface is a common mechanism for ligand-triggered dimerization of receptor kinases in animals (6). For example, the EGF-induced dimerization of EGF receptor (EGFR) is mediated by homophilic interaction of a protruding loop in the structurally rearranged ECDs of two ligand-bound EGFRs (22).
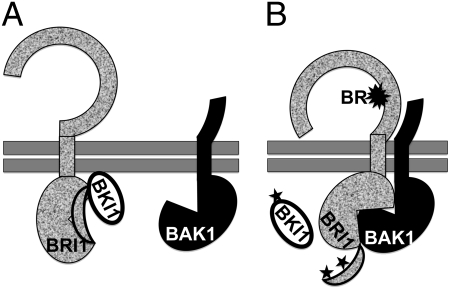
Is the LRR-mediated interaction between two LRR-RLKs sufficient to drive the formation of a stable receptor–coreceptor complex? The answer is “no” in the case of the BRI1–BAK1 complex. It was known that kinase-dead mutations had no effect on the BRI1–BAK1 interaction when coexpressed in yeast (11); however, a recent study showed that a kinase-dead mutation of BRI1 but not BAK1 inhibited the BR-dependent BRI1–BAK1 binding in transgenic plants (15). The requirement for an active BRI1 kinase to form a stable BRI1–BAK1 complex is most likely attributable to an autoinhibitory C terminus and strong binding of BRI1 with BRI1 KINASE INHIBITOR1 (BKI1) (Fig. 1A). BKI1 is a membrane-associated protein capable of blocking the in vitro BRI1-BAK1 binding and rapidly dissociates from the plasma membrane in vivo upon phosphorylation by the BR binding-activated BRI1 (23, 24) that also autophosphorylates to release the autoinhibitory C terminus (25). Therefore, the formation of a BR-induced BRI1–BAK1 complex likely involves a “double-lock” mechanism that requires participation of both ECDs and KDs of BRI1 and BAK1 (8). BR binding not only alters the conformation of the BRI1 ECD to expose a dimerization interface for low-affinity binding to the BAK1 LRR domain but rearranges the BRI1 KD structure to activate its kinase activity, causing release of the autoinhibitory BRI1
Fig. 1.
A “double-lock” model for stabilizing a BR-triggered BRI1–BAK1 complex. Details are provided in the text. Double horizontal bars indicate the plasma membrane, the crescents mark the autoinhibitory C-terminal end, and the stars mark phosphorylation. (A) In the absence of BR, neither BRI1 nor BAK1 is active. The autoinhibitory C terminus of BRI1 and BKI1 binding to BRI1 prevents the BRI1–BAK1 interaction. (B) BR binding to the ligand-binding domain of BRI1 not only triggers a conformational change in its ECD to allow its low-affinity dimerization with the BAK1 ECD but also causes a structural rearrangement in the BRI1 KD to activate its kinase activity. The slightly activated BRI1 autophosphorylates to lease its autoinhibitory C terminus and transphosphorylates to dissociate BKI1 from the plasma membrane, thus enabling physical docking of the KDs of BRI1 and BAK1 to form a stable receptor–coreceptor complex.
The formation of a BR-induced BRI1–BAK1 complex likely involves a “double-lock” mechanism.
C terminus and rapid dissociation of BKI1 to form a stable BRI1–BAK1 complex (Fig. 1B). Such a double-lock mechanism likely provides binding flexibility to allow the versatile BAK1 to be recruited to different ligand-binding LRR-RLKs yet enables the formation of a stable receptor–coreceptor complex via physical docking of their KDs to ensure efficient transphosphorylation between BAK1 and its legitimate ligand-bound partners. In addition, this dimerization model allows the formation of a receptor–coreceptor complex to be regulated by both extracellular and intracellular signals.
It remains to be determined whether the double-lock model also applies to the MAMP-triggered formation of pattern-recognition receptor (PRR)–BAK1 complexes. A recent study suggested that the flg22-triggered FLS2-BAK1 binding does not require a phosphorylation-dependent process (16). Nevertheless, it is quite possible that ligand-induced conformational change in the FLS2 KD is sufficient to enable its physical docking to the BAK1 KD and subsequent FLS2-BAK1 transphosphorylation, ensuring the full activation of the FLS2 signaling potential. It is interesting to note that although the formation of EGF-induced EGFR dimer does not require a phosphorylation event, it does involve an EGF-triggered structural rearrangement in the EGFR KD, exposing a juxtamembrane segment that latches the cytoplasmic KDs of dimerizing EGFRs (6).
Given the multifunctionality of BAK1, it is anticipated that the BRI1/FLS2–BAK1 complexes will continue to be paradigms in the coming years to study plant LRR-RLKs and cross-talk mechanisms coordinating plant growth and defense. Site-directed mutagenesis; chemical cross-linking; and structural analyses of BAK1, BRI1, and FLS2, coupled with in vivo kinetic studies of LRR-RLK heterooligomerization, will surely shed new light on the biochemical mechanisms of the ligand-induced formation of plant receptor–coreceptor complexes.
Acknowledgments
Work on BR signaling in my laboratory is supported by National Institutes of Health Grant GM060519.
Footnotes
The author declares no conflict of interest.
See companion article on page 8503.
References
- 1.Cock JM, Vanoosthuyse V, Gaude T. Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr Opin Cell Biol. 2002;14:230–236. doi: 10.1016/s0955-0674(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 2.Lehti-Shiu MD, Zou C, Hanada K, Shiu SH. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009;150:12–26. doi: 10.1104/pp.108.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 5.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torii KU. Transmembrane receptors in plants: Receptor kinases and their ligands. Annu Plant Reviews. 2008;33:1–29. [Google Scholar]
- 8.Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol. 2010;13:509–514. doi: 10.1016/j.pbi.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 11.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 12.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 13.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Schulze B, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whippo CW, Hangarter RP. A brassinosteroid-hypersensitive mutant of BAK1 indicates that a convergence of photomorphogenic and hormonal signaling modulates phototropism. Plant Physiol. 2005;139:448–457. doi: 10.1104/pp.105.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halliday K, Devlin PF, Whitelam GC, Hanhart C, Koornneef M. The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J. 1996;9:305–312. doi: 10.1046/j.1365-313x.1996.09030305.x. [DOI] [PubMed] [Google Scholar]
- 19.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 20.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar M, Sharfman M, Ron M, Avni A. BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 2010;63:791–800. doi: 10.1111/j.1365-313X.2010.04282.x. [DOI] [PubMed] [Google Scholar]
- 22.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 24.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]