Abstract
The abundance of nuclear plastid DNA-like sequences (NUPTs) in nuclear genomes can vary immensely; however, the forces responsible for this variation are poorly understood. “The limited transfer window hypothesis” predicts that species with only one plastid per cell will have fewer NUPTs than those with many plastids per cell, but a lack of genome sequence data from monoplastidic species has made this hypothesis difficult to test. Here, by analyzing newly available genome sequences from diverse mono- and polyplastidic taxa, we show that the hypothesis holds. On average, the polyplastidic species we studied had 80 times more NUPTs than those that were monoplastidic. Moreover, NUPT content was positively related to nuclear genome size, indicating that in addition to plastid number, NUPTs are influenced by the forces controlling the expansion and contraction of noncoding nuclear DNA. These findings are consistent with data on nuclear DNAs of mitochondrial origin (NUMTs), suggesting that similar processes govern the abundance of both NUPTs and NUMTs.
Keywords: chloroplast, mitochondria, NUPT, NUMT, genome architecture, noncoding DNA
The Limited Transfer Window Hypothesis
The movement of organelle DNA to the nucleus has been, and remains, a driving force in fashioning eukaryotic genomes (Timmis et al. 2004; Kleine et al. 2009). Early on in both mitochondrial and plastid evolution, there was a massive migration of organelle genes to the nuclear genome (Gray et al. 1999; Huang et al. 2003; Kleine et al. 2009); thus, present-day nuclear DNA is a mosaic of endosymbiont-derived organelle genes and “host” genes; and contemporary mitochondrial and plastid DNAs (mtDNAs and ptDNAs) are significantly more reduced than the endosymbiotic genomes from which they evolved (Gray et al. 1999; Archibald 2009). Aside from adding to the gene repertoire of nuclear genomes, organelle-to-nucleus DNA transfer events have generated, and continue to generate, forms of noncoding nuclear DNA (and occasionally exonic nuclear DNA, Noutsos et al. 2007) that share sequence identity with the coexisting organelle DNAs; these types of sequences are referred to as nuclear mitochondrial DNAs (NUMTs) and nuclear plastid DNAs (NUPTs) (Lopez et al. 1994; Richly and Leister 2004a, 2004b).
Although the nuclear genomes from at least 85 eukaryotic species have been analyzed for NUMTs (Hazkani-Covo et al. 2010 and references therein), there are relatively little data on NUPTs. This is because, until recently, there were only a small number of published nuclear genome sequences from plastid-harboring eukaryotes. Nonetheless, an intriguing observation has come from the NUPT data that are available: Species with one plastid per cell (monoplastidic) have fewer NUPTs than those with many plastids per cell (polyplastidic) (Lister et al. 2003; Martin 2003; Richly and Leister 2004b). For example, the monoplastidic protists Chlamydomonas reinhardtii and Plasmodium falciparum have <2.5 kb of NUPTs (Richly and Leister 2004b; Matsuo et al. 2005), whereas rice, which contains upwards of 100 plastids per cell, has around 900 kb of NUPTs (Guo et al. 2008). A possible explanation for these observations is that in monoplastidic species, the transfer of ptDNA to the nucleus is greatly reduced as compared with polyplastidic taxa because 1) there are fewer plastids to donate ptDNA to the nucleus and 2) lysis of the plastid would almost certainly result in death to the cell, unlike the case for polyplastidic species (Lister et al. 2003; Martin 2003; Richly and Leister 2004b). This explanation has become known as the “limited transfer window hypothesis” (Barbrook et al. 2006). When presenting this hypothesis, Barbrook et al. (2006) predicted that “the sequencing of the nuclear genomes of organisms containing a single plastid should always reveal a low abundance of NUPTs.” But a lack of nuclear DNA sequence data from monoplastidic species and from plastid-harboring taxa in general has made this prediction difficult to test.
In this study, we take advantage of newly available genomic sequence data from a series of diverse mono- and polyplastidic species to formally investigate the limited transfer window hypothesis. Altogether, we calculate the number and accumulative length of NUPTs in the nuclear DNAs of 11 polyplastidic and 19 monoplastidic (or effectively monoplastidic) eukaryotes. When possible, we also analyze these same genomes for NUMTs and compare these data with the corresponding NUPT statistics.
Testing the Limited Transfer Window Hypothesis
To assess a genome for NUPTs, at least two things are required: complete nuclear DNA and ptDNA sequence data. We found 30 species for which both these statistics are available, including 13 land plants, 7 green algae, 5 apicomplexans, 3 stramenopiles, 1 haptophyte, and 1 red alga (table 1). The sources for these genome sequence data are listed in supplementary tables S1 and S2 (Supplementary Material online). To the best of our knowledge, detailed NUPT statistics for the majority of the above-mentioned taxa have not been published elsewhere. For 20 of these species, complete mtDNA sequence data are also available, allowing for NUMT as well as NUPT analyses. Although most of these taxa have already been explored for NUMTs (Hazkani-Covo et al. 2010, and references therein), we performed our own NUMT investigations because in the past differences in search parameters among studies have led to discrepancies in NUPT/NUMT tabulations. We did try, however, to use similar search constraints as those employed in previous reports: BlastN with an expectation value of 0.0001. Another source of discrepancy in NUPT/NUMT assessments among earlier studies (Hazkani-Covo et al. 2010) were instances where one segment of nuclear DNA matched to multiple sections of organelle DNA. In our analyses, multiple organelle DNA hits to the same nuclear DNA regions were counted only once. Because many of the nuclear genomes that we scanned are only in their draft assembly stage, the NUPT/NUMT data presented here should be treated as approximations of the true values. As these genome sequences become more polished, the NUPT/NUMT estimates will change, but the major trends that we observed among the different groups should arguably remain the same.
Table 1.
Number and Total Amount (in Kilobases) of NUPTs and NUMTs in the Available Nuclear Genome Sequences from Plastid-Harboring Eukaryotes
| Taxon | Number of Plastids per Cell | Number of Mitochondria per Cell | NUPTs |
NUMTs |
||||
| Number of Blast Hitsa | Accumulative Length (kb) | Average Length (kb) | Number of Blast Hitsa | Accumulative Length (kb) | Average Length (kb) | |||
| Land plants | ||||||||
| Arabidopsis thaliana | Multiple | Multiple | 332 | 50 | 0.15 | 1,173 | 549 | 0.46 |
| Brachypodium distachyon | Multiple | Multiple | 310 | 114 | 0.37 | NA | NA | NA |
| Carica papaya | Multiple | Multiple | 839 | 291 | 0.34 | 1,528 | 467 | 0.32 |
| Cucumis sativus | Multiple | Multiple | 751 | 265 | 0.35 | NA | NA | NA |
| Glycine max | Multiple | Multiple | 3,414 | 822 | 0.24 | NA | NA | NA |
| Medicago truncatula | Multiple | Multiple | 258 | 93.3 | 0.36 | NA | NA | NA |
| Oryza sativa subsp. indica | Multiple | Multiple | 1,541 | 782 | 0.50 | 2,544 | 818 | 0.32 |
| O. sativa subsp. japonica | Multiple | Multiple | 2,036 | 1,073 | 0.52 | 3,072 | 834 | 0.27 |
| Physcomitrella patens | Effectively monoplastidicb | Multiple | 31 | 5 | 0.16 | 294 | 74 | 0.25 |
| Populus trichocarpa | Multiple | Multiple | 2,036 | 428 | 0.30 | NA | NA | NA |
| Selaginella moellendorffii | Effectively monoplastidicb | Multiple | 114 | 11.4 | 0.10 | NA | NA | NA |
| Sorghum bicolor | Multiple | Multiple | 1,574 | 329 | 0.20 | 1,957 | 406 | 0.20 |
| Vitis vinifera | Multiple | Multiple | 3,858 | 801 | 0.20 | 2,357 | 602 | 0.25 |
| Green algae | ||||||||
| Chlamydomonas reinhardtii | Single | Multiple | 35 | 1.9 | 0.05 | 35 | 3.3 | 0.09 |
| Coccomyxa sp. C-169 | Single | Single | 73 | 7.5 | 0.10 | 107 | 12 | 0.11 |
| Ostreococcus sp. RCC809 | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Ostreococcus tauri | Single | Single | 4 | 0.6 | 0.17 | 2 | 0.6 | 0.31 |
| Micromonas pusilla | Single | Single | 3 | 0.5 | 0.16 | 0 | 0 | 0 |
| Micromonas sp. RCC299 | Single | Single | 3 | 0.6 | 0.20 | 0 | 0 | 0 |
| Volvox carteri f. nagariensis | Single | Multiple | 1,100 | 65 | 0.12 | 802 | 33 | 0.09 |
| Red alga | ||||||||
| Cyanidioschyzon merolae | Single | Single | 2 | 0.37 | 0.18 | 0 | 0 | 0 |
| Apicomplexans | ||||||||
| Babesia bovis | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Eimeria tenella | Single | Single | 31 | 2.8 | 0.09 | NA | NA | NA |
| Plasmodium falciparum | Single | Single | 2 | 0.11 | 0.05 | 2 | 0.11 | 0.05 |
| Theileria parva | Single | Single | 0 | 0 | 0 | 0 | 0 | 0 |
| Toxoplasma gondii | Single | Single | 77 | 10.3 | 0.03 | NA | NA | NA |
| Haptophyte | ||||||||
| Emiliania huxleyi | 1–2 | Single | 2 | 0.15 | 0.07 | 2 | 0.1 | 0.05 |
| Stramenopiles | ||||||||
| Aureococcus anophagefferens | Single | Single | 0 | 0 | 0 | NA | NA | NA |
| Phaeodactylum tricornutum | Single | Multiple | 14 | 4 | 0.29 | NA | NA | NA |
| Thalassiosira pseudonana | 1–2 | Multiple | 8 | 1.6 | 0.20 | 0 | 0 | 0 |
NOTE.— NA—data not available (i.e., mitochondrial genome has not been sequenced; thus, we were unable to search the nuclear DNA for NUMTs).
Blast parameters were as follows: BlastN (version 2.2.23) with an expectation value of 0.0001; a word size of 11; match and mismatch scores of 2 and −3, respectively; and gap cost values of 5 (existence) and 2 (extension). Multiple organelle DNA hits to the same nuclear DNA regions were counted only once. Regions of nuclear DNA that contained tight clusters of NUPTs/NUMTs (i.e., sections of organelle-like DNA interrupted by genomic sequence that did not show sequence identity to organelle DNA) were not counted as a single NUPT/NUMT but as separate hits. See supplementary table S3 (Supplementary Material online) for references and notes on the number of organelles per cell.
S. moellendorffii and P. patens both contain cells that are polyplastidic, but for the purpose of this study, they are considered “effectively monoplastidic” because mitosis and meiosis only occurs in cells that contain a single plastid (Brown and Lemmon 1990).
Of the 30 species we investigated, 11 are polyplastidic and 19 are either monoplastidic or effectively so. Thirteen of the monoplastidic species are also monomitochondrial (i.e., they have one mitochondrion per cell). The number of organelles per cell for each species and the references used to determine these statistics are listed in table 1 and supplementary table S3 (Supplementary Material online), respectively. When possible, the decision to categorize a species as having one or multiple organelles per cell was based on published ultrastructural data. Two caveats should be noted: The haptophyte Emiliania huxleyi and the stramenopile Thalassiosira pseudonana can have up to two plastids per cell but, for simplicity, were treated here as monoplastidic (Badger et al. 1998; Dassow et al. 2008, and references therein); and the lycophyte Selaginella moellendorffii and the moss Physcomitrella patens both contain cells that are polyplastidic, but for the purpose of this study, they were considered “effectively monoplastidic” because mitosis and meiosis only occur in cells that contain a single plastid (Brown and Lemmon 1990).
Polyplastidy Means More NUPTs
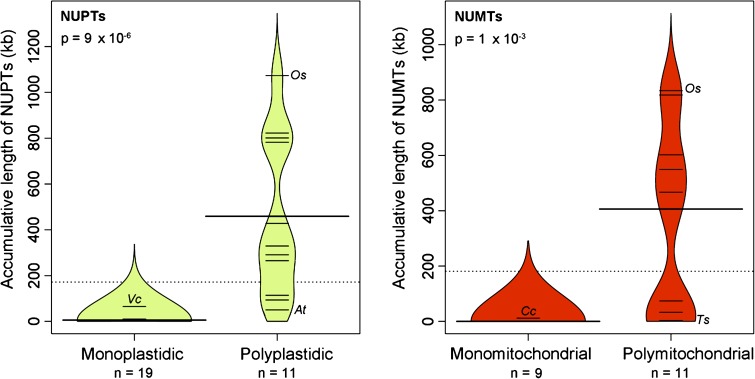
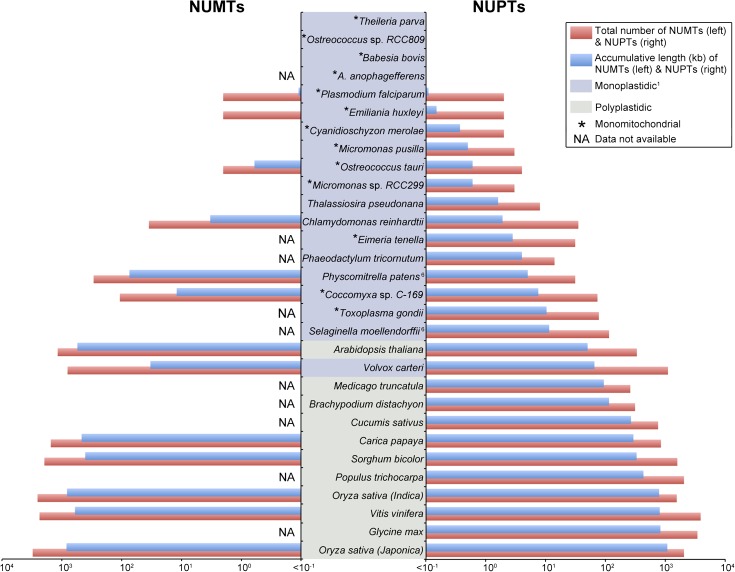
Complete NUPT and, when attainable, NUMT statistics for the various plastid-bearing species that we investigated are shown in table 1. Overall, we found the difference in NUPT content between mono- and polyplastidic species to be highly significant (fig. 1). Species with multiple plastids per cell had on average 80 times more NUPTs than those with one plastid per cell. The mean NUPT content for polyplastidic species was 460 kb as compared with only 6 kb for monoplastidic taxa. Moreover, the average number of NUPTs (based on Blast hits, not accumulative length) for polyplastidic individuals was 20 times greater than that of monoplastidic species (1,540 vs. 79 hits). In species with only a single plastid, the NUPT content ranged from undetectable levels in the protists Aureococcus anophagefferens, Babesia bovis, Ostreococcus sp. RCC809, and Theileria parva to ∼65 kb for the multicellular green alga Volvox carteri, whereas in polyplastidic species, it spanned from 50 kb for Arabidopsis thaliana to >800 kb for Glycine max, Vitis vinifera, and Oryza sativa. There is a clear separation in NUPT content between mono- and polyplastidic species, with the members of the latter group having considerably more NUPTs than the former (figs. 1 and 2). The only exceptions were V. carteri and A. thaliana (fig. 2); the reasons for this may be linked to their capacity (or deficiency) for purging bulk nuclear DNA (discussed further below). It is also noteworthy that of the 13 land plants that were explored the 2 that are effectively monoplastidic (S. moellendorffii and P. patens) had about 60 times fewer NUPTs than those that are polyplastidic (figs. 1 and 2).
FIG. 1.—
“Beanplot” depicting the difference in NUPT/NUMT content between mono- and polyplastidic (and polymitochondrial) species. Plot was generated using the beanplot package (Kampstra 2008) from R v. 2.1.1. The dashed line in the middle of each of the two plots is the overall average of the continuous variable (total NUPT or NUMT content in kilobases). The thick black line in the middle of each category (mono or poly) is the median of each continuous variable (NUPT or NUMT content) with respect to the categorical variable (mono- or polyplastidic/polymitochondrial). The colored curved beanpod surrounding the observations “beans” is the theoretical probability density distribution of these observations. If there were multiple observations with the same number (e.g., NUPTs content of 4 kb for two different taxa), then the line gets longer respective to the other measurements in the beanplot. A Wilcoxon signed rank test (nonparametric) was performed in R on all data because errors were not normally distributed. Note, the P values shown are approximations—the exact values could not be computed due to ties in the data. For polyorganellar species, the lowest and highest data points are named. However, to avoid clutter, only the highest points are labeled for mono-organellar species. Note: At, Arabidopsis thaliana, Cc, Coccomyxa sp. C-169; Os, Oryza sativa; Ts, Thalassiosira pseudonana; and Vc, Volvox carteri.
FIG. 2.—
Log-scale bar graph showing the number and accumulative length of NUPTs (right side) and NUMTs (left side) for various plastid-harboring eukaryotes. Species are ranked in ascending order based on their total NUPT content. See table 1 and supplementary table S3 (Supplementary Material online) for references and notes on the number of organelles per cell. 1Emiliania huxleyi and Thalassiosira pseudonana can have up to two plastids per cell. Selaginella moellendorffii and Physcomitrella patens are effectively monoplastidic—mitosis and meiosis only occurs in cells that contain a single plastid (Brown and Lemmon 1990).
Our NUMT analyses revealed similar trends and conclusions as those described above for NUPTs. Species with only a single mitochondrion per cell had significantly fewer NUMTs than those with many mitochondria per cell (figs. 1 and 2). The average NUMT content for monomitochondrial species (1.1 kb) was ∼300 times less than that of polymitochondrial taxa (380 kb). These data are consistent with earlier observations on NUMTs (Hazkani-Covo et al. 2010) and support the belief that organelle number influences both NUPT and NUMT content.
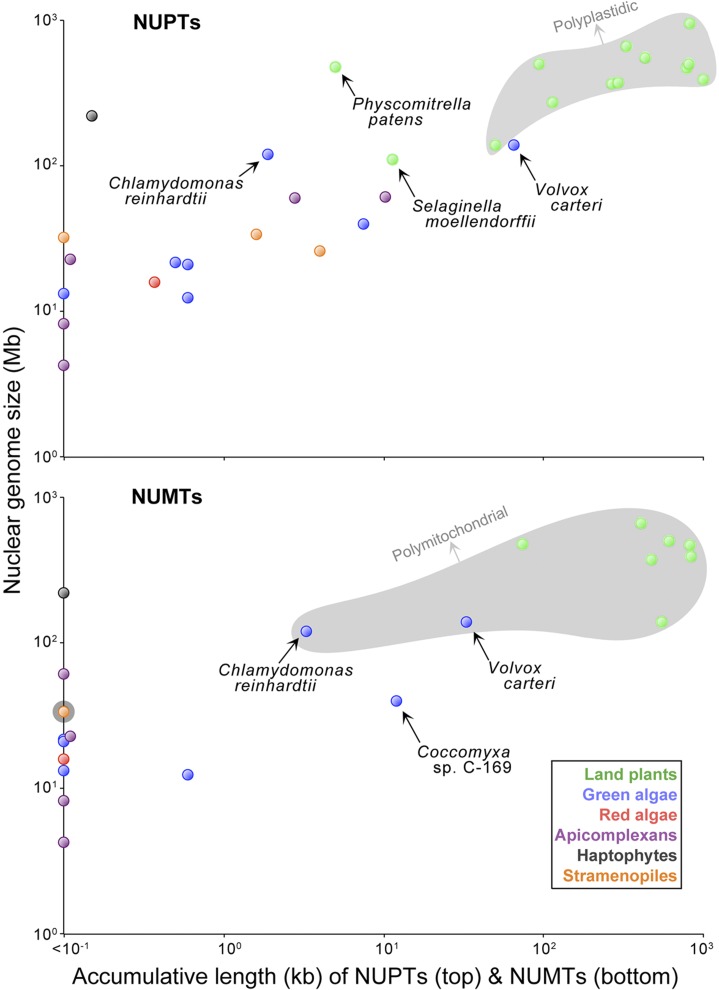
Larger Genomes, Larger NUPT Content
It was recently shown that NUMT content scales positively with nuclear genome size (Hazkani-Covo et al. 2010). Here, we found that this is true for NUPTs as well: Bloated nuclear genomes tend to have more NUPTs than those that are compact (fig. 3)—based on the 30 species investigated here, we found a reasonably strong relationship between nuclear genome size and NUPT content (R2 = 0.57, P = 8.6 × 10−7). We expected to find this relationship because NUPTs are a type of excess DNA, and it is well established that all types of excess DNA mutually expand as the number of nucleotides in a genome increases (Lynch and Conery 2003). Ultimately, this suggests that the forces governing the expansion and contraction of noncoding nuclear DNA impact the accumulative length of NUPTs in a nuclear genome. Although the nature of these forces is hotly debated, there is evidence that the tendency for excess DNA to accumulate depends on the combined effects of the mutation rate (μ) and the effective genetic population size (Ne; Lynch and Conery 2003). According to this hypothesis, one may expect species with a low Neμ to have more NUPTs than those with a high Neμ. Although there are very few reliable data on this fundamental population genetic parameter, a recent study indicates that V. carteri has a very small Neμ, especially relative to other protists (Smith and Lee 2010). This could help explain why of all the monoplastidic species that we studied, V. carteri had the largest NUPT content. Being monoplastidic, one would expect the transfer of ptDNA to the nuclear genome to be rare in V. carteri, but having a low Neμ implies that it has a reduced ability to detect and eradicate excess DNA (Smith and Lee 2010) so that the few NUPTs that do arise avoid deletion and therefore can accumulate to reasonably high levels over time. Interestingly, the Neμ estimates for the nuclear DNA of Arabidopsis spp. are about three times those of V. carteri (Wright et al. 2008); thus, one explanation for why A. thaliana, which is polyplastidic, had fewer NUPTs than the monoplastidic V. carteri could be that it is reasonably efficient at perceiving and purging noncoding nuclear DNA. It is worth mentioning that the V. carteri ptDNA, at ∼525 kb, is the largest plastid genome sequenced to date (Smith and Lee 2010), being >300 kb larger than any other ptDNA employed in our data set. And although we did not find an association between plastid genome size and NUPT content (supplementary fig. S1, Supplementary Material online), there is still the possibility that the prodigious ptDNA of V. carteri is in some way contributing to its elevated NUPT content.
FIG. 3.—
Log-log plot of NUPT (top) and NUMT (bottom) content versus nuclear genome size. Polyplastidic and polymitochondrial species are shaded gray on the top and bottom plots, respectively. The names of species of particular interest are shown on the plot. Nuclear genome size data came from GenBank’s Entrez Eukaryotic Genome Project database (http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi).
The Evolution of NUPTs: It Is a Give and Take Relationship
The data presented here provide support for the limited transfer window hypothesis and the notion that the number of plastids per cell in a eukaryotic species governs the amount of NUPTs found in its nuclear genome. We argue that the evolution of NUPTs is a “give and take” process where plastid number determines the potential for ptDNA to be donated (i.e., given) to the nuclear genome, and the probability that these ptDNA sequences will be accepted (i.e., taken) by the nuclear genome and persist as nuclear DNA is determined by a species’ ability (or lack thereof) to detect and eliminate excess DNA. In this study, being monoplastidic versus polyplastidic was used to define the number of plastids per cell in a species, and nuclear genome size was used as of a proxy for defining a species, ability to eradicate noncoding nuclear DNA. This same argument applies to NUMTs as well.
Again, it must be stressed that many of the nuclear DNA sequences that we used to calculate NUPT/NUMT abundance were in their draft assembly stage. As these genome assemblies improve, their NUPT/NUMT statistics may change, but we believe that the major trends reported here will be borne out by future investigations. Finally, given the wide diversity of eukaryotic species that we explored, there are certainly factors in addition to a limited transfer window and susceptibility to bulk DNA that are influencing NUPT and NUMT content; however, we argue these additional factors (whatever they may be) will turn out to be secondary to the forces outlined here.
Materials and Methods
The sources and references for the nuclear genome sequences employed in this study, as well the GenBank accession numbers, when available, are shown in supplementary table S1 (Supplementary Material online). All nuclear DNA data came from publicly available sources. The organelle DNA sequences (including their lengths, GenBank accession numbers, and noncoding DNA contents) used as queries for BlastN searches against nuclear genomes are listed in supplementary table S2 (Supplementary Material online). Some of the organelle DNA sequences that were used in this study are not deposited in GenBank but are available for download online from the given genome project Web site (Supplementary table S2, Supplementary Material online).
Nuclear genomes were scanned for NUPTs and NUMTs with BlastN (version 2.2.23) (Altschul et al. 1990) using the following parameters: an expectation value of 0.0001; a word size of 11; match and mismatch scores of 2 and −3, respectively; and gap-cost values of 5 (existence) and 2 (extension). Hits under 30 nt and showing <70% sequence identity to the query were ignored. Spurious hits, such as those where organelle genes matched to homologous genes in the nuclear genome (e.g., organelle ribosomal DNA [rDNA] matching to nuclear rDNA) were ignored. Instances where one segment of nuclear DNA matched to multiple sections of organelle DNA (i.e., duplicate Blast hits) were reduced to a single Blast hit; in other words, NUPTs and NUMTs matching to multiple organelle genomic regions, such as repetitive elements, were counted only once. Regions of nuclear DNA that contained tight clusters of NUPTs or NUMTs (i.e., sections of organelle-like DNA interrupted by genomic sequence that did not show sequence identity to organelle DNA) were not counted as a single NUPT or NUMT but as separate hits. Data and sources used to calculate the number of organelles per cell are shown in supplementary table S3 (Supplementary Material online).
Supplementary Material
Supplementary tables S1–S3 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a grant to R.W.L. from the Natural Sciences and Engineering Research Council of Canada.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Badger MR, Andrews TJ, Whitney SM. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO 2-concentrating mechanisms in algae. Can J Bot. 1998;76:1052–1071. [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. Monoplastidic cell division in lower land plants. Am J Bot. 1990;77:559–571. doi: 10.1002/j.1537-2197.1990.tb13588.x. [DOI] [PubMed] [Google Scholar]
- Dassow PV, Petersen TW, Chepurnov VA, Armbrust EV. Inter- and intraspecific relationship between nuclear DNA content and cell size in selected members of the centric diatom genus Thalassiosira (Bacillariophyceae) J Phycol. 2008;44:335–349. doi: 10.1111/j.1529-8817.2008.00476.x. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Guo X, Ruan S, Hu W, Cai D, Fan L. Chloroplast DNA insertions into the nuclear genome of rice: the genes, sites and ages of insertion involved. Funct Integr Genomics. 2008;8:101–108. doi: 10.1007/s10142-007-0067-2. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLos Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Kampstra P. Beanplot: a boxplot alternative for visual comparison of distributions. J Stat Softw. 2008;28:1–9. [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Lister DL, Bateman JM, Purton S, Howe CJ. DNA transfer from chloroplast to nucleus is much rarer in Chlamydomonas than in tobacco. Gene. 2003;316:33–38. doi: 10.1016/s0378-1119(03)00754-6. [DOI] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci U S A. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Ito Y, Yamauchi R, Obokata J. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell. 2005;17:665–675. doi: 10.1105/tpc.104.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutsos C, Kleine T, Armbruster U, DalCorso G, Leister D. Nuclear insertions of organellar DNA can create novel patches of functional exon sequences. Trends Genet. 2007;23:597–601. doi: 10.1016/j.tig.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004a;21:1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- Richly E, Leister D. NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol Biol Evol. 2004b;21:1972–1980. doi: 10.1093/molbev/msh210. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27:2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Wright SI, Nano N, Foxe JP, Dar VU. Effective population size and tests of neutrality at cytoplasmic genes in Arabidopsis. Genet Res. 2008;90:119–128. doi: 10.1017/S0016672307008920. [DOI] [PubMed] [Google Scholar]