Abstract
The principles of self-assembly and self-organization are major tenets of molecular and cellular biology. Governed by these principles, the eukaryotic nucleus is composed of numerous subdomains and compartments, collectively described as nuclear bodies. Emerging evidence reveals that associations within and between various nuclear bodies and genomic loci are dynamic and can change in response to cellular signals. This review will discuss recent progress in our understanding of how nuclear body components come together, what happens when they form, and what benefit these subcellular structures may provide to the tissues or organisms in which they are found.
The spatial arrangement of chromatin within the nuclear volume entails a complex interplay between factors involved in chromosome maintenance and those involved in gene expression. Understanding how genomes actually function in vivo has been termed the “Holy Grail” of genome biology and a logical next step after the sequencing projects (Misteli, 2007). To accomplish this lofty goal, we must learn in detail how the Central Dogma is applied in three dimensions over developmental time. Fundamental to this understanding will be knowledge of the relationship between the chromatin and the interchromatin space, i.e., the genome and its immediate environment.
The cell nucleus is a complex organelle whose dynamic architecture consists of numerous subcellular compartments, collectively referred to as nuclear bodies (Figure 1; Matera, 1999). These structures include nucleoli, Cajal bodies (CBs), histone locus bodies (HLBs), splicing factor compartments (a.k.a. speckles or interchromatin granule clusters), paraspeckles, promyelocytic leukemia (PML) bodies, Gemini bodies (gems), perinucleolar compartments (PNCs), polycomb group (PcG) bodies, heat shock factor 1 (HSF1) foci, SAM-68 bodies, GATA-1 foci, and many more. Important nuclear processes, such as DNA replication and repair (Hozak et al., 1993; Jackson et al., 1994; Lisby et al., 2003; Nakamura et al., 1986; Nakayasu and Berezney, 1989) or RNA transcription and processing (Carmo-Fonseca et al., 1992; Fu and Maniatis, 1990; Jackson et al., 1993; Matera and Ward, 1993; Wansink et al., 1993), are organized in discrete subdomains. One of the emergent principles of nuclear organization is that certain subnuclear domains are associated with specific gene loci. Another important rule is that associations between these subdomains and loci are dynamic and can change in response to cellular signals.
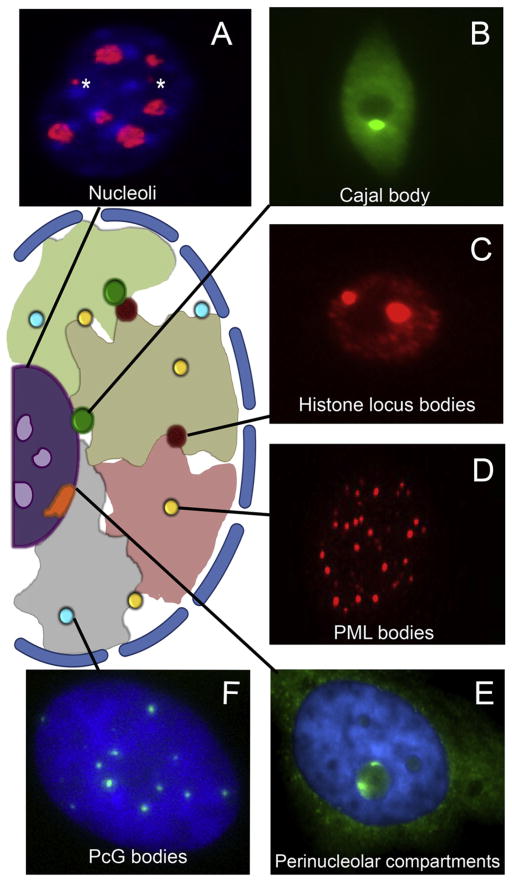
Figure 1. Diversity of Nuclear Bodies.
The cartoon in the center of the figure depicts the nucleus of a higher eukaryote. Interphase chromosomes occupy distinct territories (large irregular shapes). The interchromatin space contains numerous subdomains or bodies (colored dots).
(A) The nucleoli of a mouse embryonic fibroblast were stained with anti-fibrillarin (red) and counter-stained for DNA using DAPI (blue). Note that fibrillarin localizes primarily to nucleoli (large blobs) but is also found in Cajal bodies (asterisks).
(B) Antibodies targeting the U2B″ protein were used to identify the Cajal body (bright dot) in this Arabidopsis nucleus. Note that the nucleolus shows up as a negatively stained region within the nucleoplasmic U2B″ signal.
(C) Anti-FLASH antibodies highlight the two histone locus bodies (bright foci) within this Drosophila S2 cell.
(D) Mammalian nuclei, as illustrated by this mouse NIH 3T3 cell, typically contain 10–30 PML bodies, stained here with anti-PML.
(E) The perinucleolar compartment (PNC) is shown in this human (HeLa) cell hybridized with an oligo-nucleotide probe targeting hY1 RNA (green). The nucleus is counterstained in blue with DAPI. Note that this RNA localizes to the PNC as well as to the cytoplasm.
(F) This human U2OS cell was transfected with YFP-tagged Bmi1, a Polycomb group (PcG) protein that is used as a marker for PcG bodies (green). Counterstaining was performed using Hoechst (blue).
As suggested in the title of this review, nuclear bodies might simply be a reflection of a propensity for certain proteins to form macromolecular aggregates. Indeed, many of the signature proteins of nuclear bodies are known to self-interact (Hebert and Matera, 2000 and references therein). Protein aggregation and misfolding are cardinal features of numerous devastating diseases, including Alzheimer’s, Huntington’s, cystic fibrosis, Creutzfeldt-Jakob syndrome, and type II diabetes. However, overexpression of nuclear body signature proteins does not typically induce the formation of aberrant nuclear foci or result in an increase in the number or the size of their respective nuclear bodies. Nuclear body proteins are not known to be associated with protein folding diseases, and, in fact, there may even be a negative correlation. Comparative genome analyses have shown that natural selection acts against the aggregation of essential or self-interacting proteins (Chen and Dokholyan, 2008). Thus, if nuclear bodies are not simply aggregates of sticky proteins, what functional roles do they play? This review will focus on studies that are beginning to elucidate the molecular mechanisms underlying nuclear body assembly and function, using nucleoli, Cajal bodies, and histone locus bodies as paradigms.
Assembly of Nuclear Bodies
Historically, the term “nuclear bodies” has been reserved for structures that were characterized morphologically in the electron microscope. More recently, however, nuclear foci observed in the light microscope by immunocytochemistry have often been termed “bodies” without prior morphological evidence at the ultrastructural level. Although we are still far from understanding why most nuclear bodies form, recent progress has been made in elucidating how they are assembled in the cell.
Two distinct assembly models have been considered, both of which involve recruitment of individual subunits (or small sub-complexes thereof) from a soluble nucleoplasmic pool (Cook, 2002; Misteli, 2001, 2007). The main difference is that one model holds that the subunits are assembled in an orderly fashion, built around a central scaffolding factor, whereas the other model posits that structures are built up essentially randomly (Figure 2). Using an approach similar to the one taken by the Misteli laboratory for the study of DNA double-strand break repair foci (Soutoglou et al., 2007), Dundr and colleagues showed that essentially any Cajal body protein can nucleate formation of the entire CB structure de novo (Kaiser et al., 2008). By tethering a given CB component to a specific site in the genome using the lac repressor/operator system, the investigators showed that the tethered protein or RNA was able to recruit most, if not all, of the other CB components (Kaiser et al., 2008). The structures formed de novo had similar size to their endogenous counter-parts, and the components of the tethered structures had similar dissociation kinetics to those of endogenous CBs (Dundr et al., 2004; Kaiser et al., 2008). Moreover, the tethered CBs could be disassembled (or reassembled) by interfering with (or restoring) the lac repressor’s ability to bind to the operator (Kaiser et al., 2008). Finally, tethering non-CB components to the lac operator array failed to nucleate CB formation, whereas tethering of PML body components resulted in formation of de novo PML bodies (Kaiser et al., 2008). Taken together, these data strongly support a stochastic assembly model and argue against an ordered or hierarchical nuclear body assembly pathway (Figure 2).
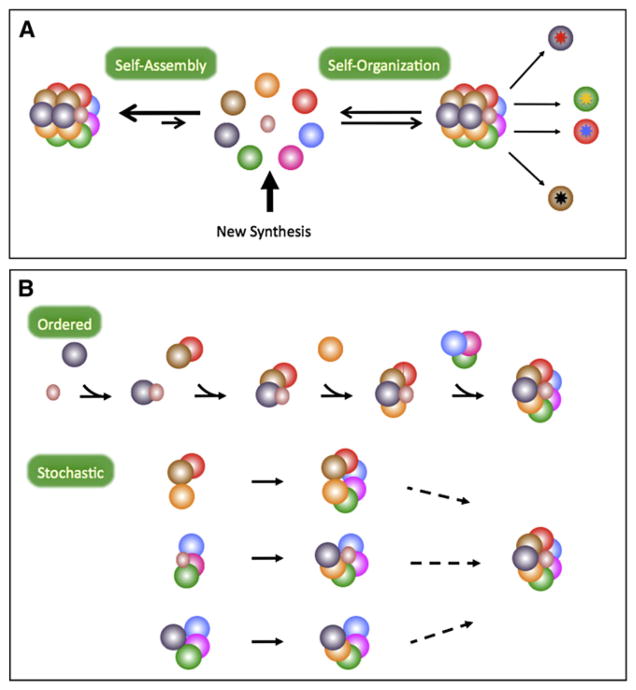
Figure 2. Mechanisms of Nuclear Body Formation.
(A) Biological systems are thought to be governed by the principle of self-organization (Camazine et al., 2001), which is distinct from the concept of self-assembly (Worrall et al., 2007). Self-assembly involves formation of stable complexes that essentially reach thermodynamic equilibrium (left). In contrast, self-organization operates on steady-state systems—those that are far from equilibrium (right). As outlined by Misteli (2001), in cell biological terms, self-organization can be defined as: “the capacity of a macromolecular complex or organelle to determine its own structure, based on the functional interactions of its components.” Through this mechanism, which requires a continuous exchange of materials, the cell is capable of generating a stable (steady-state) structure from a set of dynamic components. In the cartoon, the steady-state approximation is met because a constant flux of components is maintained. Factors enter the body from the newly synthesized pool and can exit the structure, perhaps in a modified form (sunbursts). Note that the modifications do not necessarily preclude a given component from rebinding to the structure.
(B) The assembly of a nuclear body can follow a hierarchically ordered assembly pathway (top), or components can assemble stochastically by a number of individual pathways (bottom). Note that components can enter singly or as large complexes. Although the order of assembly is random in the stochastic model, it is still predicated on molecular interactions. Thus, loss of a given component could lead to failure to incorporate another component or complex.
The Kaiser et al. (2008) study represents a kind of cellular “Field of Dreams” experiment—if you tether it, will they come? Though certainly a major step forward, the lac repressor tethering system essentially creates an artificial scaffold, raising the question of whether or not the system faithfully reflects the formation of nuclear bodies in vivo. One argument to the positive is that the de novo CBs formed by tethering are of a similar size and shape as the endogenous CBs (Kaiser et al., 2008). However, we do not know whether the size is a function of the number of lac operator repeats. What happens if you change the length of the tethering chromatin? Does it change the size of the resultant CB? Does a single component truly seed the formation of a nuclear body on its own, or must it assemble some kind of subcomplex prior to its arrival at the lac operator targeting site? A more basic question is whether or not CBs require a tether in the first place. In other words, do CBs require specific DNA or RNA sequences in order to nucleate, or can they form independently (Matera, 1998)? Previous studies can shed some light here.
In amphibian oocytes, CBs (a.k.a. “spheres”) are known to associate with the histone gene clusters at sites termed “sphere organizers” (Callan et al., 1991; Gall et al., 1981). Similarly, in interphase human cells, histone and small nuclear (sn)RNA genes associate nonrandomly with CBs (Frey and Matera, 1995; Gao et al., 1997; Jacobs et al., 1999; Shopland et al., 2001; Smith et al., 1995), and these sites have been termed “CB organizers” (Frey and Matera, 1995; Gao et al., 1997). The association of CBs and snRNA genes is not coincidental, as ectopically expressed snRNA genes can function as CB organizers (Frey et al., 1999; Frey and Matera, 2001). However, unlike the well-known rRNA genes that act as nucleolus organizers, CBs are not nucleated at snRNA gene loci following induction of transcription; rather, the snRNA genes are recruited to extant CBs (Dundr et al., 2007). Other lines of evidence against a requirement for tethering at a specific genomic locus are the findings that CBs can be assembled in vitro using Xenopus egg extracts that are completely devoid of genomic frog DNA (Bauer et al., 1994) or that microinjection of U7 snRNA can nucleate formation of mini-CBs in frog oocytes (Tuma and Roth, 1999). Thus, at least in certain circumstances, CBs can be formed independently.
Immobilization of components to a specific site in the genome can nucleate formation of a body. Alternatively, it is possible that the clustering of factors at their normal sites of action might also lead to formation of a nuclear body. We assume that the downstream, postnucleation assembly events will proceed by the same molecular mechanisms (e.g., stochastic self-organization) regardless of whether or not nuclear body formation was initiated by immobilizing a given component. However, in the absence of an appropriate assay, we cannot say for sure that the tethered bodies are functional. Experiments on another kind of nuclear body, DNA repair foci, suggest that the tethered, de novo structures are functional. Soutoglou and Misteli (2008) showed that DNA repair factors could be tethered to specific sites that could not only nucleate formation of DNA repair foci, but could also elicit the cellular DNA damage response, even in the absence of DNA lesions. These data argue strongly that the formation of subcellular compartments is governed by stochastic self-organization and that, once a nuclear body is formed, it is functional.
Nucleoli and HLBs: Where Function Meets Form
Unlike most of the nuclear bodies shown in Figure 1, which are not constitutively associated with a specific chromosomal locus, the nucleolus is intimately associated with the genes that encode the 35S preribosomal RNA. More than two decades ago, elegant work in Drosophila showed that RNA polymerase I-mediated transcription of rRNA transgenes directed formation of ectopic nucleoli, whereas expression of transgenes lacking pol I promoters did not (Karpen et al., 1988). Given that nucleolar morphology has long been shown to correlate with the relative transcriptional activity of the endogenous rRNA genes (Haaf et al., 1991; Scheer et al., 1984), it is clear that nucleoli form as a consequence of rRNA transcription and the downstream processing and ribosomal subunit assembly steps. Notably, ectopic insertion of an array of upstream binding factor (UBF, a pol I transcription factor) binding sites results in sequestration of UBF and other pol I transcription factors to the ectopic sites, although a full-blown nucleolus is not formed (Mais et al., 2005). It would be interesting to see whether lac repressor fusions of other nucleolar components might generate nucleolus-like subcompartments on lac operator arrays. Self-organization notwithstanding, given the complex nature of the pol I transcription and rRNA-processing machineries, it seems doubtful that tethering any given nucleolar protein would nucleate assembly of an entire nucleolus.
The HLB is another example of a chromatin-associated nuclear body (Figure 3). Metazoan genomes typically contain a set of histone genes that are expressed only during DNA replication (S phase) and another set of “replacement” histone genes that are constitutively expressed (reviewed in Marzluff et al., 2002). The genes encoding the replication-dependent histones are typically clustered, whereas the replacement histone genes are interspersed (Marzluff et al., 2002). HLBs associate specifically with the replication-dependent histone gene clusters and are thought to coordinate the transcription and 3′ end processing of histone pre-mRNAs (for details, see Figure 3 legend).
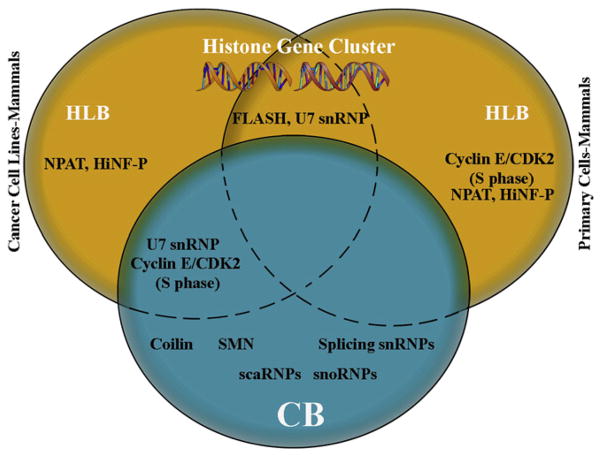
Figure 3. Components of Cajal and Histone Locus Bodies.
The replication-dependent histone genes are typically clustered in metazoan genomes. The histone locus body (HLB) can be viewed as a nuclear subdomain dedicated to the transcription and processing of histone pre-mRNA. In the Venn diagram above, factors known to localize to HLBs in both primary and cancer cell lines are shown. Factors that localize to Cajal bodies (CBs) are also listed. Note that the U7 snRNP, which is essential for processing of histone pre-mRNA 3′ ends, is localized to the CB in human cancer cell lines but to the HLB in primary cells.
Although the term HLB was only recently coined (Liu et al., 2006), the structure was probably first identified in 1981, when Gall and coworkers showed that the “sphere organelle” is bound to the histone gene clusters in the newt, Triturus (Gall et al., 1981). These structures were later termed CBs. The presence of extrachromosomal sphere organelles (also termed CBs) in amphibian oocytes and the absence of markers to distinguish between these two types of structures has also been a hindrance. In 1998, Spradling and coworkers (Calvi et al., 1998) showed that an unknown cyclin E-dependent phosphoepitope (MPM-2) localized to a nuclear domain that was subsequently identified as the HLB (White et al., 2007). The first clear demonstration of a structure located at the mammalian histone gene cluster was by Zhao and coworkers, who found that NPAT, a CDK2-cyclin E substrate, colocalized with the mammalian histone genes (Zhao et al., 2000). Subsequently, another HLB protein, called FLASH, was shown to colocalize with NPAT (Barcaroli et al., 2006).
Factors required for histone gene expression, including NPAT, HiNF-P, FLASH, and the U7 snRNP, all are concentrated within the structure we now refer to as the HLB (Barcaroli et al., 2006; Bongiorno-Borbone et al., 2008; Ghule et al., 2009; Liu et al., 2006; Yang et al., 2009; Zhao et al., 2000). Previous studies in human cancer cells had shown that the U7 snRNP primarily accumulates in CBs (Frey and Matera, 1995; Pillai et al., 2001; Shopland et al., 2001). However, in Drosophila, the U7 snRNP typically colocalizes with the histone gene cluster in HLBs, structures that are distinct from but often adjacent to CBs (Liu et al., 2006, 2009). The peculiar localization of the U7 snRNP to CBs in most human cancer cell lines (Figure 3) has therefore caused some confusion.
The recent availability of monospecific antibodies targeting Lsm10 and Lsm11 (components of U7 snRNP) has allowed investigators to reconcile work in the mammalian and invertebrate systems. The emerging picture is that, in human primary cells, U7 snRNP components colocalize precisely with the HLB marker proteins NPAT and FLASH (Ghule et al., 2009). Due to hyperphosphorylation of coilin (Hearst et al., 2009), CBs are not typically observed in human primary fibroblasts (Spector et al., 1992) but are prominent features of other primary cells such as neurons (Cajal, 1910). Thus, it remains an open question as to why U7 snRNP becomes delocalized from HLBs in cancer cell lines.
Interestingly, Duronio and coworkers have shown that at least some of the components of HLBs can form nuclear foci in the absence of the histone gene cluster (i.e., in a strain carrying an appropriate deletion; White et al., 2007). These findings are somewhat reminiscent of the “residual” Cajal bodies that form in coilin knockout cells (Tucker et al., 2001; Jady et al., 2003) and suggest that stochastic self-organization is also an important factor in the assembly of HLBs. However, unlike CBs, HLBs can be viewed as nuclear subdomains that are dedicated to the expression of replication-dependent histone genes.
Cajal and PML Bodies: Where the Ends Begin
Although they are not thought to be constitutively bound to particular chromosomal regions, two prominent nuclear subdomains (Cajal and PML bodies) are known to associate transiently with specific genomic loci. As discussed above, CBs have been shown to associate with histone, snRNA, and small nucleolar (sno)RNA genes in various human cancer cell lines (Frey and Matera, 1995; Gao et al., 1997; Jacobs et al., 1999; Schul et al., 1998; Shopland et al., 2001; Smith et al., 1995). CB association with snRNA genes requires active snRNA transcription (Frey et al., 1999; Frey and Matera, 2001) and is inhibited by over-expression of a nonpolymerizable isoform of nuclear actin (Dundr et al., 2007).
Illustrating the plurifunctionality of this nuclear organelle, CBs have also been shown to play a role in telomere length regulation. Vertebrate telomerase RNAs contain a domain that is very similar to a class of small Cajal body (sca)RNAs, which typically guide the posttranscriptional modification of other small RNAs (Darzacq et al., 2002; Kiss et al., 2002; Tycowski et al., 2004; Xie et al., 2007). During S phase, human telomerase RNA and the reverse transcriptase component hTERT colocalize within telomere-proximal CBs (Jady et al., 2004; Tomlinson et al., 2008; Zhu et al., 2004). During the gap phases of the cell cycle (G1 and G2), hTERT does not localize to CBs (Jady et al., 2006; Tomlinson et al., 2006). Targeting of a variety of scaRNAs, including telomerase RNA, to CBs requires the activity of a WD repeat protein called WDR79/TCAB1 (Tycowski et al., 2009; Venteicher et al., 2009). This protein is part of the telomerase holoenzyme, and depletion of WDR79/TCAB1 by RNAi (Venteicher et al., 2009) or mutation of its binding site on telomerase RNA (Cristofari et al., 2007) not only disrupts trafficking of telomerase and other scaRNAs to CBs, but also inhibits telomerase function. Because localization of telomerase RNA to CBs is not cell-cycle dependent, whereas hTERT accumulates in CBs only during mid-S phase, these findings strongly suggest a mechanism whereby CB-mediated RNP assembly drives the activity of the telomerase holoenzyme complex.
Of interest, in the absence of functional telomerase, another type of nuclear body appears to play a role in maintaining telomere length. Certain tumors and immortalized cell lines are telomerase negative and maintain their telomeres using a recombination-mediated alternative lengthening of telomeres (ALT) mechanism (Bryan et al., 1995; Dunham et al., 2000). In addition to the lack of telomerase activity, ALT cells share a number of common features, including a unique pattern of telomere length heterogeneity (Bryan et al., 1995) and the presence of ALT-associated PML bodies (APBs) that contain telomeric DNA sequences as well as telomere-specific binding proteins (Yeager et al., 1999).
The formation of APBs is induced by DNA damage (Fasching et al., 2007) and/or upregulation of the p53/p21 pathway and requires the heterochromatin binding protein HP1 (Jiang et al., 2009). Furthermore, live-cell imaging experiments suggest that APBs are formed in two steps. A preexisting PML body is first thought to bind to the telomere, followed by recruitment of additional PML (and presumably other proteins) from the nucleoplasm until the typical APB structure is assembled (Jegou et al., 2009). Ample evidence suggests that, when telomere repeat length is reduced below a critical threshold, the normal telomeric chromatin structure (the telosome) is disrupted, perhaps resulting in an increased mobility of the chromosome end. A current model of APB formation holds that the shortened telomere then associates with a PML body to form an APB (Jegou et al., 2009). Subsequently, telomere length is increased, allowing the telosome to reassemble. Although APBs may not be required for the recombination-mediated repair event, per se (Jiang et al., 2009), they are thought to function to protect ALT cells from the apoptotic consequences of DNA damage-induced signaling.
Heterogeneity of Nuclear Bodies: A Signature of Plurifunctionality?
The nucleolus has long been thought to carry out multiple functions (reviewed in Pederson, 1998; Pederson and Tsai, 2009), so the potential plurifunctionality of nuclear bodies is not a new idea. However, proving that a given biochemical reaction is taking place in a particular nuclear subcompartment and not in another closely related one is not an easy task. The studies of ALT-associated PML bodies outlined above suggest that, upon receipt of the appropriate cellular signal, a garden variety PML body can morph into a different type of nuclear subcompartment—or, at least, it can perform a different function. Caution must be used when interpreting such transformations, as oftentimes, a limited number of markers are monitored, and it is not always clear whether the nuclear body itself is morphing or whether one or a few of its components simply relocalize to a different structure.
The structural heterogeneity among nuclear bodies of the same class is an underexplored topic. Alternative pre-mRNA processing of PML is thought to create different binding interfaces and thereby modulate functional diversity among PML bodies (reviewed in Bernardi and Pandolfi, 2007). With regard to heterogeneity among Cajal bodies, are the CBs that appear to be floating free in the nucleoplasm structurally distinct from those located adjacent to snRNA genes, histone gene clusters, nucleoli, or telomeres? Static images of human cancer cell lines suggest that most CB components are shared among all of the CBs in a given cell (reviewed in Matera, 1999). In living cells, CB components display reasonably tight retention time profiles. In other words, there is little variation in the dissociation kinetics of a given component from one CB to another (Dundr et al., 2004). The fact that individual CBs are capable of associating with multiple DNA loci at the same time (Frey et al., 1999) also suggests an equivalency between CBs. However, a study of CB motility and dynamics provided clear evidence of unequal partitioning of components to daughter CBs upon splitting (Platani et al., 2000). A clearer understanding will require not only the identification of multiple marker proteins for a given structure, but also their simultaneous use in an experiment. Furthermore, because most of the above studies were carried out in a single human cell line (HeLa), additional studies in other cells and organisms will be needed in order to better understand how these intranuclear leopards might change their spots.
Crucibles of Macromolecular Assembly
The eukaryotic nucleus is a congested place. Macromolecular crowding is thought to play an important role in increasing the relative concentration of nuclear proteins and accelerating the rates of biochemical reactions (Hancock, 2004; Richter et al., 2008; Zhou et al., 2008). Based on the principle of mass action, higher concentrations of reactants can drive a given reaction forward. In situ hybridization and digital imaging microscopy experiments have shown that the highest concentration of spliceosomal snRNPs in a HeLa cell nucleus is in the CB (Carmo-Fonseca et al., 1992; Matera and Ward, 1993). What kinds of reactions take place within CBs? Do they slow down if the CB is disassembled? What are the cellular and organismal consequences of the loss of this structure? These and other questions are considered below.
A relatively large body of evidence points to a role for CBs in the assembly and modification of a variety of different small RNPs (Sleeman and Lamond, 1999; Darzacq et al., 2002; Jady et al., 2003; Boulon et al., 2004; Nesic et al., 2004; Schaffert et al., 2004; Shpargel and Matera, 2005; Stanek et al., 2003; Stanek and Neugebauer, 2004; Tanackovic and Kramer, 2005; Jady et al., 2006; Li et al., 2006; Tomlinson et al., 2006, 2008; Li et al., 2008). These macromolecular assembly processes apparently do not strictly require the presence of CBs, as mutations in coilin result in the disassembly of CBs, and the homozygous mutants are at least partially viable (Collier et al., 2006; Liu et al., 2009; Tucker et al., 2001). Of note, loss or depletion of coilin leads to reduced growth rates in human cultured cells (Lemm et al., 2006) and reduced viability, fertility, and fecundity in mice (Walker et al., 2009). Thus, though certain organisms can do without coilin, the protein has been conserved since before the divergence of plants and animals (estimated at > 1.5 billion years ago).
Cajal bodies have been described as waystations, meeting places, and assembly factories for RNPs (Matera and Shpargel, 2006; Stanek and Neugebauer, 2006). The developmental need for a given class of RNP may not be the same in one species as it is for another. The emerging evidence suggests that CBs have taken on different functions throughout evolution (Matera, 2006; Pontes and Pikaard, 2008). For example, the acquisition of a snoRNA-like domain in mammalian telomerase RNAs (Mitchell et al., 1999) is likely an important feature, enabling telomere length to be regulated by an RNP assembly mechanism (Tomlinson et al., 2008; Venteicher et al., 2009). Similarly, the presence of an additional Argonaute protein (Ago4) has enabled plant cells to create nuclear siRNA “processing centers” (Li et al., 2006; Pontes et al., 2006) for RNPs involved in RNA-directed DNA methylation. A related but distinct activity also exists in another type of nuclear “dicing” body (Fang and Spector, 2007; Fujioka et al., 2007; Li et al., 2008; Song et al., 2007). Thus, in addition to localizing a particular biochemical reaction to a specific genomic locus (e.g., the telomere), nuclear bodies might also function to accelerate the assembly of macromolecular complexes.
What is the evidence for this other general function? Recently, Neugebauer and colleagues carried out an important study that illustrates how a structure such as the CB can facilitate a key step in gene expression, namely the assembly of U4/U6 spliceosomal di-snRNPs (Figure 4). Using a combination of in vivo measurements and in silico modeling, these investigators showed that U4 and U6 snRNP concentrations are up to 20-fold higher in the CB than in the surrounding nucleoplasm, leading to a dramatic (11-fold) increase in the rate of U4/U6 di-snRNP assembly in cells that contained CBs versus those lacking them (Klingauf et al., 2006). The optimum number of CBs was calculated to be between three and four per cell (Figure 4); there are apparently decreasing marginal returns on the investment of more than four CBs per cell (Klingauf et al., 2006). Interestingly and perhaps not coincidentally, the number of CBs in a typical mammalian cell line is between three and five (Matera, 1999). Collectively, the experiments suggest that coilin expression and CB formation greatly facilitate RNP assembly reactions that are rate limiting for cellular proliferation.
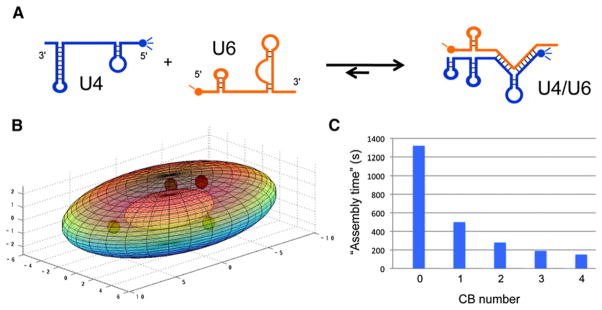
Figure 4. Modeling Macromolecular Assembly.
(A) Formation of the U4/U6 di-snRNP requires extensive base-pairing interactions between U4 and U6 snRNAs. The assembly of U4/U6 di-snRNPs is a necessary step that takes place prior to spliceosome formation. Proteins that bind to these snRNAs are not shown in the reaction scheme.
(B) Three-dimensional projection of a HeLa cell nucleus showing a single nucleolus and four Cajal bodies (CBs) within its interior (reprinted with permission from Klingauf et al., 2006). Dimensions are in microns.
(C) Through the use of simulated random walks within the nuclear space (excluding the nucleolus), Klingauf et al. (2006) showed that the time for productive assembly of U4/U6 di-snRNPs was greatly accelerated by the presence of one or more CBs; optimal assembly rates were achieved when cells contained three to four CBs.
Conclusions and Prospectus
In a broader context, it is tempting to speculate that subcellular compartments (i.e., those that are bound by membranes as well as those that are not) may generally function to concentrate reactants and thereby enhance the rates of association of various macromolecular complexes contained within their borders. Indeed, nuclear Cajal bodies have many of the same molecular features and experimental challenges as cytoplasmic processing bodies (P bodies). Both are membrane-less, RNP-rich, steady-state structures that are assembled from sets of dynamic components (Aizer et al., 2008; Dundr et al., 2004; Kedersha et al., 2005). Like CBs, P bodies can be found in close association with other compartments or foci, such as stress granules and U bodies (Buchan et al., 2008; Liu and Gall, 2007; Kedersha et al., 2005). Also, like their nuclear brethren, P bodies are not essential structures, although they contain many essential components (Anderson and Kedersha, 2009; Eulalio et al., 2007a; Parker and Sheth, 2007). Thus, CBs and P bodies are not obligate structures; molecular processes thought to take place in these domains are ongoing in the absence of the structures themselves (Eulalio et al., 2007b; Jady et al., 2003). These and other results suggest that cellular structures such as the CB or P body contribute to the overall fitness of the organism by allowing additional layers of regulation and fine-tuning.
How do cells and organisms coordinate the regulation of multiple macromolecular assembly reactions? What are the feedback mechanisms? The past few years have seen a tremendous amount of progress in elucidating the molecular mechanisms underlying the assembly and function of nuclear subcompartments. However, the examples above highlight the need for combined approaches involving genetic, cellular, and organismal studies in order to better understand how the machinery in nuclear (and cytoplasmic) bodies interfaces with various assembly and/or signaling factors to regulate gene expression. The prediction is that a given nuclear body component might localize to distinct subcellular compartments based on differential affinities for factors in those compartments. Future studies will thus require the use of biosensors that function in living cells and can give readouts of molecular states.
Acknowledgments
We are grateful to M. Chen, M. Hübner, I. Meier, K. Neugebauer, and D. Spector for providing images. We apologize to all colleagues whose work could not be cited, owing to space limitations. K.P. was supported, in part, by an American Heart Association predoctoral fellowship. Work in the Matera laboratory is supported by National Institutes of Health grants R01-GM053034 and R01-NS041617.
References
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci USA. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DW, Murphy C, Wu Z, Wu CH, Gall JG. In vitro assembly of coiled bodies in Xenopus egg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, Bertrand E. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SRy. El núcleo de las células piramidales del cerebro humano y de algunos mamíferos. Trab Lab Invest Biol (Madrid) 1910;8:27–62. [Google Scholar]
- Callan HG, Gall JG, Murphy C. Histone genes are located at the sphere loci of Xenopus lampbrush chromosomes. Chromosoma. 1991;101:245–251. doi: 10.1007/BF00365156. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camazine S, Deneubourg J-L, Franks NR, Theraulaz G, Bonabeau E. Self-Organization in Biological Systems. Princeton, NJ: Princeton University Press; 2001. [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dokholyan NV. Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm. Mol Biol Evol. 2008;25:1530–1533. doi: 10.1093/molbev/msn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw PJ. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. Predicting three-dimensional genome structure from transcriptional activity. Nat Genet. 2002;32:347–352. doi: 10.1038/ng1102-347. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, Neumann AA, Muntoni A, Yeager TR, Reddel RR. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol. 2001;154:499–509. doi: 10.1083/jcb.200105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- Gall JG, Stephenson EC, Erba HP, Diaz MO, Barsacchi-Pilone G. Histone genes are located at the sphere loci of newt lampbrush chromosomes. Chromosoma. 1981;84:159–171. doi: 10.1007/BF00399128. [DOI] [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J Struct Biol. 2004;146:281–290. doi: 10.1016/j.jsb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Sites in human nuclei where damage induced by ultraviolet light is repaired: localization relative to transcription sites and concentrations of proliferating cell nuclear antigen and the tumour suppressor protein, p53. J Cell Sci. 1994;107:1753–1760. doi: 10.1242/jcs.107.7.1753. [DOI] [PubMed] [Google Scholar]
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell. 1999;10:1653–1663. doi: 10.1091/mbc.10.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelman G, Wachsmuth M, Gorisch SM, Greulich-Bode KM, Boukamp P, Lichter P, Rippe K. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol Biol Cell. 2009;20:2070–2082. doi: 10.1091/mbc.E08-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Nguyen A, Henson JD, Toouli CD, Braithwaite AW, Reddel RR. Induction of alternative lengthening of telomeres-associated PML bodies by p53/p21 requires HP1 proteins. J Cell Biol. 2009;185:797–810. doi: 10.1083/jcb.200810084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGO-NAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li CF, Henderson IR, Song L, Fedoroff N, Lagrange T, Jacobsen SE. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Liu JL, Gall JG. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the inter-chromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG. Drosophila Cajal bodies: Accessories not included. J Cell Biol. 2006;172:791–793. doi: 10.1083/jcb.200602002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta. 2008;1783:2100–2107. doi: 10.1016/j.bbamcr.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hugle B, Hazan R, Rose KM. Drug-induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-dichloro-beta-D-ribofuranosylbenzimidazole. J Cell Biol. 1984;99:672–679. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell. 1998;9:1025–1036. doi: 10.1091/mbc.9.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell. 2001;12:565–576. doi: 10.1091/mbc.12.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Kramer A. Human Splicing Factor SF3a, but Not SF1, Is Essential for Pre-mRNA Splicing In Vivo. Mol Biol Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Roth MB. Induction of coiled body-like structures in Xenopus oocytes by U7 snRNA. Chromosoma. 1999;108:337–344. doi: 10.1007/s004120050385. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall JA, Gorna M, Pei XY, Spring DR, Nicholson RL, Luisi BF. Design and chance in the self-assembly of macromolecules. Biochem Soc Trans. 2007;35:502–507. doi: 10.1042/BST0350502. [DOI] [PubMed] [Google Scholar]
- Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. Sno/scaR-NAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007;35:D183–D187. doi: 10.1093/nar/gkl873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-c, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a pro-apoptotic protein involved in activation of caspase-8 is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]