Abstract
Eradication of smallpox was accomplished 30 yr ago, but poxviral infections still represent a public health concern due to the potential release of variola virus or the emergence of zoonotic poxviruses, such as monkeypox virus. A critical determinant of poxvirus virulence is the inhibition of interferons (IFNs) by the virus-encoded type I IFN-binding protein (IFNα/βBP). This immunomodulatory protein is secreted and has the unique property of interacting with the cell surface in order to prevent IFN-mediated antiviral responses. However, the mechanism of its attachment to the cell surface remains unknown. Using surface plasmon resonance and cell-binding assays, we report that the IFNα/βBP from vaccinia virus, the smallpox vaccine, interacts with cell surface glycosaminoglycans (GAGs). Analysis of the contribution of different regions of the protein to cell surface binding demonstrated that clusters of basic residues in the first immunoglobulin domain mediate GAG interactions. Furthermore, mutation of the GAG-interaction motifs does not affect its IFN-binding and -blocking capacity. Functional conservation of GAG-binding sites is demonstrated for the IFNα/βBP from variola and monkeypox viruses, extending our understanding of immune modulation by the most virulent human poxviruses. These results are relevant for the design of improved vaccines and intervention strategies.—Montanuy, I., Alejo, A., Alcami, A. Glycosaminoglycans mediate retention of the poxvirus type I interferon binding protein at the cell surface to locally block interferon antiviral responses.
Keywords: immune evasion, cytokine, smallpox, innate immunity
The interferons (IFNs) are a family of secreted cytokines that are expressed as a first line of defense against viral infections. They are able to inhibit virus replication and limit viral spread via direct antiviral and indirect immunoregulatory activities (1). The importance of IFNs in the host response against viral infections is underpinned by the variety of viral proteins that impair their activity (1, 2).
Viral infection initially triggers the expression of IFN through a set of molecular recognition events, which converge on the activation of key transcription factors: IRF3 and/or IRF7 and NF-κB. The secreted IFN can then act either locally or at a distance through the IFN receptors activating signaling cascades involving Janus protein tyrosine-kinase (JAK) and signal transducers and activators of transcription (STAT) pathway, and inducing the expression of proteins that put the cell in an antiviral state. Viruses have developed multiple strategies to evade this response, acting at the level of the initial recognition of the invading virus by the host cell, the inhibition of the signaling cascade triggered by IFNs, or the effector mechanisms that contribute to the antiviral state (1, 2).
The Poxviridae are a family of large cytoplasmic DNA viruses, vaccinia virus (VACV) being the most studied poxvirus. VACV was used as the live vaccine to eradicate smallpox by 1980, the first and only human viral disease eradicated as a result of a global vaccination campaign. Interestingly, the smallpox eradication was achieved with very limited knowledge of the molecular biology of poxviruses or the immune mechanisms that conferred protection (3). Two other members of the family are of special interest: the highly virulent variola virus (VARV), the causative agent of smallpox, and monkeypox virus (MPXV), an emerging zoonotic virus causing sporadic infections in humans with case-fatality rates of up to 10% (4). The increase of a nonvaccinated population that would be susceptible to a potential deliberate VARV release, or to infection by a zoonotic poxvirus such as MPXV, has strengthened the interest in poxvirus research, since many aspects of viral immunity and pathogenesis are still poorly understood, and new effective therapies for poxviral infections in humans are needed (3, 5–7).
The relevance of IFNs for protection against poxvirus infections was soon realized. Mice with a disrupted IFN system show an increased susceptibility to lethal disease (8–10). Moreover, neutralizing anti-IFN antibodies reduced clearance of the virus in a mousepox model of infection (11), whereas pretreatment of mice with IFN prevented VACV infection (12).
Accordingly, poxviruses have developed a large set of mechanisms to block the activity of IFNs. These include the E3 protein (binds dsRNA and inhibits IFN production, as well as PKR effector functions), the K3 protein (prevents eIF2α phosphorylation, inhibiting PKR), and the H1 protein (dephosphorylates STAT1). In addition, poxviruses encode inhibitors of TLR-signaling that lead to IFN production, such as A46 and A52, or proteins that inhibit antiviral effectors induced by IFN, such as K1 and C7 (13, 14).
A unique and efficient IFN evasion strategy employed by poxviruses is to encode soluble proteins that are secreted from infected cells and function as soluble IFN decoy receptors (15). By binding IFN molecules with high affinities, they are able to prevent their interaction with cellular receptors and, hence, the establishment of a potent antiviral response. The viral IFNα/β-binding protein (IFNα/βBP) (16–20) can neutralize type I IFN and is widely expressed by different poxviruses, including some VACV strains, VARV, MPXV, and ectromelia virus (ECTV), the poxvirus that causes mousepox and is used as a surrogate model of smallpox (21).
The contribution of the IFNα/βBP to poxvirus pathogenesis was first recognized using the VACV model of infection, where its absence caused a strong virus attenuation during intranasal infection of mice (16). In the case of ECTV, the IFNα/βBP has been demonstrated to be an essential virulence factor since a 107-fold attenuation has been observed in its absence (22). Notably, the ECTV IFNα/βBP was shown to represent an effective target for protective immunization, suggesting an alternative vaccination strategy against poxviral infections in humans (22).
The IFNα/βBP encoded by VACV strain Western Reserve (WR) B18R gene, known as B18, is a glycoprotein with 3 immunoglobulin (Ig)-like domains and sequence unrelated to type I IFN cellular receptors, which have fibronectin type III domains (23, 24). In contrast to the cellular receptors, the viral protein binds IFNs from a broad range of host species (16).
A special property of IFNα/βBPs encoded by VACV, VARV, and MPXV is their ability to bind to the cell surface and prevent IFN signaling at this location (17, 18, 20). It has been hypothesized that the protein is secreted so that it can reach surrounding uninfected cells, evading the induction of an antiviral state before cells become infected (18, 20). However, despite the potential relevance of the cell-binding ability of IFNα/βBP for its anti-IFN activity, the mechanism of attachment to cell surface remains unknown.
Here, we report that the VACV B18 protein attaches to the cell surface via an interaction with glycosaminoglycans (GAGs). We identified different regions comprising clusters of basic residues in the amino terminus of the B18 protein, which are important for this interaction. Moreover, we show that mutants lacking GAG-binding activity can still bind IFN molecules in solution with high affinity, but they are unable to block IFN signaling and antiviral activity at the cell surface, a critical place for anti-IFN action. Notably, we show that the interaction with the cell surface via GAGs is shared by the IFNα/βBPs encoded by VARV and MPXV viruses.
MATERIALS AND METHODS
Cells and reagents
HeLa cells were grown in 10% FCS containing DMEM. Chinese hamster ovary K1 (CHO-K1) and mutant (CHO-745, CHO-618, and CHO-677) cells were kindly provided by Dr. F. Arenzana-Seisdedos (Institut Pasteur, Paris, France) and were grown in 10% FCS containing DMEM-Ham′s F12 (1:1) medium. Molt4, Raji, U937, HL60, THP1, and EL4 cells were cultured in 10% FCS containing RPMI medium. Hi5 insect cells were cultured in TC-100 medium supplemented with 10% FCS. Vesicular stomatitis virus (VSV) strain Cocal was obtained from Dr. W. James (Oxford University, Oxford, UK). Recombinant hIFNα-2b (intron A) was from Schering-Plough (Kenilworth, NJ, USA). Heparin, heparan sulfate, and chondroitin sulfate A and B were from Sigma (Steinheim, Germany).
Cloning and expression of recombinant proteins
The plasmid containing the B18 coding sequence, pMF2, has been described previously (20). The sequences coding for amino acid residues 24–167 or 168–351 were PCR amplified from pMF2 and cloned into pAL7, a modified pFastBac vector bearing the honeybee mellitin signal peptide at the 5′ region and C-terminal V5HIS tag, to generate the plasmids pIM13 and pIM14 for expression of the tagged B18-N and B18-C fragments, respectively. The plasmids pMF1 (VARV B17) and pMF6 (MPXV B16) have been previously described (20). The substitution mutants described in this work were engineered by the overlap extension PCR method (25) or by using the QuickChange II site-directed mutagenesis kit (Stratagene, Cedar Creek, TX, USA). Plasmids were sequenced to confirm the presence of the desired mutations and the absence of unwanted mutations. Primer sequences and templates used are listed in Supplemental Tables S1 and S2. Handling of VARV DNA was performed under World Health Organization (WHO) permission and in accordance with WHO guidelines. Recombinant baculoviruses were obtained using the Bac-to-Bac system (Invitrogen, Carlsbad, CA, USA). Recombinant baculovirus expressing ECTV semaphorin homologue (SEMA) fused to V5 and 6xHis C-terminal tags will be described elsewhere (unpublished results).
Sequence analysis
Sequence alignments of B18 from VACV strain WR (GenBank accession number YP_233082), VARV strain Bangladesh 1975 (B17R; GenBank AAA60926), MPXV strain Zaire_1979-05 (B16R; GenBank AAY97385), ECTV strain Naval-176 (Viral Bioinformatics Resource Center accession number VP0042431), were performed with ClustalW (http://www.ebi.ac.uk). Plot of charge density at pH 7 was made with the DNASTAR Protean program (DNASTAR, Madison, WI, USA), using a window size of 6 residues.
Recombinant protein purification
Supernatants from baculovirus-infected Hi5 cells were concentrated on a stirred ultrafiltration cell (Amicon, Danvers, MA, USA) using YM-10 membranes (Millipore, Bedford, MA, USA), and buffer was exchanged into binding buffer (50 mM phosphate, 300 mM NaCl, and 10 mM imidazole, pH 7.4) on PD-10 desalting columns (GE Healthcare, Little Chalfont, UK). The His-tagged recombinant proteins were affinity-purified using Ni-NTA columns (Qiagen, Valencia, CA, USA), dialyzed into PBS, and concentrated using Vivaspin 500 columns (Sartorius Stedim Biotechnology, Aubagne, France). Protein purity was checked on Coomassie blue-stained SDS-PAGE and quantified by gel densitometry or BCA assay (Pierce Biotechnology, Rockford, IL, USA).
Immunofluorescent cell staining
Cells seeded on coverslips were incubated for 30 min at 4°C with supernatants (50 μl) from baculovirus-infected Hi5 cells (1 ml of supernatant was obtained from 1.3×106 Hi5 infected cells). Unbound protein was removed with several washes in cold PBS. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature and washed with 1% BSA in PBS. Immunofluorescent detection was performed without cell membrane permeabilization using a monoclonal anti-V5 antibody (Invitrogen), followed by Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen). Cell nuclei were stained with DAPI (Calbiochem, Darmstadt, Germany). Images were acquired using an Axioskop 2 Plus microscope (Carl Zeiss, Jena, Germany) and were captured with a Coolsnap FX color camera (Roper Scientific, Tucson, AZ, USA) and MetaVue 5.07 software (Molecular Devices, Downingtown, PA, USA). Images were also collected using a confocal laser scanning microscope (MicroRadiance; Bio-Rad, Hemel Hempstead, UK) and LaserSharp2000 software (Bio-Rad).
Analysis of protein binding to cells by flow cytometry
Adherent cells (3×105) were harvested with 4 mM EDTA in PBS and incubated on ice with supernatants from baculovirus-infected Hi5 cells containing the corresponding proteins for 30 min in a total volume of 100 μl. After incubation, cells were washed with 1% BSA and 1% FCS in PBS, and protein binding was assessed by flow cytometry using a monoclonal anti-PentaHis antibody (Qiagen) followed by Alexa Fluor 488 donkey anti-mouse IgG secondary antibody. The data were collected on a BD FACSCanto II system (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo 7.2.2 software (Treestar, Ashland, OR, USA).
Sodium chlorate treatment of cell cultures
Sodium chlorate was used to inhibit sulfation of GAGs (26). CHO-K1 cells were cultured in sulfate-free DMEM-Ham's F12 (1:1) medium supplemented with 10% dialyzed FCS containing different concentrations (5–50 mM) of sodium chlorate. When indicated, 10 mM of sodium sulfate was also added. After overnight culture, cells were processed as before for B18 binding analysis by flow cytometry.
Neutralization of IFN antiviral activity
In the preincubation assay, recombinant hIFNα-2b (50 U) was preincubated with supernatants containing viral proteins in 10% FCS containing DMEM (50 μl of total volume) for 30 min at 37°C and added to HeLa cells grown in 96-well plates for 16 h. In the washing assay, culture supernatants were added to HeLa cell monolayers, incubated for 30 min at 37°C, and extensively washed prior to the addition of h-IFNα-2b. In both cases, after 16 h incubation, cells were washed and infected with VSV at a multiplicity of infection of 50 PFU/cell for 72 h (20). Cell viability was then determined using the Cell Titer 96 Aqueous One Solution cell proliferation assay (Promega, Madison, WI, USA).
Inhibition of type I IFN-induced STAT-1 phosphorylation
HeLa cells (3×105) were incubated with supernatants from baculovirus-infected cells and thoroughly washed to remove unbound material. Subsequently, cells were incubated with 1000 U of hIFNα for 30 min at 37°C. Cells were lysed in Laemmli buffer and analyzed by Western blot using the anti-phospho STAT1 (Tyr701) and anti-STAT1 antibodies (Cell Signaling Technology, Danvers, MA, USA), as well as the anti-V5 (Invitrogen) and anti-α-tubulin (Sigma) antibodies.
Biomolecular interaction analysis by surface plasmon resonance (SPR)
Measurement of the kinetic and affinity binding parameters between recombinant viral proteins and heparin was performed using a BIAcore X biosensor (BIAcore Life Sciences, Uppsala, Sweden). A heparin-coated streptavidin (SA) sensor chip was used as described previously (27). Briefly, for kinetic studies, different concentrations of the purified protein were injected at a flow rate of 30 μl/min over 2 min and allowed to dissociate for 5 min. The surface was regenerated by eluting bound protein with a 30-μl injection of 2 M NaCl. BIAcore sensorgrams were analyzed using BIAevaluation 3.2 software. Bulk refractive index changes were removed by subtracting the reference flow cell responses, and the average response of a blank injection was subtracted from all analyte sensorgrams to remove systematic artifacts. Kinetic data were globally fitted to a 1:1 Langmuir model.
For the SPR competition assay, B18 protein (100 nM) was preincubated with several concentrations (0, 1, 10, and 100 μg/ml) of soluble GAGs for 30 min and then was injected over the heparin-SA sensor chip at a 10 μl/min flow rate. The response at equilibrium was recorded.
To determine the binding and affinity constant of IFN to B18-derived mutants, purified M15 and M22 proteins were amine coupled to CM4 sensor chips at low densities [Rmax<200 response units (RU)]. Different concentrations of hIFNα-2b were injected at a flow rate of 30 μl/min over a 2-min period and allowed to dissociate for an additional 5 min. The surface was regenerated after each injection by using 10 mM glycine-HCl (pH 2.5). The sensorgrams obtained were analyzed as detailed above.
RESULTS
VACV B18 protein interacts with GAGs at the cell surface
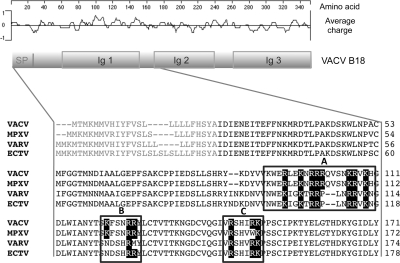
VACV B18 protein is a secreted glycoprotein that lacks a transmembrane domain but associates to the cell surface after secretion (18, 28). A previous observation showed that B18 binds with different degrees to most of the cell types tested (18). As a first step toward the identification of the B18 cell-binding mechanism, we confirmed this finding with a more quantitative assay by flow cytometry (Supplemental Fig. S1). Because of the ability of B18 to bind to a broad range of cell types, we hypothesized that it might interact with a conserved component in the cell membrane like GAGs. Analyses of the primary sequence of the VACV B18 protein revealed different regions that might be responsible for the interaction with GAGs, which are referred to here as regions A–C (Fig. 1). Region A contains a well-conserved cluster of basic residues with positive charge suitable for the interaction with the negative charges of sulfates on GAGs. This region includes two putative GAG-binding motifs in tandem, which can conform to the canonical GAG-binding motifs BBXB and BBBXXB, where B is a basic residue and X any amino acid (29). Similarly, regions B and C closely fit one such motif each (Fig. 1).
Figure 1.
Sequence analysis of B18 protein. Schematic representation of VACV B18 showing the locations of conserved domains and a plot of charge density along the protein. SP, signal peptide; Ig, immunoglobulin domain. Partial amino acid alignment of the B18 protein (first 171 aa) and its MPXV, VARV, and ECTV orthologues is shown. Boxed regions A–C indicate potential GAG-binding domains identified. Black boxes highlight R and K residues.
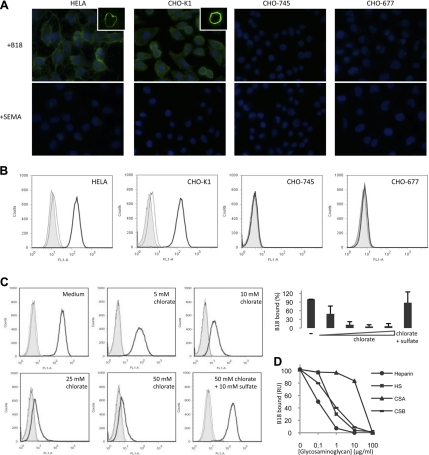
To test the potential interaction of B18 with GAGs, we used a panel of wild-type CHO-K1 and mutant CHO cells that differentially express GAGs on their surface (30). Supernatants from insect cells infected with a recombinant baculovirus expressing VACV B18 protein fused to V5 and 6xHis C-terminal tags (20) were incubated with the different cell lines, and binding was determined by immunofluorescence and flow cytometry. Binding of VACV B18 protein to both HeLa and CHO-K1 cells was readily detected by immunofluorescence, with confocal microscopy showing a clear membrane pattern in both cell lines. However, no binding was observed to the mutant cell lines lacking surface GAG expression (Fig. 2A). As a control, we detected no binding when supernatant from cells infected with a recombinant baculovirus expressing an equivalent amount of an unrelated protein, the ECTV SEMA protein fused to V5 and 6xHIS C-terminal tags, was used (Fig. 2A). Flow cytometric analyses showed specific binding of VACV B18 protein to CHO-K1 cells but no binding to CHO-745 cells (Fig. 2B), which lack xylosyltransferase, the enzyme required for biosynthesis of both heparan sulfate (HS) and chondroitin sulfate (CS) (30). Less than 10% binding was detected in a similar mutant cell line, CHO-618, which is defective in galactosyltransferase and does not synthesize GAGs (ref. 30 and data not shown). In addition, no binding was recorded in CHO-677 cells. These cells express CS but not HS on the cell surface due to the lack of specific N-acetylglucosaminyl- and glucuronosyl-transferase activities (30). The results shown indicate that the VACV B18 protein binds to the cell surface through GAGs. Moreover, the lack of binding to CHO-677 cells suggests a preferential binding of VACV B18 for HS.
Figure 2.
B18 interaction with GAGs. A) Indicated cells were incubated with 50 μl of supernatants containing equivalent amounts of the V5HIS-tagged recombinant protein B18 or control SEMA for 30 min at 4°C; after extensive washes, binding of the protein was assessed by immunofluorescence with anti-V5 antibody. Nuclei were stained with DAPI. Insets: images obtained by confocal microscopy of cells incubated as before, showing plasmatic membrane localization of the protein. One representative experiment of 3 is shown. B) Indicated cells were incubated as before, and binding was detected by flow cytometric analysis using anti-HIS antibody. One representative experiment of 4 is shown. Shading, medium only; dotted trace, unrelated SEMA protein; solid trace, B18 protein. C) Wild type CHO-K1 cells were cultured overnight in sulfate-free medium in the presence of the indicated concentration of sodium chlorate or chlorate plus sulfate. After harvesting, cells were tested for B18 binding by FACS. Shading, medium only; dotted trace, unrelated SEMA protein; solid trace, B18 protein. Histograms are from 1 representative experiment of 3; mean ± sd percentage of bound B18 protein is plotted at bottom. Untreated cells (−), cells treated with increasing concentrations of chlorate, and cells treated with chlorate and supplemented with sulfate are represented. D) Purified recombinant B18 protein (100 nM) alone or preincubated 30 min with increasing concentrations of soluble heparin, HS, CSA, or CSB was injected over a heparin-coated BIAcore SA sensor chip. SPR signal is plotted as a function of the concentration of the competing GAG.
It is frequently observed that the specificity of protein-GAG interactions is dependent on the position and degree of sulfation of GAGs (31). To evaluate whether GAG sulfation is required for cell binding of B18, we treated CHO-K1 cells with the reversible sulfation inhibitor sodium chlorate (ref. 26; Fig. 2C). Trypan blue determination of cell viability, as well as the comparable percentage of cells in the live gate for chlorate-treated and untreated cells, showed that sodium chlorate was not toxic for the cells at the concentrations used (data not shown). Binding of B18 to CHO-K1 cells was greatly reduced in a dose-dependent manner by chlorate treatment (Fig. 2C). Furthermore, this situation could be reversed by the addition of excess exogenous sodium sulfate, showing that loss of B18 binding to the cell surface was due to inhibition of GAG sulfation. Thus, these studies suggest that sulfated GAGs are involved in B18 binding to cells.
High-affinity interaction of VACV B18 protein with GAGs
To further confirm that the VACV B18 protein is able to bind to GAGs, the ability of the purified recombinant protein to bind to heparin was measured by SPR in a BIAcore biosensor, using a heparin-coated chip as an artificial GAG-containing surface (27). The purified recombinant B18 protein was found to readily bind to the heparin surface when injected over the chip. Analysis of the kinetic binding parameters allowed us to determine that the VACV B18 binds to heparin with high affinity (Table 1). The capacity of B18 to bind to GAGs other than heparin was also measured in competition experiments using SPR (Fig. 2D). B18 was preincubated without GAGs or with increasing concentrations of soluble heparin, HS, chondroitin sulfate A (CSA), or chondroitin sulfate B (CSB). Competitive inhibition by soluble GAG was detected as a reduction in the maximum binding compared with the binding of B18 alone. All of the soluble GAGs tested competed with immobilized heparin for B18 binding, with HS being the most effective competitor. These results showed that B18 is capable of interacting with a variety of sulfated GAG structures in this in vitro assay. Overall, the experiments shown indicate that binding of B18 to cells occurs through an interaction with GAGs on the cell surface.
Table 1.
Comparison of kinetic parameters and derived affinity constants for the binding of VACV B18 protein and derived mutants to heparin and hIFNα
| Protein | Heparin binding |
hIFNα binding |
||||
|---|---|---|---|---|---|---|
| Ka (M−1·s−1) | Kd (s−1) | KD (M) | Ka (M−1·s−1) | Kd (s−1) | KD (M) | |
| VACV B18 | 5.7 × 105 | 2.18 × 10−3 | 3.83 × 10−9 | 3.89 × 105a | 2.25 × 10−4a | 5.8 × 10−10a |
| M15 | 1 × 106 | 2.22 × 10−4 | 2.21 × 10−10 | |||
| M22 | 2.24 × 106 | 1.35 × 10−3 | 6.02 × 10−10 | |||
Data from Fernández de Marco et al. (20).
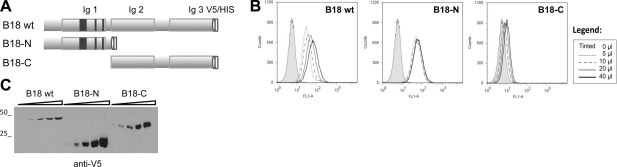
Cell surface binding of B18 is mediated by the N-terminal region
As a first approach to study the region of B18 involved in cell binding, we generated two deletion mutants expressing the amino (B18-N) or carboxy (B18-C) B18 fragments. These truncated versions of B18 were fused at the N terminus to the signal peptide from mellitin to drive the secretion of the proteins in insect cells and tagged to V5 and 6xHis at the C terminus (Fig. 3). Flow cytometry experiments using the supernatants of recombinant baculoviruses containing the corresponding proteins showed that, whereas removal of the amino-terminal part of B18 abolished its cell-binding capacity, the attachment of the B18-N fragment to cells was similar to that of the full-length B18 protein (Fig. 3B). This finding indicates that the N-terminal region, which comprises the first 167 aa of the protein, is sufficient for the attachment of B18 to the cell surface, and is in agreement with the in silico identification of putative GAG-binding sites within the first Ig domain of B18 (Fig. 1).
Figure 3.
Cell-binding ability of B18 and its N- and C-terminal fragments. A) Schematic representation of the mature recombinant B18-tagged protein comprising 3 Ig domains, and the 2 B18-N and B18-C truncated versions of B18 generated. Dark boxes illustrate the location of the putative GAG-binding regions identified. B) Flow cytometric analysis (as described in Fig. 2) of HeLa cells incubated with increasing volumes of supernatants (0–40 μl) containing the corresponding tagged proteins. One representative experiment of 2 is shown. Legend: volumes corresponding to traces. C) Western blot with anti-V5 antibody monitoring the protein amounts used in the experiment. Molecular mass markers (kDa) are at left.
Identification of residues of B18 involved in the interaction with GAGs
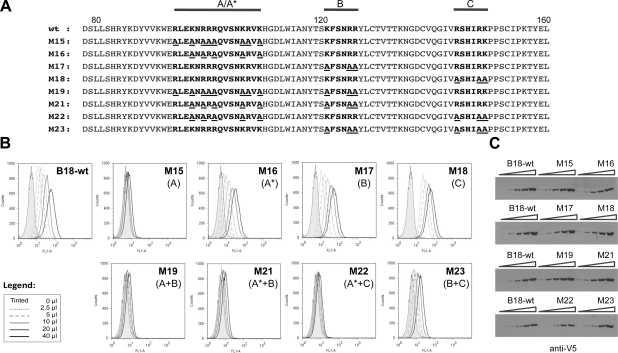
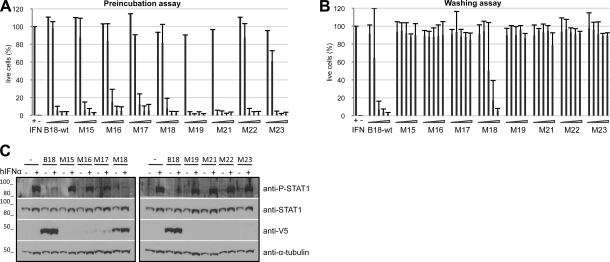
A site-directed mutagenesis approach was employed to identify amino acid residues involved in B18 interaction with GAGs. For this purpose, different mutants were designed in which basic residues arginine (R) and lysine (K) in the presumed GAG-binding regions were mutagenized to alanine (A) (Fig. 4A). To assess the effect of these mutations on the binding capacity of B18 to the cell surface, flow cytometry experiments were performed. Figure 4B shows that the substitution of all R and K amino acids in the first putative GAG-binding region of B18, region A, completely abolished its capacity to bind to cells (M15 mutant). Partial replacement of these positive residues in the region A, which was named A* (M16 mutant), as well as the mutation of all three basic amino acids in the putative GAG-binding region B (M17 mutant) had a limited effect on the ability of the protein to bind to cells, with 44 and 83% of wild-type binding, respectively (calculated as a median of the values obtained in 2 independent experiments at 10 μl of protein added). By contrast, mutation of all the basic residues in region C (M18 mutant) did not affect the ability of the B18 protein to bind to the cell. Except for M15, with 8 basic residues changed to alanine, the mutant proteins described above contained 3–5 amino acid substitutions (Fig. 4A). It is worth noting that combinations of different sets of mutations also affected cell binding, as exemplified by the other 4 mutants. For instance, the M23 mutant, a combination of M17 and M18 containing 6 amino acid changes, showed an ∼85% reduction in cell binding as compared to the parental protein. Moreover, M21 and M22 mutants, with 8 amino acid changes (5 from M16 plus 3 from M17/M18), were unable to bind to cells (Fig. 4B). In all cases, similar amounts of protein were used, as confirmed by Western blot (Fig. 4C).
Figure 4.
Cell surface binding of mutant B18 constructs. A) Amino acid sequence of wild-type B18 and mutant derivatives generated. Amino acids corresponding to 78–160 of B18 are shown: boldface indicates putative GAG-binding regions; underscore indicates substituted residues. Horizontal lines mark the putative GAG-binding regions mutagenized: regions A–C. A* indicates the mutant in which a partial substitution of basic residues in region A was carried out. B) HeLa cells were incubated with increasing volumes of supernatants (0–40 μl) containing wild-type B18 or mutant proteins, and binding was detected by flow cytometry using anti-HIS antibody. One representative experiment of 2 is shown. Legend: volumes corresponding to traces. Type of mutation introduced in each protein is indicated in parenthesis. C) Western blot monitoring of protein amounts used in the experiment, developed with anti-V5 antibody.
To confirm that GAG binding was abolished in mutants unable to attach to the cell surface, we purified two of them, M15 and M22, and analyzed by SPR their interaction with GAGs. These mutants were not able to interact with the heparin-coated surface (Table 1 and Supplemental Fig. S2).
The results presented here indicated that basic residues in the 3 regions identified are responsible for its interaction with GAGs.
Abrogation of GAG-binding ability does not impair the IFN-binding and -blocking activities of the B18 protein in solution
To evaluate whether the mutations that hindered binding of B18 to GAGs affected its capacity to interact with IFNs and block their activity, functional assays evaluating the inhibition of type I IFN antiviral activity by B18 mutants were performed. We first tested this activity in a preincubation assay, adding preformed B18-IFN complexes to the cells. As shown in Fig. 5A, all mutants tested were able to efficiently block IFN activity, indicating that the structure of the protein is not drastically affected by the mutations and that the mutations of the residues involved in GAG binding do not alter the IFN-binding capacity in solution. Moreover, by SPR experiments, we determined that the recombinant mutant proteins M15 and M22 still interacted with hIFNα molecules with high affinity, comparable to that of the wild-type B18 protein (Table 1).
Figure 5.
IFN-binding and -blocking activities of B18 and mutant proteins. A) Preincubation assay, in which increasing amounts of supernatants containing the recombinant proteins (0.078–1.25 μl) were preincubated with 50 U of type I IFN, and the mixture was added to HeLa cells. After 16 h of incubation, cells were infected with VSV, and cell viability was determined after 72 h. Controls of treated (+IFN) and untreated (−IFN) cells are indicated. Data are means ± sd of >3 independent experiments done in triplicate. B) Determination of the inhibition of type I IFN antiviral activity by B18 protein and mutants at the cell surface in a wash assay. HeLa cells were incubated with increasing volumes (2.5–40 μl) of supernatants containing the indicated recombinant proteins for 30 min at 37°C. Cells were washed, and 50 U of type I IFN was subsequently added to HeLa cell monolayers. After 16 h incubation, cells were infected with VSV, and cell viability was determined after 72 h. Controls of treated and untreated cells are indicated. Data are means ± sd of >3 independent experiments done in triplicate. C) Inhibition of type I IFN-induced signaling by wild-type B18 and mutant proteins. HeLa cells were incubated with 40 μl of supernatants containing the corresponding proteins; after several washings, cells were left untreated (−) or exposed to 1000 U of hIFNα (+) for 30 min. Samples were analyzed by Western blot using the indicated antibodies. Molecular mass markers (kDa) are at left.
We next carried out a different experiment (washing assay) in which cells were incubated with supernatants containing B18 or the indicated mutants, followed by extensive washings to remove unbound protein prior to the addition of IFN treatment. In this case, none of the mutant proteins were able to counteract the IFN activity as compared to B18, except for M18, which retained the cell-binding activity (Fig. 5B). Mutants with partially reduced cell-binding capacity, M16 and M17, were probably unable to neutralize the IFN activity because the amount of protein retained at the cell surface under these conditions may have not been sufficient to efficiently block the binding of IFN to cellular IFN receptors.
We also determined whether cell surface-associated B18 or derived mutants prevented IFN-induced signaling. As before, HeLa cells were incubated with supernatants containing the indicated proteins and extensively washed prior to hIFNα treatment. The addition of hIFNα induced STAT-1 phosphorylation in control cells. However, as described previously (20), we did not detect STAT-1 phosphorylation in cells preincubated with wild-type B18 protein, which was found by Western blot to bind to the cells and to effectively block hIFNα-induced signaling (Fig. 5C). Of the 8 mutants tested, only the M18 protein was readily detected on cells by Western blot and blocked IFN-induced signaling (Fig. 5C). Consistent with the IFN antiviral activity washing assay (Fig. 5B), the M16 and M17 proteins showed reduced cell binding by Western blot and did not block IFN-mediated signaling (Fig. 5C).
These results showed that binding of B18 to GAGs allows its retention on the cell surface and is required for its anti-IFN blocking activity on cells.
VARV and MPXV B18 orthologues bind to the cell surface through its amino-terminal region
The immunomodulatory properties of the VARV and MPXV B18 orthologues encoded by VARV and MPXV (B17 and B16, respectively) have been recently described, and it has been shown that they also bind to the cell surface (20). Given the important role that cell attachment has for the anti-IFN activity of the B18 protein, we examined in the VARV and MPXV B18 orthologues for the functional conservation of the region involved in protein-GAG interactions. Examination of the amino acid sequence showed a great conservation of the potential GAG-binding regions A–C identified for the VACV B18 protein (Fig. 1). We therefore mutagenized region A, as we did before to generate the VACV mutant M15, in both VARV B17 and MPXV B16 parental proteins. The resultant B16-mutant from MPXV is equal to M15 (denoted B18-mut here), with 8 amino acid changes. B17 has one less basic residue in this region; thus, B17-mutant from VARV encompasses 7 amino acid changes (Fig. 6A). We analyzed the cell-binding ability of these mutant proteins by flow cytometry. As shown in Fig. 6B, none of the mutant proteins were able to attach to cell surface. Nevertheless, all of them could effectively block antiviral hIFNα activities in solution (Fig. 6C), as shown before for the B18 protein.
Figure 6.
Cell-binding ability and IFN-blocking antiviral activity of parental and mutant VARV and MPXV IFNBPs. A) Sequence of parental and mutant derivatives of the VACV (B18), VARV (B17), and MPXV (B16) proteins generated. Amino acids 92–111, 97–114, and 93–112 of the B18, B17, and B16 proteins, respectively, are shown; boldface indicates putative GAG-binding regions; underscore indicates substituted residues. Horizontal lines mark the N-terminal putative GAG-binding region A. B) Flow cytometric analysis (as described in Fig. 2) of HeLa cells incubated with increasing volumes of supernatants (0–40 μl) containing the corresponding tagged proteins. Legend: volumes corresponding traces. C) Western blot monitoring of protein amounts used in the experiment, developed with anti-V5 antibody. D) Increasing volumes of supernatants containing the recombinant proteins (0.625–2.5 μl) were preincubated with 50 U of type I IFN, and the mixture was added to HeLa cells. After 16 h incubation, cells were infected with VSV, and cell viability was determined after 72 h. Controls of infected, treated (+IFN), and untreated (−IFN) cells are also indicated. Data are means ± sd of 2 independent experiments done in triplicate.
These experiments demonstrated that basic regions in the amino-terminal part of IFNα/βBPs from the highly virulent VARV and MPXV are responsible for their interaction with the cell surface, sharing the same strategy as VACV B18.
DISCUSSION
The poxvirus IFNα/βBP was initially identified in VACV, the smallpox vaccine (16, 17), and has been found to be conserved in many poxviruses, including VARV, the causative agent of human smallpox, and MPXV, a related virus that is also highly virulent in humans (20). The conservation of the IFNα/βBP among a variety of poxviruses suggests a critical role in pathogenesis, and this has been experimentally demonstrated in mouse models of VACV and ECTV pathogenesis (16, 17, 22). This viral protein has unique properties, such as a protein structure unrelated to host IFN receptors and its ability to bind IFNs from different animal species, which may reflect the evolutionary history of poxviruses. Another outstanding feature of this secreted protein is its ability to bind to noninfected cells through an unidentified mechanism. As proposed before (18), the binding to the cell membrane could be the best way to transport a protein from infected to neighboring uninfected cells, rapidly spreading inhibition of the antiviral effects of type I IFN at early times of infection and providing the virus with an initial advantage for replication within host tissues.
Therefore, we aimed to study the mechanism by which the soluble viral IFN type I receptor binds to cells (18, 20). The finding of several clusters of positive charges along the protein, particularly within its amino-terminal region, as well as the ability of the VACV B18 protein to bind to many cell types (present work and ref. 18), suggested its potential interaction with GAGs, which are ubiquitously present on the surface of cells.
GAGs are complex linear polysaccharides, usually sulfated, that are covalently attached to a core protein, constituting the proteoglycans, which are present on the surface of virtually all animal cells and in the extracellular matrix. Regardless of being ubiquitous molecules, a big structural variation exists within GAGs due to their composition, linkage, or modifications, such as the number and position of sulfate groups, creating specific and unique patterns of expression and accordingly particular docking sites for numerous proteins (31, 32). Thus, proteoglycans, and especially those harboring the more diverse HS chains, have been recognized as molecules that influence not only numerous biological events but also host-pathogen interrelations (33–36).
We report here that VACV B18 is a GAG-binding protein. The B18 protein did not bind to mutant CHO cells that do not express GAGs on the cell surface or to CHO cells in which GAG sulfation was inhibited. Moreover, recombinant VACV B18 protein was demonstrated by SPR to bind to heparin molecules with high affinity, showing a preference for HS. GAG binding may be an efficient mechanism to spread the B18 protein during infection to distant tissues, retaining it on the surface to prevent the action of IFN type I induced early after infection. The fact that B18 binds preferentially to specific GAGs may explain its differential binding capability to the cell lines tested. This specificity may have implications for the distribution of B18 to different tissues and cell types in vivo, as proposed for other viral proteins, such as the NS1 protein from dengue virus (37).
Binding to GAGs is a mechanism frequently used by viral proteins to attach to the cell surface. Several viral structural proteins interact with GAGs to allow initial binding of the incoming virion to the cell surface, to facilitate interaction with specific receptors and viral entry. Well-known examples are HIV gp120 (38) or glycoproteins gB, gC, and gD from herpes simplex virus 1 and 2 (39). In the case of VACV, A27, D8, and H3 proteins have been reported to interact with GAGs (40–43). In addition, other poxviral immunomodulatory proteins have been found to use the same mechanism for cell attachment: the ECTV chemokine-binding protein E163 (27), the IL-18 binding protein from molluscum contagiosum virus and VARV (44, 45), and the complement control protein from VACV and VARV (46, 47). In all cases, the affinities for protein-GAG interaction observed were in the range of 0.5 to 55 nM, similar to that determined here for VACV B18 (3.8 nM). This suggests that the GAG-mediated attachment to the cell surface may potentiate the immunomodulatory activity of secreted poxvirus proteins, most likely locating them in its place of action.
GAG-binding motifs in proteins are different in sequence and not exclusively linear motifs. Moreover, distant amino acids can contribute to form a defined cluster at the surface of the folded protein (31, 32). We have identified specific residues involved in B18-GAG interaction and have shown that the overall positive charge in the first Ig domain of the B18 protein determines its capacity of attachment to the cell surface, with a number of interactions between the sulfate GAG structure and the key residues present in the specific positive charged clusters in the protein being required to achieve efficient binding. Overall, it seems that the binding of B18 to GAGs is mainly dominated by electrostatic interactions, although additional specific interactions between carbohydrate sequences and key amino acids in the protein may be determinant, as observed for the heparin-binding regions of other proteins, like for VACV A27 (48).
Our mutagenesis studies of the residues involved in GAG binding indicate that the amino-terminal part of the IFNα/βBP is crucial for its cell-binding ability. Experimental evidence for the involvement of the amino-terminal region in cell binding has also been described recently (49). These results are consistent with previous studies with the VACV strain Wyeth, which encodes a truncated IFNα/βBP lacking the third Ig domain that retains the ability to attach to the cell surface but has lost some affinity for IFN (18). VACV Wyeth, also known as DryVax, was a commonly used smallpox vaccine, and a plaque-purified VACV Wyeth constitutes the second-generation smallpox vaccine VACV ACAM2000. Interestingly, VACV-modified virus Ankara (MVA) is a promising nonreplicating third-generation smallpox vaccine and also encodes a truncated version of the IFNα/βBP lacking the third Ig domain (50). The expression of a truncated IFNα/βBP by these smallpox vaccines may partly account for their attenuated phenotype. It should also be considered that immunity to the IFNα/βBP has been shown to mediate protection from mousepox, a smallpox-like disease caused by the highly virulent ECTV (22). Thus, the expression of a full-length B18 polypeptide, mutagenized to lose IFN inhibitory activity, may improve the potency of these VACV vaccines against virulent poxviruses expressing the IFNα/βBP, such as VARV or MPXV.
The secreted IFNα/βBP (Y136) encoded by the more distantly related yaba-like disease virus blocks not only type I IFN but also type III IFN and has been found to attach to the cell surface (51). It will be interesting to determine whether this variant IFNα/βBP interacts with GAGs. Examination of Y136 sequence did not reveal a conservation of the amino acid motifs described here to be involved in the interaction of IFNα/βBP from VACV, VARV, or MPXV with GAGs (data not shown). However, it does not imply that binding does not occur via GAGs, as GAG-binding motifs are not necessarily conserved between proteins (32). For example, the canonical GAG-binding motif BBXB is found in the second Ig domain (HAKH) and the C terminus (KKIK) of the Y136 protein sequence (data not shown).
The results presented here also demonstrate that the capacity of the IFNα/βBP to bind IFN is not affected by its interaction with GAGs, as mutant proteins that are not able to bind to the cell surface still retain their structure to interact with IFN with the same affinity and protect from IFN-mediated antiviral effects in tissue culture. The fact that these mutant proteins cannot inhibit IFN activity at the cell surface, as they are easily washed away from it, could have important consequences in vivo. Thus, the construction of VACV or ECTV recombinants expressing an IFNα/βBP mutant unable to interact with GAGs but fully competent to bind and inhibit type I IFNs will be of interest to determine the importance of cell surface binding for viral pathogenesis and immune modulation. The interaction with GAGs may confer to the IFNα/βBP the ability to cover distant uninfected areas and to generate an IFN-free environment that will help the virus to replicate (18). If this were the case, new avenues for the treatment of poxviral disease would be opened through the use of GAG and/or peptide analogues to impair binding of B18 to the cell surface in vivo. This strategy has been tested before. For example, a modified heparin octasaccharide successfully inhibits herpes simplex virus 1 entry by competing gD-GAG interactions (52), and selective derivatives of sulfated K5, a polysaccharide of Escherichia coli with a heparin-like structure, blocks HIV Tat protein function by inhibiting its binding to the cell surface GAGs (53). The results described here lead to future work that will be of interest for the generation of safer smallpox vaccines.
Notably, we show that the IFNα/βBPs encoded by the highly virulent human pathogens VARV and MPXV bind to the cell surface using similar GAG-binding motifs. Site-directed mutagenesis of VARV and MPXV basic residues in the first Ig domain of the protein rendered IFNα/βBPs unable to interact with the cell surface, while retaining IFN inhibitory activity. These results suggest that GAG binding is a conserved feature and likely to be essential for the in vivo activity of the IFNα/βBP encoded by VARV and MPXV.
By using a combination of site-directed mutagenesis, SPR technology, cell-binding experiments, and IFN functional assays, we have identified specific binding sites involved in the GAG-mediated interaction of the poxvirus IFNα/βBP with the cell surface. The results described here uncover the molecular events that confer to the viral IFNα/βBP a unique property that retains the IFN inhibitor in infected tissues. Unraveling the mechanisms of immune modulation by poxviral virulence factors is important to understand the mechanisms of pathogenesis of human smallpox and monkeypox, and is of relevance for the development of safer VACV vaccines, as well as new therapeutic strategies.
Supplementary Material
Acknowledgments
The authors thank members of the laboratory for helpful suggestions and R. Martín for technical assistance.
This work was supported by U.S. National Institute of Allergy and Infectious Diseases grant U54AI057160-07 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases and grants from the Wellcome Trust and Comunidad de Madrid.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Randall R. E., Goodbourn S. (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 2. Versteeg G. A., Garcia-Sastre A. (2010) Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 13, 508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith G. L., McFadden G. (2002) Smallpox: anything to declare? Nat. Rev. 2, 521–527 [DOI] [PubMed] [Google Scholar]
- 4. Likos A. M., Sammons S. A., Olson V. A., Frace A. M., Li Y., Olsen-Rasmussen M., Davidson W., Galloway R., Khristova M. L., Reynolds M. G., Zhao H., Carroll D. S., Curns A., Formenty P., Esposito J. J., Regnery R. L., Damon I. K. (2005) A tale of two clades: monkeypox viruses. J. Gen. Virol. 86, 2661–2672 [DOI] [PubMed] [Google Scholar]
- 5. Di Giulio D. B., Eckburg P. B. (2004) Human monkeypox: an emerging zoonosis. Lancet Infect. Dis. 4, 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reed K. D., Melski J. W., Graham M. B., Regnery R. L., Sotir M. J., Wegner M. V., Kazmierczak J. J., Stratman E. J., Li Y., Fairley J. A., Swain G. R., Olson V. A., Sargent E. K., Kehl S. C., Frace M. A., Kline R., Foldy S. L., Davis J. P., Damon I. K. (2004) The detection of monkeypox in humans in the Western Hemisphere. New Engl. J. Med. 350, 342–350 [DOI] [PubMed] [Google Scholar]
- 7. Rimoin A. W., Mulembakani P. M., Johnston S. C., Lloyd Smith J. O., Kisalu N. K., Kinkela T. L., Blumberg S., Thomassen H. A., Pike B. L., Fair J. N., Wolfe N. D., Shongo R. L., Graham B. S., Formenty P., Okitolonda E., Hensley L. E., Meyer H., Wright L. L., Muyembe J. J. (2010) Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107, 16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muller U., Steinhoff U., Reis L. F., Hemmi S., Pavlovic J., Zinkernagel R. M., Aguet M. (1994) Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 9. Van den Broek M. F., Muller U., Huang S., Aguet M., Zinkernagel R. M. (1995) Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J. Virol. 69, 4792–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deonarain R., Alcami A., Alexiou M., Dallman M. J., Gewert D. R., Porter A. C. (2000) Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 74, 3404–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karupiah G., Fredrickson T. N., Holmes K. L., Khairallah L. H., Buller R. M. (1993) Importance of interferons in recovery from mousepox. J. Virol. 67, 4214–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodriguez J. R., Rodriguez D., Esteban M. (1991) Interferon treatment inhibits early events in vaccinia virus gene expression in infected mice. Virology 185, 929–933 [DOI] [PubMed] [Google Scholar]
- 13. Perdiguero B., Esteban M. (2009) The interferon system and vaccinia virus evasion mechanisms. J. Interferon Cytokine Res. 29, 581–598 [DOI] [PubMed] [Google Scholar]
- 14. Meng X., Jiang C., Arsenio J., Dick K., Cao J., Xiang Y. (2009) Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J. Virol. 83, 10627–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alcami A. (2003) Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. 3, 36–50 [DOI] [PubMed] [Google Scholar]
- 16. Symons J. A., Alcami A., Smith G. L. (1995) Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81, 551–560 [DOI] [PubMed] [Google Scholar]
- 17. Colamonici O. R., Domanski P., Sweitzer S. M., Larner A., Buller R. M. (1995) Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 270, 15974–15978 [DOI] [PubMed] [Google Scholar]
- 18. Alcami A., Symons J. A., Smith G. L. (2000) The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74, 11230–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith V. P., Alcami A. (2002) Inhibition of interferons by ectromelia virus. J. Virol. 76, 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez de Marco M. M., Alejo A., Hudson P., Damon I. K., Alcami A. (2010) The highly virulent variola and monkeypox viruses express secreted inhibitors of type I interferon. FASEB J. 24, 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esteban D. J., Buller R. M. (2005) Ectromelia virus: the causative agent of mousepox. J. Gen. Virol. 86, 2645–2659 [DOI] [PubMed] [Google Scholar]
- 22. Xu R. H., Cohen M., Tang Y., Lazear E., Whitbeck J. C., Eisenberg R. J., Cohen G. H., Sigal L. J. (2008) The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. J. Exp. Med. 205, 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith G. L., Chan Y. S. (1991) Two vaccinia virus proteins structurally related to the interleukin-1 receptor and the immunoglobulin superfamily. J. Gen. Virol. 72, 511–518 [DOI] [PubMed] [Google Scholar]
- 24. Alcami A., Smith G. L. (1992) A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell 71, 153–167 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J., Russell D. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA [Google Scholar]
- 26. Baeuerle P. A., Huttner W. B. (1986) Chlorate—a potent inhibitor of protein sulfation in intact cells. Biochem. Biophys. Res. Commun. 141, 870–877 [DOI] [PubMed] [Google Scholar]
- 27. Ruiz-Arguello M. B., Smith V. P., Campanella G. S., Baleux F., Arenzana-Seisdedos F., Luster A. D., Alcami A. (2008) An ectromelia virus protein that interacts with chemokines through their glycosaminoglycan binding domain. J. Virol. 82, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morikawa S., Ueda Y. (1993) Characterization of vaccinia surface antigen expressed by recombinant baculovirus. Virology 193, 753–761 [DOI] [PubMed] [Google Scholar]
- 29. Cardin A. D., Weintraub H. J. (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Lawrence R., Frazier B. A., Esko J. D. (2006) CHO glycosylation mutants: proteoglycans. Methods Enzymol. 416, 205–221 [DOI] [PubMed] [Google Scholar]
- 31. Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. (1998) Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20, 156–167 [DOI] [PubMed] [Google Scholar]
- 32. Gandhi N. S., Mancera R. L. (2008) The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Design 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 33. Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 34. Taylor K. R., Gallo R. L. (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 [DOI] [PubMed] [Google Scholar]
- 35. Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 36. Bartlett A. H., Park P. W. (2010) Proteoglycans in host-pathogen interactions: molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 12, e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avirutnan P., Zhang L., Punyadee N., Manuyakorn A., Puttikhunt C., Kasinrerk W., Malasit P., Atkinson J. P., Diamond M. S. (2007) Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathogens 3, e183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saphire A. C., Bobardt M. D., Zhang Z., David G., Gallay P. A. (2001) Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 75, 9187–9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shukla D., Spear P. G. (2001) Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chung C. S., Hsiao J. C., Chang Y. S., Chang W. (1998) A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72, 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsiao J. C., Chung C. S., Chang W. (1998) Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72, 8374–8379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hsiao J. C., Chung C. S., Chang W. (1999) Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73, 8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin C. L., Chung C. S., Heine H. G., Chang W. (2000) Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74, 3353–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiang Y., Moss B. (2003) Molluscum contagiosum virus interleukin-18 (IL-18) binding protein is secreted as a full-length form that binds cell surface glycosaminoglycans through the C-terminal tail and a furin-cleaved form with only the IL-18 binding domain. J. Virol. 77, 2623–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Esteban D. J., Nuara A. A., Buller R. M. (2004) Interleukin-18 and glycosaminoglycan binding by a protein encoded by Variola virus. J. Gen. Virol. 85, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 46. Smith S. A., Mullin N. P., Parkinson J., Shchelkunov S. N., Totmenin A. V., Loparev V. N., Srisatjaluk R., Reynolds D. N., Keeling K. L., Justus D. E., Barlow P. N., Kotwal G. J. (2000) Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 74, 5659–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liszewski M. K., Bertram P., Leung M. K., Hauhart R., Zhang L., Atkinson J. P. (2008) Smallpox inhibitor of complement enzymes (SPICE): regulation of complement activation on cells and mechanism of its cellular attachment. J. Immunol. 181, 4199–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shih P. C., Yang M. S., Lin S. C., Ho Y., Hsiao J. C., Wang D. R., Yu S. S., Chang W., Tzou D. L. (2009) A turn-like structure “KKPE” segment mediates the specific binding of viral protein A27 to heparin and heparan sulfate on cell surfaces. J. Biol. Chem. 284, 36535–36546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Golden J. W., Hooper J. W. Evaluating the orthopoxvirus type I interferon-binding molecule as a vaccine target in the vaccinia virus intranasal murine challenge model. Clin. Vaccine Immunol. 17, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Antoine G., Scheiflinger F., Dorner F., Falkner F. G. (1998) The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244, 365–396 [DOI] [PubMed] [Google Scholar]
- 51. Huang J., Smirnov S. V., Lewis-Antes A., Balan M., Li W., Tang S., Silke G. V., Putz M. M., Smith G. L., Kotenko S. V. (2007) Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc. Natl. Acad. Sci. U. S. A. 104, 9822–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Copeland R., Balasubramaniam A., Tiwari V., Zhang F., Bridges A., Linhardt R. J., Shukla D., Liu J. (2008) Using a 3-O-sulfated heparin octasaccharide to inhibit the entry of herpes simplex virus type 1. Biochemistry 47, 5774–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Urbinati C., Bugatti A., Oreste P., Zoppetti G., Waltenberger J., Mitola S., Ribatti D., Presta M., Rusnati M. (2004) Chemically sulfated Escherichia coli K5 polysaccharide derivatives as extracellular HIV-1 Tat protein antagonists. FEBS Lett. 568, 171–177 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.