BRCA1 interacts with ERM proteins at leading edges and focal adhesion sites and modulates motility via its ubiquitin ligase activity.
Abstract
BRCA1 C-terminal (BRCT) domains in BRCA1 are essential for tumor suppressor function, though the underlying mechanisms remain unclear. We identified ezrin, radixin, and moesin as BRCA1 BRCT domain–interacting proteins. Ezrin–radixin–moesin (ERM) and F-actin colocalized with BRCA1 at the plasma membrane (PM) of cancer cells, especially at leading edges and focal adhesion sites. In stably expressing cancer cells, high levels of enhanced green fluorescent protein (EGFP)-BRCA11634–1863 acted as a dominant-negative factor, displacing endogenous BRCA1 from the PM. This led to delayed cell spreading, increased spontaneous motility, and irregular monolayer wound healing. MCF-7 cells (intact BRCA1) showed lower motility than HCC1937 cells (truncated BRCA1), but expression of EGFP-BRCA11634–1863 in MCF-7 increased motility. Conversely, full-length BRCA1 expression in HCC1937 decreased motility but only if the protein retained ubiquitin ligase activity. We conclude that full-length BRCA1 is important for complete tumor suppressor activity via interaction of its BRCT domains with ERM at the PM, controlling spreading and motility of cancer cells via ubiquitin ligase activity.
Introduction
The identification of Mendelian inheritance of increased vulnerability to breast and ovarian cancer drove a search for the genes involved, culminating in the discovery of two tumor suppressor genes, BRCA1 and BRCA2. Germline mutations in BRCA1 are associated with an increased susceptibility to breast and ovarian cancer. Yet the biological functions underlying its role in carcinogenesis are still unclear. BRCA1 encodes a 1,863-aa tumor suppressor protein that contains several functional motifs, including an N-terminal RING (really interesting new gene) domain and a pair of ∼90 amino acid repeats at the C terminus called BRCA1 C-terminal (BRCT) domains (Fig. S1; Koonin et al., 1996).
BRCT domains have since been identified in other proteins known to play a role in regulation of DNA damage responses and cell cycle checkpoint-mediated repair (Bork et al., 1997; Glover et al., 2004). The paired BRCT domains have been shown to be important for nuclear targeting of BRCA1 (Rodriguez et al., 2004), transcription regulation (Monteiro, 2000), binding to DNA strand breaks, multimer formation at DNA fragment ends (Yamane et al., 2000), and altering chromatin structure (Hu et al., 1999). BRCT domains are essential for the tumor suppression activity of BRCA1 as protein truncations, and missense variants within these domains are associated with human breast and ovarian cancers (Glover, 2006). Moreover, they function as a phosphopeptide-binding module with a pSer-X-X-Phe phosphoserine specificity (Glover et al., 2004) and may thus recruit phosphorylated signaling factors or complexes related to the DNA damage response.
Despite its central importance to tumor suppression activity, it is still uncertain how exactly BRCA1 works. The only known enzymatic activity for BRCA1 is that of an E3 ubiquitin ligase of the RING subclass when in a heterodimer with BARD1 (Starita et al., 2004; Starita and Parvin, 2006). This complex can auto-ubiquitinate the BRCA1 component, yet other functionally relevant targets have not been systematically studied.
We have previously shown that hyperphosphorylated BRCA1 is present in mitochondria, which is consistent with a function in mitochondrial DNA maintenance (Coene et al., 2005). To further investigate mitochondrial functions of BRCA1, we searched for interaction partners of the BRCT domains of BRCA1 using ligand overlay blotting and coimmunoprecipitation. HeLa postnuclear membrane pellet fractions were used to exclude the large family of nuclear BRCT-containing proteins and their interacting partners. This approach identified components of a mitochondrial import pathway, which will be described elsewhere (unpublished data). These experiments also uncovered an unexpected association between BRCT domains and a family of cytoskeletal regulatory proteins, prompting the study of motility in cells in which these BRCA1 interactions were reduced by competition or mutation. These analyses strongly implicate BRCA1 in the regulation of motility and migration of cancer cells.
Results
BRCA1 BRCT domains interact with ezrin, radixin, and moesin
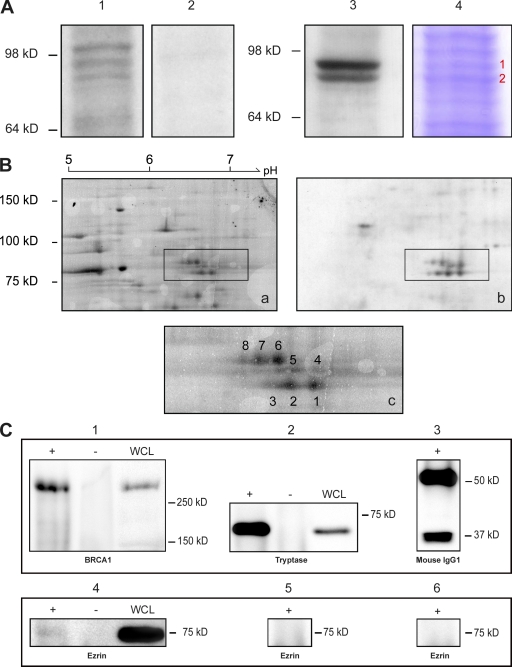
To identify BRCT-interacting proteins, HeLa postnuclear membrane pellet (P2) samples were resolved by 1D SDS-PAGE followed by ligand overlay blotting using as a probe an EGFP-tagged BRCT region (1,634–1,863 aa) containing both BRCT domains (henceforth referred to as EGFP-CTD; Fig. S1). A doublet of 75 and 80 kD was reproducibly detected using an EGFP-CTD–expressing HeLa lysate (Fig. 1 A, lane 3), whereas the detection of a third higher molecular mass band was not (Fig. 1 A, lane 1). The doublet was absent when incubating with either an EGFP–N-terminal acidic domain of cortactin (EGFP-NTA) control lysate (Fig. 1 A, lane 2) or an untransfected lysate (not depicted). The 75–80-kD doublet can therefore be regarded as a result of a specific interaction with the EGFP-CTD protein. The 80- and 75-kD bands (indicated by 1 and 2 in red in Fig. 1 A, lane 4) were excised, digested, and sequenced by liquid chromatography and tandem mass spectrometry (LC-MS/MS), identifying ezrin, radixin, and moesin among other proteins with corresponding high Mascot score values (Table S1, showing results for bands 1 and 2).
Figure 1.
Direct BRCA1–ERM interaction detected by ligand overlay and coimmunoprecipitation. (A and B) HeLa P2 fractions were resolved by 1D SDS-PAGE (A) or 2D electrophoresis (B), blotted, and probed with EGFP-CTD detected with anti-EGFP antibody. (A) A 75–80-kD doublet was reproducibly detected (lane 3), whereas a third higher molecular mass band was not (lane 1). No signal was detected in a parallel blot using the control HeLa lysate containing EGFP-NTA (lane 2). The 75–80-kD doublet (lane 4) was excised, trypsin digested, and sequenced by LC-MS/MS (Table S1). The red 1 and 2 indicate the 80- and 75-kD bands, respectively. (B) 2D gel electrophoresis: multiple spots were detected in the 75–80-kD region. (a) SYPRO ruby staining; (b) Western blot; (c) detail of SYPRO ruby spots (1–8) cut out for LC-MS/MS analysis (Table S2, showing results for each spot). (C) Coimmunoprecipitation of ezrin with BRCA1. (top, 1) Lysate + beads + mouse anti-BRCA1 Ab1 (+), lysate + beads with no antibody (−), and whole cell lysate (WCL) probed with mouse anti-BRCA1 Ab1; (2) lysate + beads + mouse antitryptase (+), lysate + beads with no antibody (−), and whole cell lysate probed with mouse antitryptase; (3) beads + mouse IgG1 probed with anti–mouse IgG1. (bottom) The same samples as in the top immunoblotted for ezrin. Ezrin protein was only detected in the BRCA1 sample (4) when Ab1 was coupled to the beads (+). Beads with no antibody (−) and both the tryptase and mouse IgG1 control samples (5 and 6, respectively) were negative for ezrin. In replicate experiments, ezrin was reproducibly detected (mean band intensity in the BRCA1 pull-down 20.0 [experiment 1] or 29.9 [experiment 2] vs. intensity in control pull-down 3.25 [experiment 1] or 1.03 [experiment 2]).
To refine the identification of BRCT-binding proteins present within the 75–80-kD doublet, 2D electrophoresis ligand overlay blotting on HeLa P2 fractions was performed. The overlay blot revealed multiple spots in the region of 75–80 kD (Fig. 1 B, spots marked from 1 to 8). LC-MS/MS analysis demonstrated the presence of different isoforms of ezrin, radixin, and moesin with high Mascot score values (Table S2, showing results for spots 1–8), confirming the 1D ligand overlay results. Indeed, for all but one of these reactive spots, ezrin, radixin, or moesin represented the highest scoring and often sole protein identified. Under these denatured conditions, the BRCT domains of BRCA1 interact independently with ezrin, radixin, and moesin.
Ezrin coimmunoprecipitates with BRCA1
To confirm a direct interaction between BRCA1 and ezrin in native conditions, we performed coimmunoprecipitation on HeLa cell lysates. The BRCA1 protein was pulled down using anti-BRCA1 antibody–coupled beads (Fig. 1 C). A control protein (tryptase) was pulled down using antitryptase-coated beads, whereas mouse IgG1–coated beads were used as a negative control (Fig. 1 C). When the same samples were probed with an antiezrin antibody, the ezrin protein was only detected in the BRCA1 pull-down sample, whereas no signal was detected in either the tryptase or mouse IgG1 control samples (Fig. 1 C, bottom) or beads incubated with lysate but no antibody. These results confirmed a direct interaction between BRCA1 and ezrin in a native context.
BRCA1 colocalizes with the ezrin–radixin–moesin (ERM) complex
ERM proteins are known to act as cross-linkers between the plasma membrane (PM) and the underlying F-actin cytoskeleton. To confirm the biochemical results and to investigate a potential involvement of BRCT domains in the ERM–F-actin interaction, immunofluorescence labeling was performed on cancer cells transiently expressing EGFP-CTD. All cell lines showed light microscope colocalization of the EGFP-CTD protein with both the ERM complex and F-actin at the PM (Figs. 2 and S2, A and B). Higher magnification of membrane extensions (Fig. 2 D, insets) frequently revealed the EGFP-CTD protein at sites where the ERM complex associated with F-actin. We next asked whether endogenous (full length) BRCA1 is also found at sites targeted by BRCT domains alone. Again, a similar distribution at the PM was seen by both light microscopy (Figs. S2 C and S3) and electron cryomicroscopy (Fig. S1 B). These results confirm the association of these proteins in a cellular context.
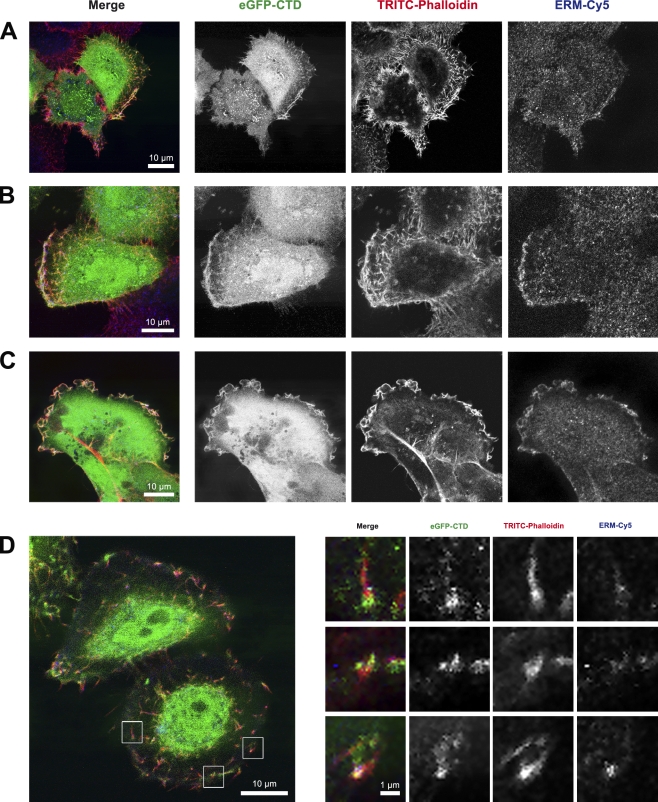
Figure 2.
EGFP-CTD colocalization with ERM and F-actin. (A–D) CLSM of transiently EGFP-CTD–expressing HeLa (A, B, and D) and HCC1937 (C) cells labeled with TRITC-phalloidin (for F-actin) and anti-ezrin–radixin–moesin (ERM) antibody detected with Cy5. Colocalization was observed at the PM (A), leading edge (B), membrane ruffles (C), and PM extensions (D). (D) Higher magnifications of three regions (insets) show EGFP-CTD localization with respect to ERM and F-actin.
Colocalization of EGFP-CTD with F-actin and the ERM complex was also observed at focal adhesions in residual membrane sheets after mechanical shearing (Fig. 3 A), suggesting that all three proteins are present at sites of cell substrate adhesion. Immunofluorescence labeling of transiently transfected pEGFP-CTD HeLa cells for F-actin and anti-FAK demonstrated the colocalization of all three proteins at PM ruffles (Fig. S4 A), focal adhesion plaques (Fig. S4 B), and at cellular footprints after mechanical cell removal (Fig. S4 C). A line scan across the PM ruffles showed the individual distribution of EGFP-CTD, F-actin, and ERM proteins (Fig. 3 B). Together with consecutive confocal xz sections (Fig. 3 C), we conclude that all three proteins colocalized in membrane extensions (microvilli) and adhesion points.
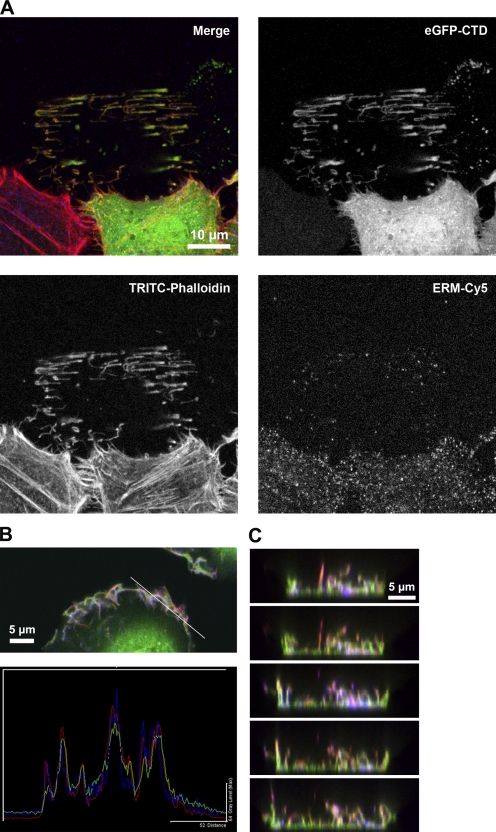
Figure 3.
EGFP-CTD localization in focal adhesion plaques. (A) HeLa cells transiently expressing EGFP-CTD were mechanically sheared and labeled with TRITC-phalloidin (for F-actin) and anti-ERM antibody (detected with Cy5). Colocalization of all three proteins was observed in the focal adhesion plaques and remaining structures. (B) Line scan across PM ruffles of a PMA-stimulated HeLa cell showing EGFP-CTD (green), F-actin (red), and ERM (blue) distribution. The white line in the top image indicates the position of the line scan shown beneath. (C) Consecutive CLSM xz sections of the PM region of a transiently EGFP-CTD–expressing HeLa cell labeled with TRITC-phalloidin and anti-ERM antibody (Cy5). Colocalization of all three labels was observed in membrane extensions and cell substrate adhesion points, resulting in a white color.
To exclude the possibility that high expression levels of a GFP-tagged protein were giving rise to nonspecific cytoplasmic filling extending into membrane ruffles, we analyzed two EGFP control proteins. HeLa cells transiently transfected with pEGFP-neo or pEGFP-centrin were double labeled for F-actin and ERM. At high expression levels, we observed a cytoplasmic distribution of both soluble EGFP and EGFP-centrin extending into membrane ruffles. Removing the soluble pool of each EGFP fusion protein by saponin permeabilization before fixation and staining leaves behind only stably bound protein; focally targeted EGFP-CTD persisted after this treatment (Fig. S5 A), whereas EGFP-centrin is found at centrioles only (Fig. S5 B), and the control soluble EGFP is completely removed (Fig. S5 C). Saponin treatment damaged the membrane ruffles, resulting in the loss of both EGFP-CTD and ERM labeling at the PM (Fig. S5 A). The punctate structures labeled by EGFP-CTD under these conditions were shown to be focal adhesion plaques in an experiment using an anti-FAK label to detect FAK (Fig. S6). Colocalization of EGFP-CTD, F-actin, and FAK was detected in these focal adhesion plaques. No such signal was observed for either soluble EGFP or EGFP-centrin, confirming the specificity of the EGFP-CTD construct and the reliability of its subcellular localization. BRCA1 is thus positioned to have a role in membrane–cytoskeletal interactions with possible functional consequences.
BRCA1 BRCT domains interact with native ezrin, radixin, and moesin: FLIM/FRET
To confirm a direct BRCA1–ERM cellular interaction, we used two resonant energy transfer methods. We performed acceptor photobleach fluorescence resonance energy transfer (FRET) on both transiently EGFP-CTD–expressing HeLa cells and HeLa cells labeled for endogenous BRCA1. Significant FRET was detected between labeled endogenous BRCA1 and ERM at the PM (mean FRET efficiency of 5.6%, P < 0.05; Fig. 4, A–C), indicating the two proteins are within nanometers of each other and most probably interacting. This finding was confirmed by fluorescence lifetime imaging microscopy (FLIM; Peter and Ameer-Beg, 2004; Peter et al., 2005) on transiently EGFP-CTD–expressing HeLa cells, in which FRET was confirmed by measuring a shortened lifetime of the donor (EGFP-CTD) in the presence of an acceptor (Cy3, which was used to detect ERM; Fig. 4, D–F).
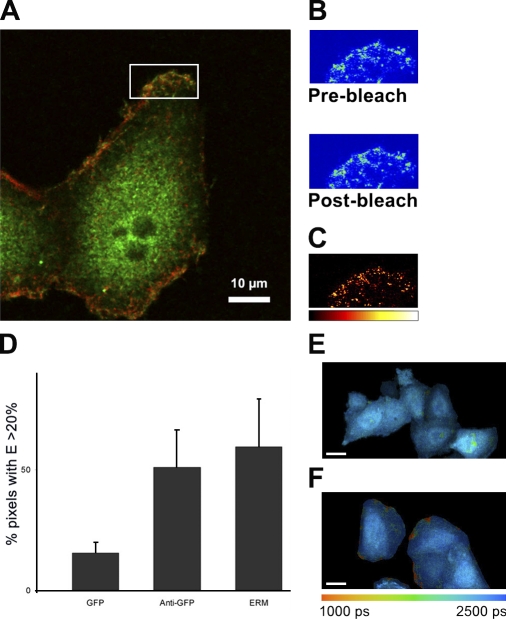
Figure 4.
FRET analyses of BRCA1 and its interacting partners at the PM. (A) Acceptor photobleach FRET on PMA-stimulated HeLa cells. Endogenous BRCA1 was detected using Alexa Fluor 488 (donor); ERM was detected using Cy3 (acceptor). Increase in donor fluorescence was measured after photobleaching of the acceptor. (B) Pre- and postbleach images are shown for a representative ROI (white box in A). (C) Absolute increase in donor fluorescence mean FRET efficiency. 5.79% ± SEM = 0.85 (n = 23). A negative control (Cy3 acceptor targeted to the transferrin receptor) gave a mean FRET efficiency of 1.37 ± SEM = 0.97 (n = 15). P < 0.001 (t test). (D–F) FLIM-based FRET between EGFP-CTD and ERM (detected using Cy3) in untreated HeLa cells. Graph shows the percentage of pixels with a lifetime shortened enough to indicate a FRET efficiency (E) >20% (means ± SEM; n = 5). There is a significant difference (P < 0.05) between EGFP lifetime in the absence of an acceptor (representative cells shown in E) and when ERM-Cy3 is present as an acceptor (representative cells shown in F). A positive control in which the Cy3 acceptor was targeted to EGFP-CTD using an anti-GFP antibody and Cy3 is also shown (anti-GFP). (E and F) Bars, 10 µm.
PMA and cytochalasin B affect F-actin, ERM, and EGFP-CTD behavior
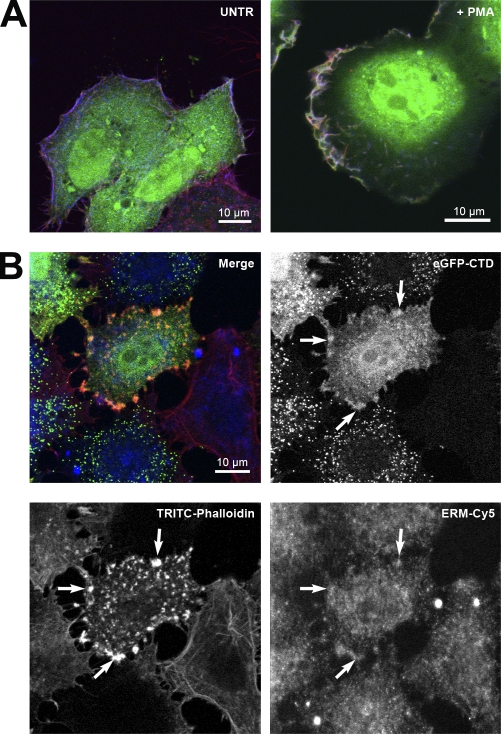
To study the influence of motility on the behavior of BRCA1 at the PM, we induced cell motility by PMA stimulation (Nomura et al., 2007) and inhibited cell motility by cytochalasin B treatment (Haynes and Weller, 1978). Double labeling for ERM and F-actin in HeLa cells transiently expressing EGFP-CTD (Fig. 5 A) showed an apparent increase in colocalization of all three proteins at the PM ruffles after PMA treatment, suggesting that BRCT domain recruitment may be increased by this treatment, and thus, BRCA1 may have an involvement in cell motility. To discard the possibility of a fixation artifact, we used native GFP fluorescence from our fusion protein for time-lapse imaging in living cells. These experiments confirmed an association of EGFP-CTD with highly dynamic membrane ruffles (Video 1).
Figure 5.
EGFP-CTD localization after PMA or cytochalasin B treatment. (A) Untreated (UNTR) transiently EGFP-CTD–expressing HeLa cells labeled with TRITC-phalloidin (for F-actin) and anti-ERM antibody (detected with Cy5) showed discrete colocalization at the PM (left), whereas enhanced colocalization was observed at the leading edges of the cell after PMA stimulation, resulting in a white color (right). (B) Cytochalasin B treatment of transiently EGFP-CTD–expressing HeLa cells followed by labeling with TRITC-phalloidin for F-actin and anti-ERM antibody (Cy5). The merged CLSM image confirms the presence of distinct structures containing all three proteins at the PM (arrows).
After cytochalasin B treatment, double labeling of HeLa cells transiently expressing EGFP-CTD showed colocalization of EGFP-CTD with F-actin and ERM at distinct patches at the PM (Fig. 5 B, arrows). The F-actin signal was absent except in those patches, suggesting that this pool may be protected from depolymerization by formation of a complex including EGFP-CTD and ERM.
EGFP-CTD competes with endogenous BRCA1
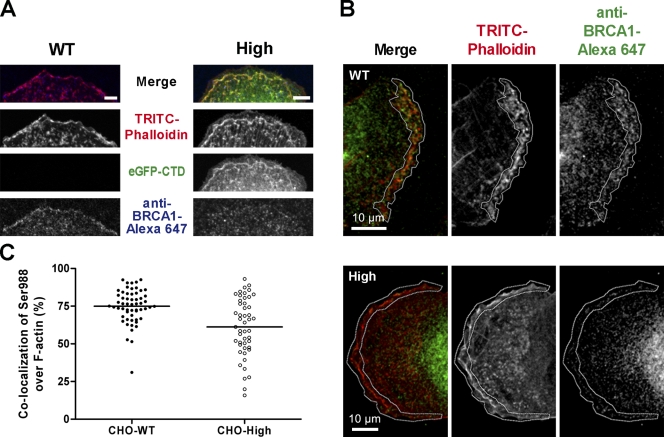
Both endogenous BRCA1 and EGFP-CTD proteins associated with the ERM complex and F-actin. This prompted us to speculate that the recombinant C-terminal fragment might compete for ERM and/or F-actin binding and displace endogenous BRCA1 protein. To test this, we used TRITC-phalloidin to label F-actin (to prevent possible steric hindrance by an additional antibody pair) and an anti-pSer988 BRCA1 antibody on CHO–wild-type (WT) and CHO cells highly expressing EGFP-CTD (CHO-High). The detection of BRCA1 in CHO cells was confirmed by Western blotting (Fig. S1 C). Single planes from confocal z stacks showed that, in CHO-WT, endogenous BRCA1 was concentrated along the PM, colocalizing with F-actin (Fig. 6 A). In the CHO-High cells, however, endogenous BRCA1 was absent in most places along the PM where EGFP-CTD was strongly expressed.
Figure 6.
Displacement of endogenous BRCA1. (A) CLSM of PMA-stimulated CHO–wild type (CHO-WT) and CHO cells stably expressing high levels of EGFP-CTD (CHO-High) labeled with TRITC-phalloidin and anti-Ser988 BRCA1 antibody (Alexa Fluor 647). Pronounced PM colocalization of endogenous BRCA1 and F-actin was observed in CHO-WT (left). In CHO-High, PM colocalization of EGFP-CTD and F-actin was observed, whereas endogenous BRCA1 was absent (right). The endogenous BRCA1 distribution in the cytoplasm remained unchanged. Bars, 5 µm. (B) Mean intensity z projection of representative TRITC-phalloidin– and anti-Ser988 BRCA1 antibody (Alexa Fluor 647)–labeled CHO-WT and CHO-High z stacks. PM colocalization of endogenous BRCA1 and F-actin was prominent in CHO-WT cells but clearly reduced in CHO-High cells. The marked white outlines indicate the selected area for calculation of colocalization parameters. (C) Quantitative displacement graph showing the distribution of all colocalization percentage values of Ser988 BRCA1 over F-actin from each plane of seven individual z stacks per CHO cell type. Overlap of Ser988 BRCA1 with F-actin is higher in CHO-WT than in CHO-High: CHO-WT, mean = 74.97 ± SEM = 1.49 (n = 57); CHO-High, mean = 61.17 ± SEM = 2.791 (n = 48). Horizontal lines indicate means. Nonparametric two-tailed Mann–Whitney test, P < 0.001; unpaired two-tailed t test with Welch’s correction, P < 0.001.
We then quantified the displacement of endogenous BRCA1 protein using a MetaMorph macro to calculate colocalization in every plane of multiple z stacks. Mean intensity projection of z stacks showed an obvious endogenous BRCA1 signal at the defined PM region for CHO-WT, whereas a clearly diminished signal was seen for CHO-High (Fig. 6 B). After analyzing the colocalization overlap parameters for every image plane in multiple z stacks for both CHO-WT and CHO-High, a mean of 75% of endogenous BRCA1 was found to overlap with F-actin for CHO-WT, whereas for CHO-High a mean value of only 60% was found (Fig. 6 C). The individual colocalization values were mainly concentrated around the mean for CHO-WT, whereas the distribution was more dispersed with lower colocalization values for CHO-High. This difference was highly significant (P < 0.001 by either a nonparametric two-tailed Mann–Whitney test or an unpaired two-tailed t test with Welch’s correction).
These data demonstrate competition between the EGFP-CTD chimera and endogenous BRCA1, resulting in displacement of endogenous BRCA1 protein. The specificity of this competition and displacement is further addressed later in this paper (see BRCA1 plays a role in single-cell motility). Because EGFP-CTD lacks the other domains of BRCA1, such a displacement event might have functional consequences. To assess this, we performed three different functional assays to screen for potential abnormalities in cellular behavior.
BRCA1 plays a role in cell spreading
We investigated the role of BRCA1 in cell spreading by determining the percentage of spread out cells shortly after seeding. The results showed that, after seeding and a 3-h attachment time period, more CHO-WT cells were spread out than CHO-High (P < 0.01; Fig. S1 D) and CHO cells expressing low levels of EGFP-CTD (CHO-Low; P < 0.01; Fig. 7 A). Cells fixed 24 h after seeding no longer showed differences in spreading, suggesting there is no absolute deficit but rather a kinetic difference. We next measured each cell area in populations of the different CHO cell types. The mean area of rounded up cells in CHO-WT, CHO-Low, and CHO-High was 351 (± SEM = 7), 319 (± SEM = 6), and 320 (± SEM = 5) pixels, respectively, whereas the mean area of the spread out cells in CHO-WT, CHO-Low, and CHO-High was 635 (± SEM = 40), 564 (± SEM = 18), and 547 (± SEM = 23) pixels, respectively. These results suggest that BRCA1 may be involved in regulating the initial spreading of transformed cells.
Figure 7.
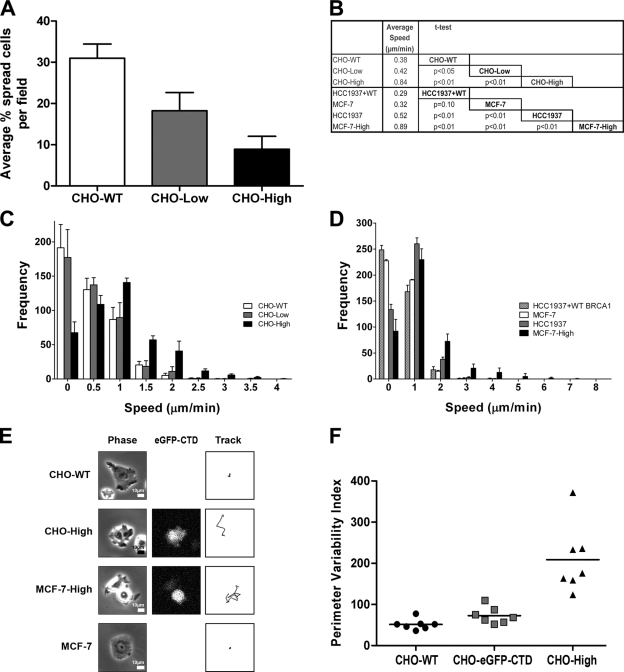
Functional assays on CHO, MCF-7, HCC1937, and HCC1937 + WT BRCA1 cell lines. (A) Cell spreading: the percentage of spread out cells per field at 3 h after seeding for CHO–wild type (CHO-WT) and CHO stably expressing low (CHO-Low) or high (CHO-High) levels of EGFP-CTD (means ± SD; n = 5). Each pairwise comparison by t test gave P < 0.01. (B) Table comparing mean speeds for different cell populations. CHO-WT and MCF-7 moved slower than their high expressing counterparts CHO-High and MCF-7–High. MCF-7–High moved faster than HCC1937. HCC1937 cells rescued with full-length BRCA1 (HCC1937 + WT BRCA1) displayed a highly significant lower mean speed than their native HCC1937 counterpart. All but one of the mean speed differences were highly significant (P < 0.01). There is no difference in the mean speed of HCC1937 + WT BRCA1 versus MCF-7 (P = 0.1). Means ± SEM for CHO-WT, CHO-Low, CHO-High, HCC1937 + WT, MCF-7, HCC1937, and MCF-7–High are 0.38 ± 0.01, 0.42 ± 0.01, 0.84 ± 0.02, 0.29 ± 0.01, 0.32 ± 0.01, 0.52 ± 0.01, and 0.89 ± 0.02, respectively (n = 4). (C) Motility of CHO cell populations: a speed frequency histogram is shown for CHO-WT, CHO-High, and CHO-Low cell types (means ± SD; n = 4). The CHO-High moved faster than CHO-WT and CHO-Low. CHO-WT and CHO-Low behaved similarly. (D) Motility of MCF-7, HCC1937, HCC1937 + WT BRCA1, and MCF-7–High cell populations: a speed frequency histogram is shown for MCF-7, HCC1937, HCC1937 + WT BRCA1, and MCF-7–High cells (means ± SD; n = 4). HCC1937 and MCF-7–High cells moved faster than HCC1937 + WT BRCA1 and MCF-7. MCF-7–High cells moved faster than HCC1937. Restoring full-length BRCA1 in HCC1937 reversed the hypermotility pattern. X-axis values are upper bounds for each speed bin. (E) Cell tracks for CHO-WT, CHO-High, MCF-7, and MCF-7–High cells are shown, including phase contrast and EGFP-CTD expression. CHO-WT and MCF-7 moved less far than their high expressing counterparts over this 1-h time period imaged at 2-min intervals. (F) Wound healing of CHO-WT, unsorted CHO–EGFP-CTD, and CHO-High cell monolayers. PVIs from seven independent wounds are presented for each cell type. The CHO-High population displayed the highest PVI values, which were correlated with a highly irregular wound margin. CHO-WT and unsorted CHO–EGFP-CTD showed a more regular behavior. Means for CHO-WT, unsorted CHO–EGFP-CTD, and CHO-High were 51, 73, and 209, respectively (horizontal lines). Nonparametric two-tailed Mann–Whitney U tests gave P < 0.05 (CHO-WT vs. unsorted CHO–EGFP-CTD) and P < 0.001 (CHO-WT vs. CHO-High; n = 7).
BRCA1 plays a role in single-cell motility
We next studied the spontaneous motility behavior of individual cells. CHO-WT, CHO-High, and CHO-Low cells were manually tracked in time-lapse videos of low density cultures. Mean speed values suggested a dose effect with CHO-WT moving slower than CHO-Low (P < 0.05) and significantly slower than CHO-High (P < 0.01) cell populations (Fig. 7 B). CHO-Low also moved significantly slower than the CHO-High cell line (P < 0.01).
Speed frequency histograms showed that CHO-WT and CHO-Low had similar motility behavior, whereas CHO-High exhibited a different speed pattern (Fig. 7 C). In addition, we analyzed motility of the unsorted CHO–EGFP-CTD cell population. This showed a similar mean speed to CHO-High (P = 0.5) and a comparable speed frequency histogram (unpublished data). Overall, these results showed that CHO-High move faster than CHO-WT and CHO-Low cells. This strongly suggests that interfering with endogenous BRCA1 leads to enhanced cell motility and that BRCA1 may play a role in controlling cell movement.
To confirm this finding in human breast cancer cell lines with (MCF-7) or without (HCC1937) functional BRCA1, we repeated the motility experiment on HCC1937, MCF-7, and MCF-7 cells highly expressing EGFP-CTD (MCF-7–High). MCF-7 moved slower than either HCC1937 (P < 0.01) or MCF-7–High cells (P < 0.01), whereas HCC1937 moved slower than MCF-7–High cells (P < 0.01; Fig. 7 B). In addition, MCF-7–High and HCC1937 had a similar speed distribution to CHO-High, whereas the MCF-7 cells showed a similar distribution to CHO-WT and CHO-Low (Fig. 7 D). Thus, the effects of EGFP-CTD expression seen in CHO cell lines can be extrapolated to a human cell line expressing endogenous BRCA1.
To ensure the dominant-negative effect of the EGFP-CTD fusion is solely caused by targeting and displacing BRCA1 and not another BRCT domain–containing protein, we analyzed the motility behavior of both native HCC1937 cells and HCC1937 cells stably expressing full-length BRCA1 (HCC1937 + WT BRCA1). HCC1937 cells have been shown to have one BRCA1-null allele and to carry a BRCA1 mutation (5382insC) in the second allele, which leads to a premature stop codon truncating the protein at residue 1,829 (Fig. S1 A; Tomlinson et al., 1998). This mutation resulted in very aggressive early onset breast cancer. Comparing mean speeds for both cell types, we found that the rescued HCC1937 + WT BRCA1 cells moved significantly slower than the native HCC1937 cell line, which only contained the truncated BRCA1 protein (P < 0.01; Figs. 7, B and D; and 8 C, top). Thus, restoring full-length BRCA1 in HCC1937 cells clearly reversed its original highly motile phenotype into a more static one, which is now comparable with cells expressing WT BRCA1, including the low motile nonmetastatic MCF-7 cell line. We conclude that the observed changes in phenotype upon overexpression of the EGFP-CTD fragment are caused by specific displacement of the BRCA1 protein.
Figure 8.
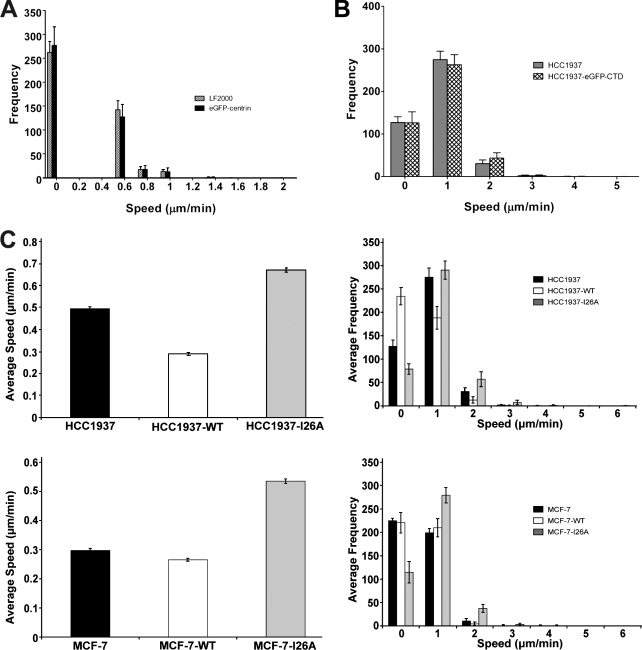
Functional assays on different CHO, HCC1937, and HeLa cell populations. (A) Speed frequency histogram for HeLa versus HeLa–EGFP-centrin cells (means ± SEM; n = 12). X-axis values are upper bounds for each speed bin. Overexpression of EGFP-centrin did not affect the motility behavior of HeLa cells. (B) Speed frequency histogram for HCC1937 versus HCC1937–EGFP-CTD (means ± SEM; n = 8). Both cell lines showed indistinguishable motility. (C) Single-cell motility assay on human breast cancer cell lines. Graphs showing the mean speed (means ± SEM; n = 120) for different cell populations (left). HCC1937 rescued with full-length BRCA1 (HCC1937-WT) displayed a highly significant lower mean speed than their native HCC1937 counterpart. On the contrary, HCC1937 rescued with full-length ubiquitin ligase–dead BRCA1 (HCC1937-I26A) displayed a highly significant increase in mean speed over HCC1937. No change in mean speed is seen in MCF-7 cell lines expressing full-length BRCA1, but MCF-7 expressing the ubiquitin ligase–dead mutation (MCF-7–I26A) displayed a highly significant higher mean speed than their native MCF-7 counterpart. All of the mean speed differences for HCC1937 cell types rendered P < 0.01; all of the mean speed differences for MCF-7 cell types rendered P < 0.01; the mean speed difference of HCC1937-WT versus MCF-7 was not significant (P = 0.3). Speed frequency histograms for different cell populations (right). HCC1937 shows higher motility than MCF-7; expressing full-length BRCA1 in HCC1937 resulted in a left shift of the motility pattern to resemble that of MCF-7 cells. Expression of the I26A mutant in either cell type gave a right shift to increased motility, higher than the native or BRCA1-expressing cells. X-axis values are upper bounds for each speed bin.
A corollary of this conclusion is that there should be no dominant-negative effect from the expression of EGFP-CTD in HCC1937 cells, which lack a functional BRCA1 protein for displacement. Therefore, we compared the motility behavior of HCC1937 cells with that of a stable HCC1937–EGFP-CTD cell line. HCC1937 and HCC1937–EGFP-CTD showed similar speed distribution and indistinguishable mean speeds of 0.50 (± SEM < 0.01) and 0.52 µm/min (± SEM < 0.01), respectively (Fig. 8 B).
Cell tracks monitored over a time period of 1 h for both CHO and MCF-7 (WT vs. High) can be seen in Fig. 7 E. The stable high-expressing cell lines showed enhanced motility versus their WT counterparts. As an additional control, we included a single-cell motility analysis on cells expressing unrelated EGFP fusion proteins, for example HeLa cells transiently transfected to express EGFP-centrin. Cytoplasmic accumulation of this fusion protein, even at high expression levels, did not interfere with the motility behavior of the cells (Fig. 8 A). The mean speed of HeLa cells treated with the transfection reagent only and HeLa cells transiently expressing EGFP-centrin were 0.2 (± SEM < 0.01) and 0.18 µm/min (± SEM < 0.01), respectively.
Finally, to achieve a more mechanistic insight, we focused on the only known enzymatic activity of BRCA1 as an E3 ubiquitin ligase, making use of a point mutation (I26A) that abrogates this activity. We compared the motility behavior of HCC1937, HCC1937-WT, and HCC1937-I26A. Cells were manually tracked in time-lapse videos of low density cultures and analyzed as described for CHO cells. Mean speed measurements revealed that restoring HCC1937 with a full-length, but ubiquitin ligase–dead, BRCA1 mutant (HCC1937-I26A) resulted in an even higher mean speed than HCC1937 (Fig. 8 C, top left). Rescued HCC1937-WT moved slower than HCC1937 cells, whereas HCC1937-I26A moved even faster than the already highly motile native HCC1937 cell line (Fig. 8 C, top right).
To test whether or not we could now reverse the low motility phenotype of MCF-7, we made stable lines of MCF-7 reconstituted with the full-length BRCA1 (MCF-7–WT) and the I26A mutation in full-length BRCA1 (MCF-7–I26A) and repeated the same experiment. MCF-7 and MCF-7–WT showed similar slow motility, whereas MCF-7–I26A showed increased motility (Fig. 8 C, bottom). Thus, additional expression of WT BRCA1 in MCF-7 cells already expressing functional endogenous BRCA1 did not affect motility, whereas expression of BRCA1 bearing an inactivating point mutation lead to a phenotype of increased motility, suggesting a dominant-negative effect of the enzymatically inactive I26A mutant protein. These observations suggest that interfering with either the subcellular targeting or E3 ubiquitin ligase activity of BRCA1 protein leads to enhanced cell motility, which is consistent with an important role for BRCA1-dependent ubiquitination in the regulation of cell spreading and motility.
BRCA1 plays a role in wound healing
To examine the consequences of increased mobility on cell populations rather than single cells, we turned to a widely used in vitro wound-healing model. By growing cells to a near confluent monolayer, scraping a wound in the monolayer, and following the healing of the wound by time-lapse microscopy, we were able to assess BRCA1 function in modulating the influence of cell–cell contact and external stimuli on cell motility. The simplest outcome measure in this assay is an assessment of wound closure at a predefined end point. No significant difference was observed between the different CHO populations when determining the percentage of gap closure after 12 h (unpublished data). Nonetheless, some individual cells expressing EGFP-CTD (either from unsorted CHO–EGFP-CTD or flow-sorted CHO-High cell populations) exhibited strikingly abnormal behavior. Their movement was haphazard and faster than other cells in the population. Of those presenting abnormal motility behavior, some were located in deeper cell layers relative to the wound and exhibited enhanced invasive capacity (Video 2, orange cell track). Other cells seemed to have lost cell–cell contact and attachment with the underlying cell layers and moved into the gap as single cells (Video 2, pink cell tracks). The latter behavior was rare in the CHO-WT population. Loss of cell–cell contact and independent migration of separated cells into the wound space was most apparent in CHO-High cells. As a result, the perimeter of wounds in CHO-High monolayers appeared discontinuous and irregular in contrast to the more uniform boundary of wounds in CHO-WT (Video 3).
To quantify these wound edge differences, we defined a perimeter variability index (PVI) based on changes in the length of a semiautomated trace along the wound perimeter over time. We wrote a second journal (macro) to handle this calculation for all 300 images of each time-lapse video (Fig. 7 F). The PVI values for CHO-WT replicates were closely grouped together with a low mean of 51.4, reflecting a highly reproducible regular wound perimeter morphology, whereas the CHO-High cells presented more variability in the wound perimeter over time, hence a higher mean PVI value of 208.9. The unsorted CHO–EGFP-CTD cell type showed a distribution similar to CHO-WT with a mean value of 72.9. Nonparametric two-tailed Mann–Whitney U tests gave P < 0.05 for the comparison between CHO-WT and unsorted CHO–EGFP-CTD and P < 0.001 for the comparison between CHO-WT and CHO-High. These findings suggest that the EGFP-CTD fusion acts as a dominant-negative inhibitor that competes with the endogenous BRCA1 protein for ERM and/or F-actin binding sites, leading to abnormalities in cell spreading and cell motility, changing the cell behavior.
Discussion
Since the discovery of the BRCA1 gene, multiple functions have been assigned to the protein it encodes. Yet the biological functions underlying its role in carcinogenesis remain to be fully elucidated. Here, we reveal some unexpected, yet potentially very important, functions for the BRCA1 protein in regulating cell spreading and motility, which have significant implications for tumor invasion and metastasis.
Partners interacting with BRCA1 BRCT domains
We demonstrated a specific interaction between the paired BRCT domains of BRCA1 and three proteins from the ERM family, namely ezrin, radixin, and moesin. The paired BRCT domain fragment recognizes each of these proteins independently rather than a complex between them. The proteins of the ERM family have structural resemblances but are still distinct. However, they all share the FERM (band 4.1, ezrin, radixin, and moesin) domain, an N-terminal domain highly conserved in this band 4.1 subclass (Chishti et al., 1998; Bretscher et al., 2002), which includes talin and FAK, both of which are involved in cell motility regulation (Diakowski et al., 2006). We do not know whether the BRCT domains of BRCA1 interact with the FERM domain in these proteins, but note that we did not detect other FERM domain–containing proteins in our ligand overlay experiments.
Intracellular colocalization of BRCA1 with ERM and F-actin
In contrast to adult tissues, cultured cell lines usually express all three ERM proteins, which are concentrated in actin-rich cell surface structures, such as microvilli, focal adhesions, and membrane ruffles (Sato et al., 1992). Confocal laser-scanning microscopy (CLSM) revealed a widespread subcellular distribution of the BRCT domain fusion protein in multiple cell types but also confirmed a significant colocalization with ERM family proteins at PM sites. Subsequent examination of full-length BRCA1 distribution showed the endogenous protein also present at these ERM- and F-actin–enriched PM sites. Although intracellular association between BRCA1 and cytoskeletal features had been described previously (Coene et al., 2005), the data reported here characterize a new subcellular localization for BRCA1 at the PM and reveal associations with PM ruffles, leading edges, microvilli, and focal adhesion plaques. Intracellular FRET results strongly suggest an association with the ERM complex and F-actin.
The ERM complex controls and regulates cell adhesion, spreading, and motility, and ezrin has been shown to be a key component in tumor metastasis (Hunter, 2004) because of its interaction with adhesion molecules such as CD43, CD44, ICAM-1, and ICAM-2 (Heiska et al., 1998; Yonemura et al., 1998) implicated in cell migration and metastasis (Tang and Honn, 1994–1995; Rosette et al., 2005). Ezrin and potentially other members of the ERM family have key roles in transferring signals from metastasis-associated cell surface molecules to other downstream signal transduction components (Bretscher et al., 2002). The association of BRCA1 with the key regulators of cell adhesion, spreading, and motility suggests that BRCA1 may act up- or downstream in their signaling pathways.
Consequences of displacing endogenous BRCA1
Endogenous BRCA1 and the EGFP-CTD fusion protein compete for the same binding sites on the ERM–F-actin complex at the PM. Expressed EGFP-CTD displaces endogenous BRCA1 and behaves as a dominant-negative inhibitor, leading to disturbances in cell spreading and motility and giving rise to a change in wound-healing behavior. The use of early passage number stable EGFP-CTD cell lines with a stable EGFP-CTD expression level excluded the possibility that these altered phenotypes were a consequence of an acute displacement caused by a temporary EGFP-CTD overexpression during transient transfection.
However, BRCT domains are not unique to BRCA1 but have also been described in other proteins involved in cell cycle checkpoint-mediated repair and DNA damage response pathways. Thus, it is possible that the spreading, motility, and wound-healing phenotypes might result from EGFP-CTD effects on another BRCT domain–containing protein. To address this issue, we turned to human breast cancer cell lines with well-characterized BRCA1 status. We studied HCC1937 cells containing only a truncated BRCA1 protein, HCC1937 cells expressing the EGFP-CTD fusion protein, and restored HCC1937 cells with full-length BRCA1. Only in this latter setup were we able to dramatically reverse its behavior from a high to a low motility phenotype similar to that of cells naturally containing functional BRCA1, including CHO and HeLa as well as another breast cancer cell line MCF-7, known to be nonmetastatic. Finally, we repeated our observations on a stable MCF-7 cell line expressing EGFP-CTD and found as expected that this resulted in a hypermotile phenotype. Collectively, these results suggest that EGFP-CTD does indeed interfere with BRCA1 and accounts for the phenotypic changes observed. Furthermore, we conclude that both the N and C termini of the BRCA1 protein are needed to perform its role in regulating cell spreading and motility correctly.
Both N-terminal RING and C-terminal BRCT domains function as interaction modules and interact with proteins involved in multiple pathways. The N-terminal RING finger of BRCA1 is known to interact with BARD1, its heterodimeric partner. If BRCA1 and BARD1 cooperate in the regulation of cell motility, one would expect that tumorigenic mutations within either BRCA1 or BARD1, impairing the BRCA1–BARD1 interaction, will influence cell spreading and motility. Interestingly, repression of Bard1 induced complex phenotypic changes in mammary epithelial cells, which were suggestive of a premalignant phenotype. Bard1-suppressed murine mammary epithelial cells overcame contact inhibition, formed multiple cell layers, and expressed an invasive phenotype (Irminger-Finger et al., 1998; Soriano et al., 2000). Mouse Brca1 mutants that lack both BRCT domains developed various tumors, including breast cancer. The breast tumor phenotype showed invasive to highly invasive borders as well as pushing margins (Ludwig et al., 2001). These findings suggest that, at least in mice, Bard1 and Brca1 seem to participate in regulating cell spreading and motility but also support our results and hypothesis that BRCT domains of BRCA1 are needed to regulate cell motility successfully.
Furthermore, in population-based experiments, it was observed that the majority of the BRCA1 breast cancers displayed histological grade III (Brekelmans et al., 2007). This is an important finding because high histological grade is usually associated with a greater propensity to distant metastasis.
Unselected invasive breast cancers have showed both reduced BRCA1 mRNA and protein expressions when compared with noninvasive cancers and benign tissues (Thompson et al., 1995; Taylor et al., 1998; Seery et al., 1999). A significant association between low BRCA1 expression levels and the development of distant metastases and acquisition of an invasive phenotype has also been reported (Seery et al., 1999; Xu et al., 1999). This suggests a role for BRCA1 as a suppressor for invasion. Again, all these observations are consistent with our hypothesis that BRCA1 is needed to control cell motility and inhibit metastasis and invasion.
Invasion and metastasis are known to be complex biological processes. The first event to happen is the detachment of cancer cells from the original tumor followed in a later stage by invasion. Our results confirmed these single-cell detachment and loss of cell–cell contact events as well as the invasion capacity when we interfered with the endogenous BRCA1 protein using the EGFP-CTD protein as a dominant-negative factor (Videos 2 and 3). These time-lapse videos provide evidence that interfering with endogenous BRCA1 promotes an invasive phenotype.
BRCA1 function at the PM
The only enzymatic role that has been identified for BRCA1 is that of E3 ubiquitin ligase activity in a heterodimer composed of BRCA1 and BARD1 (a protein structurally and functionally related to BRCA1), and experiments support a role for BRCA1 in the ubiquitination pathway (Starita and Parvin, 2006). The importance of an intact BRCA1 N terminus, where the RING domain ligase active site is found, in tumor suppression has been shown before, but the underlying mechanisms are unclear. We demonstrate that intact E3 ubiquitin ligase activity of BRCA1 is needed to control motility. Moreover, this activity needs to be targeted to ERM and F-actin–containing sites at the PM. Loss of BRCA1 protein from these sites or loss of ubiquitin ligase activity can promote increased motility with implications for tumorigenesis.
Different forms of ubiquitin modification exist, each of them playing roles in diverse cellular processes (Weissman, 2001). Monoubiquitination is regarded as a major signaling event that can mediate endocytosis and, thus, regulate turnover of PM proteins. The interaction of ERM proteins with the BRCT domains of BRCA1 raises the intriguing possibility that activated ERM proteins could themselves be normal targets of BRCA1 E3 ubiquitin ligase activity. If so, loss of this activity might lead to an accumulation of ERM proteins at the cell surface; we note that high expression of ezrin has been linked to high metastatic potential (Akisawa et al., 1999), supporting this hypothesis. This hypothesis, together with the fact that both intact N and C termini of BRCA1 are needed to successfully perform its ubiquitination function (Starita et al., 2004), fits closely with our displacement assay and the consequences for cell spreading and cell motility. The single-cell motility assays with HCC1937 versus MCF-7 add further strength to this hypothesis. Loss of even a few C-terminal residues of BRCA1 might lead to an unstable BARD1–BRCA1 heterodimer, causing irregularities in the E3 ubiquitin ligase activity, which in turn could lead to a deregulation of the turnover/degradation of PM proteins. This would explain why BRCA1-truncated HCC1937 cells displayed a higher mean speed than the MCF-7 containing intact BRCA1.
An alternative, but not mutually exclusive, possibility is that PM-localized BRCA1 functions via an association with ERM proteins that interact with PM receptors. Ubiquitin ligase activity could target machinery or cargo in the endocytic pathway and, thus, influence trafficking of PM receptors.
Until now, it remained unknown how BRCA1 might exert an effect on the invasion and migration of breast cancer cells. In light of the results reported here, we suggest a potential role for BRCA1 at the PM as an E3 ubiquitin ligase controlling the trafficking and/or turnover of specific PM receptors or proteins, involved in focal adhesion, cell–cell, and cell–matrix contacts. Modulation of the levels of such proteins, or their related downstream signaling pathways, may consequently regulate the invasive and metastatic properties of tumor cells. Further research is needed both to pinpoint the relevant protein targets and to identify the signaling pathway involved.
Materials and methods
Cell lines
HeLa, a human cervix carcinoma cell line, and ECV304, a vascular endothelial cell line, were cultured in DME with high glucose (4.5 g/liter), l-glutamine, and sodium pyruvate (PAA Laboratories GmbH) supplemented with 10% (vol/vol) FCS. HCC1937 is a primary breast ductal carcinoma cell line cultured in RPMI 1640 medium with l-glutamine and 1 mM sodium pyruvate supplemented with 10% (vol/vol) FCS. CHO is a CHO cell line cultured in Ham’s F12 medium with l-glutamine and supplemented with 10% (vol/vol) FCS. MCF-7 is a human breast adenocarcinoma cell line cultured in MEM with Earle’s salts, without l-glutamine, supplemented with 1% (vol/vol) nonessential amino acids and 10% (vol/vol) FCS. All cell lines were obtained from the American Type Culture Collection.
Stable cell lines
CHO and MCF-7 cell lines were transfected with 5 µg pEGFP-BRCA11634-–1863 (pEGFP-CTD) DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. G418 (Sigma-Aldrich) was used as a selection drug at 800 µg/ml. Established stable CHO–EGFP-CTD and MCF-7–EGFP-CTD cell lines were cell sorted according to expression levels and referred to as CHO-High and MCF-7–High for high and CHO-Low for low expression levels.
The MCF-7 cell line was transfected with 5 µg of full-length BRCA1-pIRES2-EGFP and full-length BRCA1 with point mutation I26A-pIRES2-EGFP plasmid DNA using Lipofectamine 2000 according to the manufacturer’s instructions. G418 was used as a selection drug at 600 µg/ml.
To generate stable BRCA1-reconstituted HCC1937 cells, HCC1937 cells were electroporated with full-length BRCA1-pIRES2-EGFP, full-length BRCA1 with point mutation I26A-pIRES2-EGFP (provided by X. Yu, Mayo Clinic, Rochester, MN), or pEGFP-CTD DNA using an electroporation system (Gene Pulser II; Bio-Rad Laboratories) at 250 V and 960 µF for 51.1 ms. A few days after electroporation, the cells were put under 600 µg/ml G418 selection.
Membrane fraction preparation
HeLa P2 fractions were prepared as previously described (Coene et al., 2005) with minor modifications. Nearly confluent HeLa cells were washed with ice-cold PBS, scraped, and harvested. The pellet was resuspended in 1 ml of isolation buffer (10 mM Tris-HCl, 10 mM NaCl, 1.5 mM MgCl2-6H2O, 1 mM EDTA, 70 mM sucrose, and 210 mM mannitol, pH 7.5) with protease inhibitors (Roche) and homogenized with 40 strokes using a Balch homogenizer (custom made by EMBL workshop) on ice. The membrane fraction was collected by differential centrifugation, dissolved in 25 mM Tris-HCl, pH 8.0, and 10 mM CaCl2-6H2O, and treated with 50 ng/ml proteinase K (Fluka; Sigma-Aldrich) in 10 mM Tris-HCl, pH 8.0, and 1 mM CaCl2-6H2O for 30 min at RT. The reaction was stopped with 10 mM PMSF for 10 min at RT. For 1D SDS-PAGE, samples were denatured by adding sample buffer with 100 mM DTT (Sigma-Aldrich) and heated for 5 min at 95°C before loading. For 2D electrophoresis, samples were acetone precipitated and redissolved in IEF solubilization buffer (7 M urea, 2 M thiourea, 2% [wt/vol] CHAPS, 65 mM DTT, 2.0% [wt/vol] ampholine [IPG buffer; GE Healthcare], and trace bromophenol blue [Sigma-Aldrich]).
1D and 2D ligand overlay blotting
For 1D SDS-PAGE, membrane fractions (equivalent to 106 cells per lane) were resolved by SDS-PAGE using standard techniques. The gel was either stained with Imperial Protein Stain (Perbio Science) and processed for LC-MS/MS or blotted. Nitrocellulose membrane was incubated overnight with HeLa lysate. Untransfected, pEGFP-CTD–transfected (3.2 µg DNA per well of a 6-well plate), and pEGFP-NTA–transfected (3 µg DNA per well of a 6-well plate) HeLa cell lysates were made using radioimmunoprecipitation assay (RIPA) buffer (Millipore) and protease inhibitor cocktail tablets (Roche) according to the manufacturer’s instructions. Goat polyclonal anti-GFP antibody (dilution 1:5,000; Abcam) and donkey anti–goat HRP-labeled secondary antibody (dilution 1:4,000; Jackson ImmunoResearch Laboratories, Inc.) were used for detection.
For 2D electrophoresis, immobilized gel strips (Immobiline Drystrip; GE Healthcare) of 13 cm and a linear pH gradient range, pH 3–10, were rehydrated in IEF solubilization buffer. Protein (150 µg/strip) was loaded from a cup on the cathodic end. Proteins were electrofocused as follows: 30 min at 250 V, 3-h linear gradient at 250–8,000 V, and 14 h at 8,000 V, all at 20°C. IEF gel strips were equilibrated in 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) SDS, and 65 mM DTT for 15 min followed by incubation in SDS equilibration buffer supplemented with 135 mM iodoacetamide (Fluka) for 15 min. Gel strips were placed onto the top of 10% SDS-PAGE and run at 8 W/gel for 2.5 h. 2D gels were either fixed in 10% (vol/vol) methanol and 7% (vol/vol) acetic acid and stained with SYPRO ruby (Sigma-Aldrich) for further imaging and spot excision or semidry transferred to nitrocellulose membrane and processed as described in the previous paragraph. For BRCA1 detection in CHO lysate on a Western blot, we used Ser988 (1:100) and Ser1497 (1:200) anti-BRCA1 antibodies (Santa Cruz Biotechnology, Inc.) as primary antibodies and donkey anti–goat HRP-labeled secondary antibody (dilution 1:4,000) for detection.
Coimmunoprecipitation
HeLa cells were treated with 150 nM PMA (Sigma-Aldrich) for 90 min at 37°C and lysed on ice using RIPA buffer (Sigma-Aldrich) supplemented with 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 mM NaF, 1 mM PMSF, and 1 mM Na3VO4. The lysate was rotated for 15 min at 4°C on an orbital rocker and centrifuged for 15 min at 14,000 g at 4°C. The supernatant was precleared by adding a 50% slurry of Protein A/G PLUS beads (Tebu-Bio) and rotating for 30 min at 4°C. 5 mg/ml of cell lysate was incubated with 2 µg each of mouse anti-BRCA1 Ab1 (MS110; Oncogene Science–Merck Chemicals), mouse antitryptase (AbCys), and mouse IgG1 pure (BD) antibodies and no antibody for 90 min at 4°C on an orbital shaker. A 50% slurry of Protein A/G PLUS beads was added overnight. Beads were washed three times with ice-cold RIPA buffer. Immunoprecipitated proteins were denatured by the addition of sample buffer, boiled for 5 min, resolved by 6% Tris-glycine SDS-PAGE, and analyzed by immunoblotting using mouse anti-BRCA1 Ab1 (1:50), mouse antitryptase (1:500), and rabbit antiezrin (1:1,000; Cell Signaling Technology) antibodies. For detection, we used goat anti–mouse HRP (Thermo Fisher Scientific) and goat anti–rabbit HRP (Dako) antibodies in combination with the SuperSignal West Dura Extended Duration Substrate kit (Thermo Fisher Scientific). Images were taken on an imaging system (ULTima16si Pro; Isogen Life Science).
Plasmids
The tubulin sequence was excised from an EGFP-tubulin vector (Takara Bio Inc.) by XhoI and XbaI digest (Roche). BRCA1 primers, including XhoI and XbaI restriction side ends, used in this study were BRCA1 (1,634–1,863 aa), 5′-GGCTGTCTAGAGTCAGTAGTGGCTGTG-3′, and BRCA1 (1,634–1,863 aa), 5′-AGATCTCGAGAGAAGCCAGAATTGACAGC-3′. DNA template pFLAG-YFP–full-length BRCA1 plasmid (gift from B. Henderson, Westmead Hospital, Sydney, Australia) was used. Inserts were digested with XhoI and XbaI, ligated in the vector, and transformed in Top10F’, and minipreps of the obtained colonies were prepared according to routine procedures. Sequence analysis was performed using the following primers: GFP forward, 5′-CATGGTCCTGCTGGAGTTCGTG-3′, and SV40 reverse, 5′-GGACAAACCACAACTAGAATGC-3′. The control pEGFP-NTA plasmid was a gift from Q. Sattentau (Dunn School of Pathology, Oxford, England, UK) and J. Komano (AIDS Research Center, Tokyo, Japan). Plasmids were transformed in DH5-α, and maxipreps of obtained colonies were prepared according to routine procedures. The control EGFP-neo was obtained from B. Wickstead (Dunn School of Pathology, Oxford, England, UK) by cutting pVP22 (Invitrogen) with HindIII–KpnI and ligating in EGFP from pGad8–VSG-S8 (Wickstead et al., 2003). The EGFP-centrin plasmid was made in house (D.J. Vaux).
LC-MS/MS
LC-MS/MS sample preparation and analysis were performed as follows: bands/spots were excised and diced. Gel fragments were destained using 25 mM ammonium bicarbonate (Fluka) in 50:50 double-distilled water/acetonitrile (HPLC grade; Sigma-Aldrich) followed by dehydration in 100% acetonitrile and vacuum drying (Speedvac). Proteins were reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide. 200 ng trypsin (Promega) in 20 µl of 25-mM ammonium bicarbonate was added to the gel pieces, and proteins were digested at 37°C overnight. Digestion was stopped with formic acid (Fluka) to a concentration of ∼0.1%, and peptides were extracted. Supernatants were pooled and evaporated to dryness. Peptides were resuspended in 0.1% formic acid and stored at −20°C before analysis. LC-MS/MS was performed on a mass spectrometer (Q-TOF Micro; Micromass) coupled to a capillary HPLC system (Waters). Proteins were identified by searching data files against Mascot (Matrix Science).
Immunofluorescence
HeLa, ECV304, and HCC1937 cells were either cultured and transfected with 5 µg pEGFP-CTD/pEGFP-centrin/pEGFP-neo DNA using Lipofectamine 2000 according to the manufacturer’s instructions or cultured and prepared for immunofluorescence as previously described (Coene et al., 2005). In brief, cells were seeded onto glass coverslips, grown until 60% confluence, transfected or not transfected, and fixed in 4% (vol/vol) PFA and 250 mM Hepes, pH 7.4, for 20 min at RT. Coverslips were quenched in 50 mM NH4Cl in PBS for 10 min, permeabilized in 0.5% (vol/vol) Triton X-100 in PBS for 5 min, and blocked in 0.4% (vol/vol) fish skin gelatin (FSG; Sigma-Aldrich) in PBS for 30 min at RT. The primary antibodies were diluted in 0.4% (vol/vol) FSG in PBS and incubated for 1 h at RT. The Ser988 and Ser1497 anti-BRCA1 antibodies were incubated at 37°C for 1 h. Secondary antibodies were diluted in 0.4% (vol/vol) FSG in PBS and incubated for 1 h at RT. Washes were performed in PBS. Coverslips were rinsed in distilled water and mounted in Mowiol (Mowiol 4–88; Hoechst AG) in 0.2-M Tris-HCl, pH 8.5, supplemented with 0.1 µg/ml DAPI. The images were taken on a laser-scanning microscope (LSM 510 META; Carl Zeiss, Inc.) using a 63× oil lens, NA 1.4. Adjustments of brightness and contrast were applied using Photoshop (Adobe). For the competition and displacement assay, z stacks were taken with a 0.3-µm-thick optical section and a typical pixel size of 0.1 or 0.2 µm.
For some experiments, coverslips were rinsed with a jet of PBS directed from a wash bottle to provide mechanical shear stress that disrupted a proportion of the cells, leaving adherent membrane sheets behind. For other experiments, saponin treatment was performed as previously described (Beranger et al., 1994).
Primary antibody dilutions used in this study were anti-BRCA1 antibodies Ab5 (1:50), Ser988 (1:50), Ser1497 (1:50), anti-ERM antibody (1:100; Cell Signaling Technology), 0.1 µg/ml TRITC-phalloidin (Sigma-Aldrich), and anti-FAK (1:50; BD). Secondary antibodies used were donkey anti–mouse Alexa Fluor 488 (1:500), donkey anti–goat Alexa Fluor 488 (1:500; both are in-house–labeled Jackson ImmunoResearch Laboratories, Inc. secondary antibodies using an Alexa Fluor 488–labeling kit from Invitrogen), donkey anti–rabbit Cy5 (1:500), donkey anti–mouse Cy5 (1:300; Jackson ImmunoResearch Laboratories, Inc.), and donkey anti–goat Alexa Fluor 647 (1:300; Invitrogen). For FRET, we used anti-pSer988 BRCA1 (1:50; Santa Cruz Biotechnology, Inc.), anti-ERM (1:100), and anti–human transferrin receptor (1:100; Invitrogen) antibodies labeled with donkey anti–goat Alexa Fluor 488 (1:500), donkey anti–rabbit Cy3 (1:300; Jackson ImmunoResearch Laboratories, Inc.), and donkey anti–mouse Cy3 (1:300; Jackson ImmunoResearch Laboratories, Inc.), respectively. As a positive control for FLIM, we used goat polyclonal anti-GFP antibody (1:1,000; Abcam) and donkey anti–goat Cy3 (1:300; Jackson ImmunoResearch Laboratories, Inc.).
Cryoimmuno-EM
HeLa cells were seeded and incubated at 37°C for a few hours. When they started to spread, cells were treated with 150 nM PMA for 90 min at 37°C, fixed, and processed for cryoimmuno-EM as previously described (Coene et al., 2005). In brief, cells were fixed in 4% PFA/250 mM Hepes, pH 7.4, for 1 h at RT, embedded in 10% gelatin, and processed for cryosectioning. Primary antibodies were rabbit anti-ERM (1:10; Cell Signaling Technology) and mouse anti-BRCA1 Ab4 (1:5; Oncogene Science–Merck Chemicals). Secondary antibodies were goat anti–rabbit 10-nm gold and goat anti–mouse 5-nm gold conjugates (BB International). Images were collected on an electron microscope (Tecnai 12; FEI).
FLIM analysis
A microscope (Eclipse TE300; Nikon) incorporated in an imaging system (Radiance 2000 MP; Bio-Rad Laboratories) was used. A pulsed laser beam of 950 nm was obtained from a mode-locked Ti:Sapphire laser (Mira 900; Coherent, Inc.) for multiphoton excitation. Fluorescence lifetimes of EGFP were recorded using the direct detection system (Bio-Rad Laboratories) with a photomultiplier tube (H5783P; Hamamatsu) optimized for photon counting and FLIM and a fast-time–correlated single-photon–counting acquisition board (SPC-830; Becker & Hickl). Lifetime images were collected at 512 × 512 pixels with a 100-s scanning time and analyzed with SPCImage software (Becker & Hickl). Within SPCImage, mask polygons were drawn around regions of interest (ROIs), and binning was set to give an initial photon count level of ≥1,000. EGFP lifetimes within the cells were analyzed by matrix calculation on a pixel-by-pixel basis. A multiexponential decay model was used, as this gave the lowest χ2 value and the best fit. Lifetime histograms (t1) were exported to and analyzed in Excel (Microsoft). The EGFP lifetime in the absence of an acceptor was used to determine FRET efficiencies using the formula 100 × [1 − (t1/tunquenched EGFP)]. The acceptor used was Cy3, which was used to detect EGFP (positive control) or ERM.
FRET analysis
FRET using the acceptor photobleaching method (Kenworthy, 2001) was performed as previously described (Malhas et al., 2009). This was performed on PMA-treated fixed cells after immunolabeling of ERM using anti-ERM antibody (1:100) and donkey anti–rabbit Cy3 as the acceptor and endogenous BRCA1 using Ser988 and donkey anti–goat conjugated to Alexa Fluor 488 as the donor. As a negative control, we used the same BRCA1–Alexa Fluor 488 donor but a mouse anti–human transferrin receptor primary antibody labeled with a Cy3-conjugated secondary antibody as an acceptor. An ROI containing membrane ruffles was selected, and both the donor and acceptor were detected for five scans. The ROI was then bleached with the 543-nm laser at 100% laser power. This was followed by scanning to detect any changes in the acceptor and donor intensities. FRET efficiency was then calculated using the following formula: 100 × [1 − (Ib/Ia)], in which Ib and Ia are the intensities of the donor before and after bleaching, respectively. Difference maps were calculated by subtracting “before” pixel values from ”after” pixel values and rescaling to fill the 8-bit histogram range (which required a scaling factor of 2.0).
PMA and cytochalasin B treatment
HeLa cells were transfected with pEGFP-CTD as described (see Immunofluorescence) and treated with 150 nM PMA for 90 min at 37°C. 20 µM cytochalasin B (Sigma-Aldrich) was added for 2 h at 37°C. Coverslips were fixed and further processed for immunofluorescence as described (see Immunofluorescence).
Competition and displacement
CHO-WT and CHO-High cells were fixed and immunostained with TRITC-phalloidin and anti-Ser988 BRCA1 antibody as described in Immunofluorescence. Confocal z stacks were obtained using an LSM 510 META and a 63× oil lens, NA 1.4. Adjustments of brightness and contrast were applied using Photoshop. To quantify the displacement, a journal (macro) was written in which we measured the proportion of overlapping actin and anti-BRCA1 staining in dual-channel immunofluorescence images using simple overlap parameters derived by Manders et al. (1993). We used the standard implementation of this parameter in the colocalization function of an image-processing program (MetaMorph; MDS Analytical Technologies; see MetaMorph colocalization macro for colocalization journal details). The percentage of values for endogenous BRCA1 overlap with F-actin was obtained for each plane in a z stack. For each z stack, only the colocalization values above a threshold cutoff (10% of the maximum value of all IF-actin values in a specific z stack) were taken into account. Colocalization values of seven individual z stacks per cell type were pooled and plotted with corresponding means. Unpaired two-tailed t test with Welch’s correction and nonparametric two-tailed Mann–Whitney test were performed. For the images in Fig. 5 B, z stacks resulting from this analysis were displayed as mean intensity z projections.
Cell spreading
CHO-WT, CHO-High, and CHO-Low cells were seeded in equal densities (1–3 × 105 cells per well), fixed 3 h after seeding, mounted in Mowiol supplemented with DAPI, and imaged on a laser-scanning microscope (LSM 5 Pascal; Carl Zeiss, Inc.) using a 10× objective with NA 0.45. Phase contrast and EGFP images of five random fields on the coverslip were taken. The percentage of spread out cells per field was calculated. The percentage values for five fields from each of three independent experiments were pooled, averaged, and plotted with corresponding SDs after confirming no significant difference in variance for a condition between experiments using an F test. t tests were calculated. The area of the spread out cells versus rounded-up cells was measured using the Trace Region tool and Show Region Statistics option in MetaMorph software.
Single-cell motility
All cell lines were seeded in low densities in dishes (MatTek Corporation). 24 h after seeding, the cells were imaged on an LSM 5 Pascal system using a 10× objective with NA 0.45. Phase contrast and EGFP images of four random fields in the dish were taken per cell type. Each field was imaged over a time period of 1 h at 2-min intervals. For each cell line, 15 cells per field were manually tracked over 30 time points using the Track Points option in MetaMorph software. The mean speed of all 60 cells over 30 time points was calculated per cell type with corresponding t tests. For Fig. 8 C, the mean speed of all cells from two independent setups (120 cells) over 30 time points was calculated per cell type. The individual speed values per field, all in micrometers per minute, were grouped in bins ranging from 1 to 5 at 0.5 intervals for CHO cell lines, ranging from 1 to 10 at single intervals for MCF-7, MCF-7–-High, MCF-7–WT, MCF-7–I26A, HCC1937, HCC1937 + WT BRCA1, and HCC1937-I26A cell lines, ranging from 1 to 5 at single intervals for HCC1937–EGFP-CTD, and ranging from 0 to 2 at 0.2 intervals for HeLa-EGFP-centrin, and the frequency per bin was determined.
The mean of all frequencies per bin per cell type and corresponding SDs were calculated and plotted. To display the individual cell tracks, the Track Objects option in MetaMorph software was used.
Wound healing
CHO-WT, unsorted CHO–EGFP-CTD, and CHO-High cells were seeded in MatTek dishes and grown to confluence. Each cell layer was scrape wounded and imaged on an LSM 510 META microscope using the Multi Time Series software option. Phase contrast and EGFP images of four random wounds in the dish were recorded over a time period of 12 h at 2-min intervals. The wound perimeter was analyzed over time in a standardized way using an in-house analysis subroutine (see MetaMorph wound-healing macro for wound-healing MetaMorph journal details). For each two consecutive time points, the change in wound perimeter (width and length corrected) was calculated, summed, and referred to as the PVI. For each cell type, seven wounds from different experiments were analyzed using this journal, and the obtained PVI values per cell type were plotted in a scatter dot plot with the corresponding means. Nonparametric two-tailed Mann–Whitney U tests were performed.
MetaMorph colocalization macro
This is a description of the algorithm used in the MetaMorph journal Coloc_003_02.jnl. The method gives two values showing the percentage of fluorescence (above a defined threshold) in each channel (of a two-channel z-stack image) that occupies the same volume (pixel by pixel) as fluorescence (above a defined threshold) in the other channel. (a) It is assumed that there is no significant bleed through between channels. (b) It is assumed images have been filtered to reduce any significant noise. (c) It is assumed that each pixel represents a sum of background + fluorescence. (d) The background distribution is obtained from a background region in each section: MeanB = background mean in each section averaged over all sections; STDB = background SD in each section averaged over all sections. (e) The fluorescence distribution is obtained over the whole image in each section: minimum = (MeanB + n × STDB); n = 3 (channel A) and 1 (channel B; selectable by user); maximum = highest intensity in image; MeanF = mean of fluorescence distribution averaged over all sections; STDF = SD of fluorescence distribution averaged over all sections. (f) A fluorescence threshold is used to include just the bright focal signal: ThresholdF = MeanF + m × STDF; m = 0.5 (all channels). (g) ThresholdF is subtracted from the image. (h) The two image channels are merged after subtracting each threshold. (i) A 2D region for colocalization measurement is drawn and applied to all sections. (j) The colocalization percentage is obtained within the defined region for each image: IA = total intensity of all pixels in channel A; IAB = total intensity of all pixels in channel A with nonzero values in channel B; IAB/IA × 100% = percentage of channel A overlying nonzero channel B intensity. (k) Colocalization of channel B on channel A is estimated as IBA, IB, and IBA/IB. (l) The colocalization percentage as a function of focus (z) is found for the entire stack: IAB(z)/IA(z) = colocalization percentage for section (z). (m) Threshold values and all the colocalization parameters are recorded in text files.
MetaMorph wound-healing macro
This is a description of the algorithm used in the MetaMorph journal IWA_1_02.jnl to estimate closure and wound edge roughness in phase contrast time-lapse images of a closing in vitro wound. The method gives two values that are estimates for (a) the mean width of the wound and (b) the combined length of lines traced around both wound edges (reduced perimeter). Values are derived for each time point. (a) The wound orientation was determined interactively (one angle for the whole stack). (b) The image stack was rotated to bring the axis horizontal (aligned in X). (c) A four-pass filter was applied to all time points to enhance cell edges using the following four kernels sequentially: kernel 1 (5, 5, 5; −3, 0, −3; −3, −3, −3), kernel 2 (5, 5, −3; 5, 0, −3; −3, −3, −3), kernel 3 (5, −3, −3; 5, −3, −3; 5, −3, −3), and kernel 4 (−3, −3, −3; 5, 0, −3; 5, 5, −3). (d) A (sequential) Lomo filter (2-pixel-diam circle) reduced noise and joined touching cells. (e) A background region was defined within the wound area. (f) A threshold was determined from intensities in the background of each time point: threshold = (1 + n) × (mean background); n = 1 was empirically determined to clearly define the wound edge and kept constant in all wounds analyzed. (g) The wound area in each time point was found by the MetaMorph immunofluorescence tool. (h) Immunofluorescence always found a single object >10,000 pixels in area above the threshold. (i) The mean wound width and a reduced perimeter of the wound area were determined: width = (wound area)/(wound length); (reduced perimeter) = (perimeter of wound area) − 2 × width. (j) The threshold value, width, and reduced perimeter were written into an ASCII (American Standard Code for Information Interchange) file.
Online supplemental material
Fig. S1 shows a schematic presentation of the BRCA1 BRCT domains, cryoimmuno-EM of PMA-stimulated HeLa cells, a Western blot of CHO cells, and spreading of CHO-WT versus CHO-High cells. Fig. S2 shows EGFP-CTD and endogenous BRCA1 colocalization with F-actin and ERM. Fig. S3 shows endogenous BRCA1 colocalization with F-actin and ERM. Fig. S4 shows that EGFP-CTD is present in focal adhesion plaques. Fig. S5 shows EGFP-protein distribution after saponin treatment. Fig. S6 shows colocalization of EGFP-CTD, F-actin, and FAK after saponin treatment. Video 1 shows PM dynamics. Video 2 shows loss of cell–cell contacts and enhanced invasion capacity in wound healing. Video 3 shows wound healing in CHO-WT versus CHO-High. Table S1 shows protein identification by mass spectrometry from SDS-PAGE. Table S2 shows protein identification by mass spectrometry from 2D gel spots. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004136/DC1.
Acknowledgments
The authors thank colleagues from the University of Oxford: B. Wickstead, C. Davison, C.F. Lee, and R. Gould for data analysis; K. Gull for 2D electrophoresis apparatus; and P. Yajnik, N. Rust, L. Jean, H. Womersley, and K. Dewar for technical assistance. We also thank B. Henderson for the pFLAG-YFP–full-length BRCA1 plasmid, Q. Sattentau and J. Komano for the pEGFP-NTA plasmid, and X. Yu for the full-length BRCA1-pIRES2-EGFP and I26A-pIRES2-EGFP plasmids.
This work was supported by grants from Scientific Research Flanders (Fonds Wetenschappelijk Onderzoek [FWO]–Vlaanderen, 1.2.900.07.N.01 and 1.5.180.07) to E.D. Coene; the Human Frontier Science Program (RG/P0058/2004) and the Wellcome Trust (066839/Z/02/Z) to C. Gadelha; and departmental support to D.J. Vaux. E.D. Coene is a Postdoctoral Researcher funded by the Fund of Scientific Research Flanders (FWO-Vlaanderen).
Footnotes
Abbreviations used in this paper:
- BRCT
- BRCA1 C-terminal
- CLSM
- confocal laser-scanning microscopy
- ERM
- ezrin–radixin–moesin
- FLIM
- fluorescence lifetime imaging microscopy
- FRET
- fluorescence resonance energy transfer
- FSG
- fish skin gelatin
- LC-MS/MS
- liquid chromatography and tandem mass spectrometry
- PM
- plasma membrane
- PVI
- perimeter variability index
- RIPA
- radioimmunoprecipitation assay
- ROI
- region of interest
- WT
- wild type
References
- Akisawa N., Nishimori I., Iwamura T., Onishi S., Hollingsworth M.A. 1999. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem. Biophys. Res. Commun. 258:395–400 10.1006/bbrc.1999.0653 [DOI] [PubMed] [Google Scholar]
- Beranger F., Paterson H., Powers S., de Gunzburg J., Hancock J.F. 1994. The effector domain of Rab6, plus a highly hydrophobic C terminus, is required for Golgi apparatus localization. Mol. Cell. Biol. 14:744–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Hofmann K., Bucher P., Neuwald A.F., Altschul S.F., Koonin E.V. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68–76 [PubMed] [Google Scholar]
- Brekelmans C.T., Tilanus-Linthorst M.M., Seynaeve C., vd Ouweland A., Menke-Pluymers M.B., Bartels C.C., Kriege M., van Geel A.N., Burger C.W., Eggermont A.M., et al. 2007. Tumour characteristics, survival and prognostic factors of hereditary breast cancer from BRCA2-, BRCA1- and non-BRCA1/2 families as compared to sporadic breast cancer cases. Eur. J. Cancer. 43:867–876 10.1016/j.ejca.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Bretscher A., Edwards K., Fehon R.G. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3:586–599 10.1038/nrm882 [DOI] [PubMed] [Google Scholar]
- Chishti A.H., Kim A.C., Marfatia S.M., Lutchman M., Hanspal M., Jindal H., Liu S.C., Low P.S., Rouleau G.A., Mohandas N., et al. 1998. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci. 23:281–282 10.1016/S0968-0004(98)01237-7 [DOI] [PubMed] [Google Scholar]
- Coene E.D., Hollinshead M.S., Waeytens A.A., Schelfhout V.R., Eechaute W.P., Shaw M.K., Van Oostveldt P.M., Vaux D.J. 2005. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol. Biol. Cell. 16:997–1010 10.1091/mbc.E04-10-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakowski W., Grzybek M., Sikorski A.F. 2006. Protein 4.1, a component of the erythrocyte membrane skeleton and its related homologue proteins forming the protein 4.1/FERM superfamily. Folia Histochem. Cytobiol. 44:231–248 [PubMed] [Google Scholar]
- Glover J.N. 2006. Insights into the molecular basis of human hereditary breast cancer from studies of the BRCA1 BRCT domain. Fam. Cancer. 5:89–93 10.1007/s10689-005-2579-z [DOI] [PubMed] [Google Scholar]
- Glover J.N., Williams R.S., Lee M.S. 2004. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29:579–585 10.1016/j.tibs.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Haynes L.W., Weller R.O. 1978. The effects of cytochalasin B and colchicine on cell motility and ultrastructure in primary cultures of malignant gliomas. Acta Neuropathol. 44:21–30 10.1007/BF00691635 [DOI] [PubMed] [Google Scholar]
- Heiska L., Alfthan K., Grönholm M., Vilja P., Vaheri A., Carpén O. 1998. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 273:21893–21900 10.1074/jbc.273.34.21893 [DOI] [PubMed] [Google Scholar]
- Hu Y.F., Hao Z.L., Li R. 1999. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 13:637–642 10.1101/gad.13.6.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter K.W. 2004. Ezrin, a key component in tumor metastasis. Trends Mol. Med. 10:201–204 10.1016/j.molmed.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I., Soriano J.V., Vaudan G., Montesano R., Sappino A.P. 1998. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 143:1329–1339 10.1083/jcb.143.5.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy A.K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 24:289–296 10.1006/meth.2001.1189 [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Altschul S.F., Bork P. 1996. BRCA1 protein products…Functional motifs…. Nat. Genet. 13:266–268 10.1038/ng0796-266 [DOI] [PubMed] [Google Scholar]
- Ludwig T., Fisher P., Ganesan S., Efstratiadis A. 2001. Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 15:1188–1193 10.1101/gad.879201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A.N., Lee C.-F., Vaux D.J. 2009. Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184:45–55 10.1083/jcb.200804155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., Aten J.A. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375–382 [DOI] [PubMed] [Google Scholar]
- Monteiro A.N. 2000. BRCA1: exploring the links to transcription. Trends Biochem. Sci. 25:469–474 10.1016/S0968-0004(00)01632-7 [DOI] [PubMed] [Google Scholar]
- Nomura N., Nomura M., Sugiyama K., Hamada J. 2007. Src regulates phorbol 12-myristate 13-acetate-activated PKC-induced migration via Cas/Crk/Rac1 signaling pathway in glioblastoma cells. Int. J. Mol. Med. 20:511–519 [PubMed] [Google Scholar]
- Peter M., Ameer-Beg S.M. 2004. Imaging molecular interactions by multiphoton FLIM. Biol. Cell. 96:231–236 10.1016/j.biolcel.2003.12.006 [DOI] [PubMed] [Google Scholar]
- Peter M., Ameer-Beg S.M., Hughes M.K.Y., Keppler M.D., Prag S., Marsh M., Vojnovic B., Ng T. 2005. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 88:1224–1237 10.1529/biophysj.104.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.A., Au W.W., Henderson B.R. 2004. Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp. Cell Res. 293:14–21 10.1016/j.yexcr.2003.09.027 [DOI] [PubMed] [Google Scholar]
- Rosette C., Roth R.B., Oeth P., Braun A., Kammerer S., Ekblom J., Denissenko M.F. 2005. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis. 26:943–950 10.1093/carcin/bgi070 [DOI] [PubMed] [Google Scholar]
- Sato N., Funayama N., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. 1992. A gene family consisting of ezrin, radixin and moesin. Its specific localization at actin filament/plasma membrane association sites. J. Cell Sci. 103:131–143 [DOI] [PubMed] [Google Scholar]
- Seery L.T., Knowlden J.M., Gee J.M., Robertson J.F., Kenny F.S., Ellis I.O., Nicholson R.I. 1999. BRCA1 expression levels predict distant metastasis of sporadic breast cancers. Int. J. Cancer. 84:258–262 [DOI] [PubMed] [Google Scholar]
- Soriano J.V., Irminger-Finger I., Uyttendaele H., Vaudan G., Kitajewski J., Sappino A.P., Montesano R. 2000. Repression of the putative tumor suppressor gene Bard1 or expression of Notch4(int-3) oncogene subvert the morphogenetic properties of mammary epithelial cells. Adv. Exp. Med. Biol. 480:175–184 10.1007/0-306-46832-8_22 [DOI] [PubMed] [Google Scholar]
- Starita L.M., Parvin J.D. 2006. Substrates of the BRCA1-dependent ubiquitin ligase. Cancer Biol. Ther. 5:137–141 10.4161/cbt.5.2.2479 [DOI] [PubMed] [Google Scholar]
- Starita L.M., Machida Y., Sankaran S., Elias J.E., Griffin K., Schlegel B.P., Gygi S.P., Parvin J.D. 2004. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 24:8457–8466 10.1128/MCB.24.19.8457-8466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D.G., Honn K.V. 1994-1995. Adhesion molecules and tumor metastasis: an update. Invasion Metastasis. 14:109–122 [PubMed] [Google Scholar]
- Taylor J., Lymboura M., Pace P.E., A’hern R.P., Desai A.J., Shousha S., Coombes R.C., Ali S. 1998. An important role for BRCA1 in breast cancer progression is indicated by its loss in a large proportion of non-familial breast cancers. Int. J. Cancer. 79:334–342 [DOI] [PubMed] [Google Scholar]
- Thompson M.E., Jensen R.A., Obermiller P.S., Page D.L., Holt J.T. 1995. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 9:444–450 10.1038/ng0495-444 [DOI] [PubMed] [Google Scholar]
- Tomlinson G.E., Chen T.T., Stastny V.A., Virmani A.K., Spillman M.A., Tonk V., Blum J.L., Schneider N.R., Wistuba I.I., Shay J.W., et al. 1998. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 58:3237–3242 [PubMed] [Google Scholar]
- Weissman A.M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169–178 10.1038/35056563 [DOI] [PubMed] [Google Scholar]
- Wickstead B., Ersfeld K., Gull K. 2003. The frequency of gene targeting in Trypanosoma brucei is independent of target site copy number. Nucleic Acids Res. 31:3993–4000 10.1093/nar/gkg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wagner K.U., Larson D., Weaver Z., Li C., Ried T., Hennighausen L., Wynshaw-Boris A., Deng C.X. 1999. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat. Genet. 22:37–43 10.1038/8743 [DOI] [PubMed] [Google Scholar]
- Yamane K., Katayama E., Tsuruo T. 2000. The BRCT regions of tumor suppressor BRCA1 and of XRCC1 show DNA end binding activity with a multimerizing feature. Biochem. Biophys. Res. Commun. 279:678–684 10.1006/bbrc.2000.3983 [DOI] [PubMed] [Google Scholar]
- Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S., Tsukita S. 1998. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140:885–895 10.1083/jcb.140.4.885 [DOI] [PMC free article] [PubMed] [Google Scholar]