The PDGF/VEGF-related receptor and the EGF receptor control the direction of collective cell migration by regulating the persistence and productivity of protrusions.
Abstract
Border cells perform a collective, invasive, and directed migration during Drosophila melanogaster oogenesis. Two receptor tyrosine kinases (RTKs), the platelet-derived growth factor/vascular endothelial growth factor–related receptor (PVR) and the epidermal growth factor receptor (EGFR), are important for reading guidance cues, but how these cues steer migration is not well understood. During collective migration, front, back, and side extensions dynamically project from individual cells within the group. We find that guidance input from both RTKs affects the presence and size of these extensions, primarily by favoring the persistence of front extensions. Guidance cues also control the productivity of extensions, specifically rendering back extensions nonproductive. Early and late phases of border cell migration differ in efficiency of forward cluster movement, although motility of individual cells appears constant. This is caused by differences in behavioral effects of the RTKs: PVR dominantly induces large persistent front extensions and efficient streamlined group movement, whereas EGFR does not. Thus, guidance receptors steer movement of this cell group by differentially affecting multiple migration-related features.
Introduction
Eukaryotic cell motility is based on an interdependent set of migratory features: formation of forward protrusions, which usually depends on an underlying dynamic actin cytoskeleton and requires membrane expansion, probes new territory. Adhesion to the substratum, which can be extracellular matrix or other cells, allows traction; deadhesion must also occur in the back of the cell. Forces acting on substrate adhesions and on cellular components, including the cytoskeleton, promote translocation (Ridley et al., 1992; Lauffenburger and Horwitz, 1996). Finally, for the migration of animal cells in vivo, an ability to invade the target tissue may also be required. In vivo, migratory cells are also generally guided to their destination by guidance cues to perform their physiological function. Guidance cues may, in principle, spatially bias any of the migratory features to give directional bias and thereby steer cell movement. Visual inspection in simple systems shows that guidance signaling affects the formation and directionality of cellular protrusions (Van Haastert and Devreotes, 2004; Berzat and Hall, 2010). This, in turn, has been related to polarized regulation of the actin cytoskeleton, elevated local actin polymerization to regulate lamellipodia, or other effects to form cell blebs (Pollard and Borisy, 2003; Insall and Machesky, 2009). Guidance input can also control the selective maintenance of one cellular protrusion over another in an all-or-none manner (Andrew and Insall, 2007; Martini et al., 2009). Finally, guidance receptors can affect adhesion to the substratum (Miao et al., 2000; Ren et al., 2004). Whether force generation is directly regulated is less clear.
In the physiological setting of a multicellular animal, some cells migrate as singular entities, whereas others migrate collectively. Cells can be considered to migrate collectively if they migrate together and affect each other while doing so. Collectively migrating cells can be epithelial or mesenchymal in nature and may migrate as small groups or large sheets, as discussed in recent reviews (Friedl and Gilmour, 2009; Rørth, 2009; Weijer, 2009). With respect to directionality, cells within a migrating group could each be steered individually, exactly as single cells. However, there is also evidence that guidance entails a collective response: response to guidance cues requires interactions between migrating cells (Theveneau et al., 2010), and differential effects on cells at distinct positions within the group may steer movement (Bianco et al., 2007; Rørth, 2007). Individual and collective guidance responses are not mutually exclusive. Understanding how collective migrations are directed is of broad interest, as such migrations are key to many aspects of tissue morphogenesis. Collective migration may also be responsible for the dissemination of tumors; in particular, those derived from epithelia (Friedl et al., 2004; Christiansen and Rajasekaran, 2006).
Border cells are a small group of cells that perform a collective, directed migration during Drosophila melanogaster oogenesis (Montell, 2003). These cells delaminate from a simple epithelium and remain tightly associated as they invade the germline tissue, squeezing in between the giant nurse cells to reach the oocyte (Fig. 1 A). The nurse cells act as substratum for the migration; adhesion between migrating cells and their substrate depends on E-cadherin (Niewiadomska et al., 1999). Two receptor tyrosine kinases (RTKs), PDGF/VEGF-related receptor (PVR) and EGF receptor (EGFR), function in border cells to guide them posteriorly to the oocyte and, finally, dorsally, close to the oocyte nucleus (Duchek and Rørth, 2001; Duchek et al., 2001). Ligands for these receptors, principally PVF1 and Gurken, respectively, are expressed by the oocyte. Either of the two RTKs can direct border cells to the oocyte, but EGFR and its ligand Gurken are required for the final dorsal migration. Genetic analysis indicated that the RTKs may use multiple pathways to direct border cell migration (Bianco et al., 2007). Interestingly, recent experiments using a photoactivatable form of the small GTPase Rac have shown that differential activity of Rac can be sufficient to direct movement of the cluster (Wang et al., 2010). Finally, live imaging of border cell migration showed that inactivation of the RTKs led to extensions being formed in all directions (Prasad and Montell, 2007). Live analysis also revealed some difference in migration behavior between the early and the late part of the process (Bianco et al., 2007). Thus, we have some information about how RTK activity may direct movement of the border cell cluster, but an overall view is lacking. PVR is also important for directed migration of other cell types in Drosophila, both for individual cells (Cho et al., 2002; Learte et al., 2008) and for epithelial sheet movement in wound closure (Wu et al., 2009). In this study, we use live imaging of border cell migration to obtain quantitative information about the behavior of border cell clusters and the relationship between migratory features, to determine the difference between the two phases of migration, and, finally, to gain further insight into how guidance input shapes border cell behavior.
Figure 1.
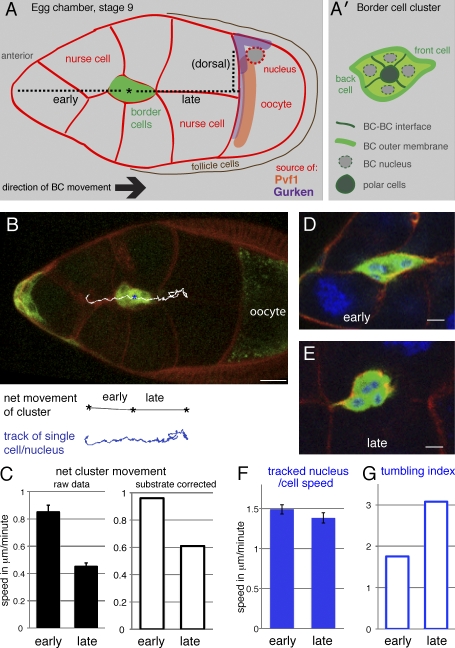
Speed and directionality of border cell clusters during early and late phases of migration. (A) Schematic drawing of a stage 9 egg chamber corresponding to the image in B. Border cells (BCs) move posteriorly to the oocyte in all images shown as left to right. The attractive ligands are made by the oocyte: Pvf1 for PVR and Gurken, enriched dorsally near the oocyte nucleus, for EGFR. The dotted line corresponds to the early, late, and dorsal segments as indicated. (A’) Details of the border cell cluster. (B) Image from a wild-type video (UAS-CD8-GFP/+;slboGal4/+) showing the border cell cluster between early and late migration and the track of one nucleus (cell) over 2 h (white in overlay; blue below). All cells are outlined by FM 4–64 (red). Bar, 20 µm. (A and B) Asterisks indicate the midpoint of the posterior migration. (C) Net (point to point) speed of the border cell cluster center in micrometers per minute in early and late phase (n = 37 and 41). Error bars indicate SEM; the difference is significant (P < 10−9). The right graph shows the mean net speed corrected for mean backward sliding of the substrate. (D and E) Early migrating border cell cluster (D) and late cluster (E) expressing 10×GFP (green) stained with phalloidin (red) and DAPI (blue). Bars, 10 µm. (F) Speed of a single tracked nuclei (representing single cells) in a 2D projection; the difference is not statistically significant. (G) Tumbling index: path of a tracked single nucleus per cell over the net cluster path. (F and G) n = 17 and 18 cells.
Results
Quantification of border cell migration during early and late phases
A schematic illustration of the border cell cluster and their migratory path is shown in Fig. 1 A. In this analysis, we consider only migration to the oocyte (posterior migration) and define the early and late phases in terms of the path: the early phase is from detachment until halfway to the oocyte; the late phase is from halfway until the oocyte is first touched. For live analysis of migration, border cells were labeled by expression of a neutral cytoplasmic GFP marker, 10×GFP or CD8-GFP, and all cells were labeled in red by a vital membrane dye (Fig. 1 B). As reported previously (Bianco et al., 2007), net migration of border cell clusters (point to point) was on average twice as fast during the early phase as during the late phase (Fig. 1 C). However, the migration substrate was not static: because of oocyte and egg chamber growth, it showed a mean backward movement relative to the imaging grid of 0.11 (early)–0.16 (late) µm/min. Thus, our use of fixed xy coordinates underestimates the cluster movement relative to the substrate and slightly enhances the difference between early and late migration (Fig. 1 C). Early movement was more streamlined and sliding (Videos 1 and 2), and clusters were elongated (Fig. 1 D). Late movement was more disordered, with clusters sometimes rotating in place (Videos 2 and 3) and rounder in shape (Fig. 1 E). The variation in behavior between individual border cell clusters was considerable and shifts could occur at any point, emphasizing the need for a systematic analysis of many videos.
To determine whether the change in net movement from early to late phase reflected a change in cell motility, we estimated the movement of individual border cells. For each cluster, the center of a single nucleus was manually tracked in 3D. The track was projected onto the xy axis, and the total distance traversed was measured (example in Fig. 1 B). The tracked movements covered wide angles (Fig. S1, 12 traces), and as border cell nuclei usually remain centrally located in the cell, most of this must be actual cell movement. Analysis of a video with only one border cell labeled confirmed this and indicated that manual nuclear tracking overestimates cell motility somewhat but does so systematically. Using tracked nuclei, apparent single-cell movement revealed only a slight (<10%) decrease from early to late phase (Fig. 1 F). This can be partially accounted for by the increase in late backward substrate sliding. Thus, the main difference from early to late migration was a decrease in apparent directionality. One complication is that, as the border cell cluster moves by invading germline tissue, cluster rotation could also be caused by lack of tissue penetration. We therefore formally describe the change from early to late phase as an increase in tumbling, which is measured as the path of tracked nuclei divided by the path of the cluster (Fig. 1 G). In conclusion, border cell clusters show constant cell motility but increased tumbling and decreased directionality as they move toward the oocyte.
Quantification of cellular extensions and movement
Cellular protrusions of different types are generally important for eukaryotic cell movement. To relate border cell cluster migration to cellular protrusions in a systematic way, we developed a method for automatically defining the body of the cluster and its extensions in all time points of a video and assembling this data into overall profiles (see Materials and methods). In brief, the border cell cluster was aligned with egg chamber geometry, the volume of all cells was projected onto the xy plane (Fig. 2 A), and the image was segmented to represent the full outline. The body of the cluster (Fig. 2 A′, blue) was defined as the area within this original area that could be explored by a ball slightly larger than a border cell nucleus. Extensions were defined as the rest of the area (Fig. 2 A′, red); that is, protrusions away from the cluster body. Videos 8 and 9 show examples of automated extension definition overlaid onto original videos. Snapshots of the extension were linked by an overlap in subsequent time points, allowing measurement of persistence time in addition to size and direction of an extension. Extensions present at only one time point were set to have a lifetime of the imaging interval (1–1.5 min) but may be even shorter lived. Also, very small cellular extensions, including filopodia, were not captured in this analysis. Generally, extensions defined in this manner each come from one cell, the one facing the direction of the extension (observations of videos; Fulga and Rørth, 2002). For example, a front cell forms a forward extension, and a back cell forms a backward extension. Direct measurements confirmed that back extensions were mainly actively formed extensions and not nonretracted tails: in 77% of cases they clearly extended further backward than the area occupied by the cell in the previous time point. This overall procedure allowed an automatic definition of the cluster center of mass (in projection), the movement of which was followed in time, coupled with a systematic characterization of cellular extensions from the cluster.
Figure 2.
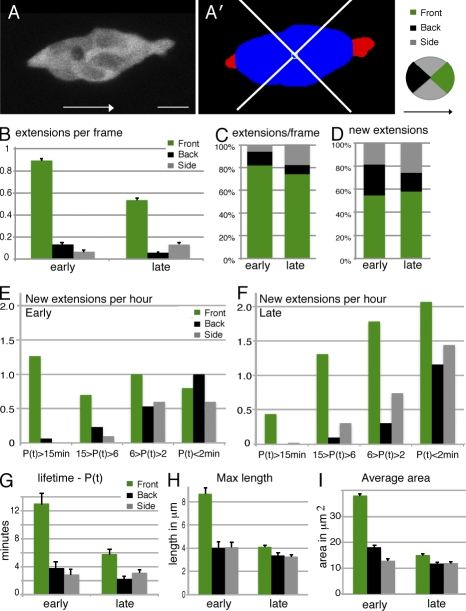
Analysis of directional bias, persistence, and size of cellular extensions from normal border cell clusters. (A) Projected GFP image of a wild-type border cell cluster (slboGal4, UAS-10xGFP/+); arrows indicate direction to the oocyte. Bar, 10 µm. (A’) Same as A, but after segmentation and automatic definition of the cluster body (blue) and extensions (red). (B) Mean number of extensions observed per frame (snapshots) in each direction (n = 1,336 [early] and 2,350 [late] frames). All differences, including between early and late back and side extensions, are statistically significant (P < 0.001). (C) Data from B in percentages. (D) The percentage of distribution of new extensions in each direction (n = 207 [early] and 509 [late] extensions). (E and F) Early phase extensions (E) and late phase extensions (F) per hour of video, binned by direction and lifetime per persistence (P(t) in minutes). P(t) < 2 min are extensions observed in one frame only. (G) Mean extension lifetime, data as in E and F. Low P(t) means are not accurate because of the high contribution from a single-frame extension. (H) Maximal extension lengths. (E–H) n = 44–295. (I) Mean extension areas (in 2D projection) in micrometers squared, based on all snapshots (n = 88–1,064). All differences between front and other extensions are statistically significant; P < 0.01. Error bars indicate SEM.
Analysis of all images in early and late videos for the presence of extensions and their direction showed an overall prominent front bias (Fig. 2, B and C) and, overall, more extensions early. To determine whether this reflected a difference in extension outgrowth frequency or in their persistence, the directional bias per new extension was determined (Fig. 2 D) as well as the lifetime of extensions in each direction (Fig. 2, E–G). Both early and late stages revealed a front bias for extension formation, with ∼60% appearing in the front quadrant. The most obvious difference between the phases was a higher frequency of long-lived (persisting over 15 min) front extensions in the early phase (Fig. 2 E). There was an increase in short-lived extensions later on (Fig. 2 F). Even at the late stage, front extensions were on average more stable than other extensions (Fig. 2 G). There were also statistically significant changes in nonfront extensions: more (and slightly more stable) early back extensions and more late side extensions. The mean maximal length of extensions was close to 4 µm for all categories except for the longer early front extension (Fig. 2 H). The mean size of extensions showed the same trend (Fig. 2 I). Extension lifetimes and maximal size were quite well correlated (Fig. S2). Thus, the early phase of migration is characterized by large and persistent front extensions, which correspond to extensions from the front cell, whereas the bias in making front extensions over other extensions is similar throughout.
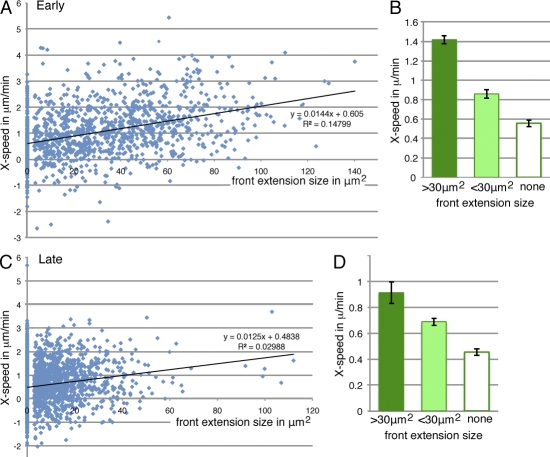
We next asked whether the two characteristics defining early migratory clusters, efficient forward movement and the presence of large persistent front extensions, were functionally related. This was first explored by determining the relationship between the size of the front extension and the forward-directed movement of the cluster to the next time point. We observed a moderate positive correlation between these features for both early (Fig. 3 A) and late migration (Fig. 3 B). Binning data according to front extension size (Fig. 3, C and D) showed the velocity differences to be highly significant (P < 0.001). The high variability observed may, in part, reflect biological noise. But coupled with the finding that most clusters with no detectable front extensions had significant forward-directed movement, it is also consistent with other variables influencing cluster movement. The relationship between front extension size and forward velocity was similar in the two phases, with large front extensions (size >30 µm2) giving a mean forward-directed speed two- to threefold higher than no front extensions and moderate front extensions (size <30 µm2) in between. When data with matched front extension sizes were compared, there was a modest (20%) decrease in forward speed in the late phase. Part of this reflects an increased backward substrate sliding as discussed in the first section of the Results. Thus, the ability to grow and maintain a large forward extension may be the major difference between early and late phases of migration. Furthermore, the data indicate that net forward movement of the cluster is directly related to how big a forward extension the front cell can produce or maintain.
Figure 3.
Relationship between front extension size and forward movement. (A) Scatter plot of all early events (frames) according to an area in a 2D projection of the front extension and instantaneous movement (speed in micrometers per minute) of the cluster toward the oocyte to the next time point, measured in the x axis only. Dataset as in Fig. 2 B. (C) Scatter plot of all late events, as in A. Best-fit linear regression lines are shown. (B and D) Data from A and C, respectively, binned according to front extension size. None indicates no front extension above the technical cutoff of 3 µm2. SEM is indicated. All differences are significant (P < 0.001).
Perturbing individual guidance receptors: PVR promotes early behavior
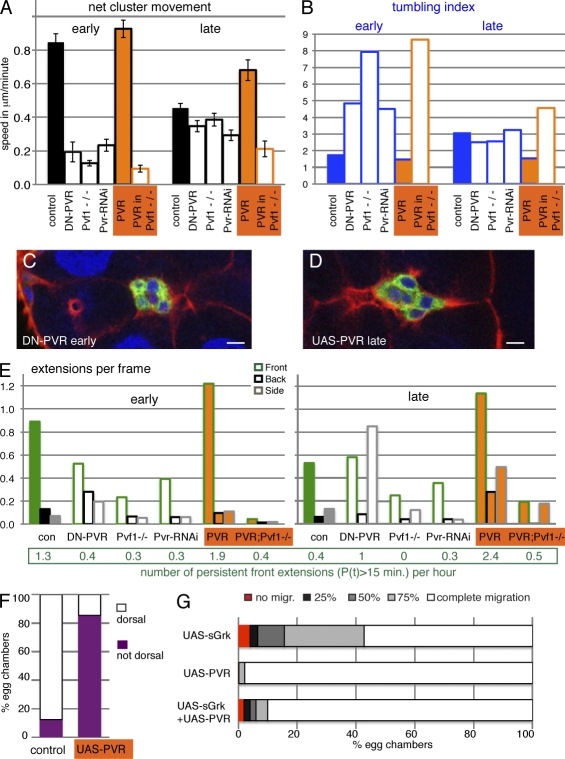
The early and late phases of migration differ mainly in apparent directionality. As this is a guided migration, directionality is related to the perception of guidance cues. To determine the role of the two known guidance receptors in early and late behaviors, we examined the detailed effects of perturbations of PVR or EGFR function. Expression of a dominant-negative (DN) version of PVR (DN-PVR) in border cells, reducing Pvr expression by RNAi, or removing the PVR ligand Pvf1 from the tissue all deceased the net forward speed of the cluster, in particular in the early phase (Fig. 4 A). In agreement with PVR having a guidance effect, apparent single-cell motility was barely affected, but tumbling was increased (Fig. 4 B). A separate set of analyses using CD8-GFP as a marker and including clonal analysis of border cells with a loss-of-function mutation for Pvr confirmed these findings (Fig. S3). Early phase movement in these genotypes resembled the normal late movement (Video 4 and Fig. 4 C). Front extensions were observed at a lower frequency when PVR activity was reduced (Fig. 4 E), including loss of the most persistent front extensions (Fig. 4 E, bottom). DN-PVR expression had a slightly different effect on cellular extensions than the other PVR loss-of-function situations, as it also increased nonfront extensions. But overall, the results indicated that PVR activity is required for the characteristic early cluster behavior.
Figure 4.
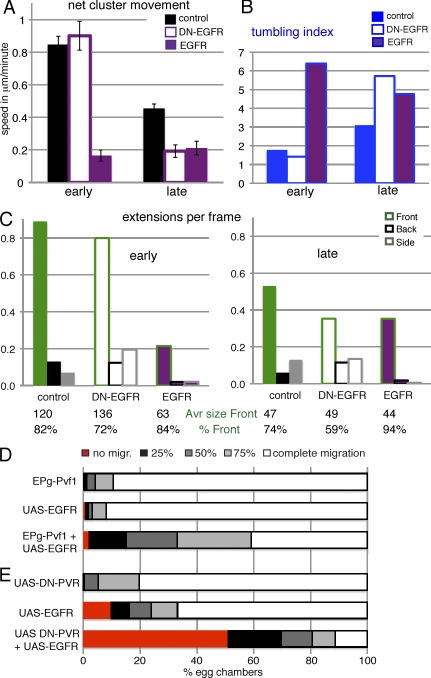
Effects of PVR on the behavior of border cell clusters. (A) Net movement of border cell clusters. Genotypes: slboGal4, UAS-10×GFP/+ (control [con]) and indicated UAS transgenes; Pvf1−/− is a Pvf11624 homozygous female, PVR in Pvf1−/− is slboGal4, UAS-PVR in a Pvf11624 mutant. SEM is indicated. All differences to control are significant (P < 0.02), except early PVR and late Pvf1−/−. n = 5–16. (B) Tumbling index (n = 3–12); genotypes as in A. (C) Early border cell cluster DN-PVR/+;slboGal4, UAS-10×GFP. (D) Late border cell cluster UAS-PVR/+;slboGal4, UAS-10×GFP/+. GFP marks border cells; phalloidin (red) marks F-actin, and DAPI (blue) marks nuclei. Bars, 10 µm. (E) Extensions per frame (n > 230 per genotype and stage). All values that appear modestly different are statistically significant (P < 0.02). The number of long-lived (P(t) > 15 min) front extension per hour is indicated below. These are rare, and most differences are not statistically significant. (F) Dorsal EGFR-dependent migration; slboGal4, UAS-10×GFP/+ (control) and UAS-PVR/+;slboGal4, UAS-10×GFP/+ (n = 78 and 100). (G) Position of border cells in stage 10 egg chambers. Genotypes: slboGal4, slbo1310-lacZ/+ and the indicated transgenes (n = 100–800). The control showed a 99% complete migration (migr.).
To determine whether PVR was sufficient to induce early behavior, we overexpressed the receptor in border cells (PVR; Fig. 4, orange). This had a mild effect on early migration, slightly increasing net speed and the lifetime of front extension. Late migration, however, was transformed to a behavior more similar to early clusters, showing sliding movement (Video 5), reduced tumbling (Fig. 4 B), and elongated shapes (Fig. 4 D). PVR overexpression induced more extensions, particularly more persistent front extensions (Fig. 4 E). All these effects required the presence of the endogenous ligand Pvf1. This indicated that PVR signaling was not only required but also sufficient for early type migration behavior. It also indicated that PVR was functional as a guidance receptor over a large expression range, with an ∼10-fold overexpression in this case. Increased expression of PVR caused it to be dominant over EGFR. Whereas wild-type clusters move dorsally upon reaching the oocyte in response to the dorsal EGFR ligand Gurken (Video 3; Duchek and Rørth, 2001), PVR-overexpressing ones do not (Fig. 4 F and Video 6). Also, the inhibitory effect of misexpressing secreted Gurken was alleviated by coexpressing PVR (Fig. 4 G), as shown previously for another EGFR ligand, Vein (Duchek et al., 2001). Thus, elevated PVR levels made border cells insensitive to EGFR ligands, ultimately perturbing the migration path.
We next analyzed the role of EGFR (Fig. 5 and Fig. S3). Loss of EGFR (DN-EGFR) had little effect on early movement but made late movement even less efficient, enhancing the difference between the two phases (Fig. 5 A). The directional movement was replaced with increased tumbling (Fig. 5 B), which was consistent with a guidance role. Apparently, endogenous EGFR signaling becomes increasingly important as border cell clusters approach the oocyte. But EGFR can act earlier: when the endogenous PVR level was reduced to half (in Pvr1/+), even early migration was affected by DN-EGFR (net speed of 0.21 µm/min; SEM of 0.03). Expression of DN-EGFR during late migration also caused fewer front extensions (Fig. 5 C). Surprisingly, clusters with an increased expression of EGFR had significantly impaired forward movement at both phases with extensive shuffling (Fig. 5, A and B; and Video 7). Fewer cellular extensions were observed overall, and the front extension was reduced, but a strong front bias was retained (Fig. 5 C, bottom). Finally, these clusters showed an increased sensitivity to misexpression of Pvf1 (Fig. 5 D) and to a suppression of signaling by DN-PVR (Fig. 5 E), indicating that the endogenous PVR pathway was still functional. This implies that EGFR, in contrast to PVR, does not function effectively as a guidance receptor over a large expression range. So, although PVR and EGFR are both RTKs and have partially redundant guidance roles, they affect the migrating cells differently.
Figure 5.
Effects of EGFR on the behavior of border cell clusters. (A) Net movement of border cell clusters. Genotypes: slboGal4, UAS-10×GFP/+ (control) and indicated UAS transgenes. SEM is indicated; differences are significant at P < 0.001, except for control versus early DN-EGFR. (B) Tumbling index. (A and B) n = 6–14. (C) Extensions per frame (n > 170 per genotype and stage); differences between genotypes are significant (P < 0.001), except for control versus early DN-EGFR. (bottom) Average (Avr) size of front extensions and percentage of total extensions (in snapshots) that are front. (D and E) Position of border cells in stage 10 egg chambers. Genotypes: slboGal4, slbo1310-lacZ/+ and the indicated transgenes; EPgPvf1 drives Pvf1 expression (n = 100–900). The control showed a 99% complete migration (migr.).
PVR and EGFR may have different effects caused by use of alternate downstream pathways. The small GTPase Rac plays a central role in border cell migration and guidance (Murphy and Montell, 1996; Duchek et al., 2001; Geisbrecht and Montell, 2004; Bianco et al., 2007; Wang et al., 2010). Also, our previous analysis indicated that the Rac exchange factor consisting of DOCK180/Mbc and Elmo was most critical for early migration, and MAPK and PLC-γ were critical for late migration (Bianco et al., 2007). The strong perturbation of Rac blocked border cell migration, but a mild reduction of Elmo levels selectively affected early behavior (Fig. S4). In other systems, Elmo/DOCK180 activation can depend on input from upstream Rac–guanine nucleotide exchange factors (Katoh and Negishi, 2003; deBakker et al., 2004). The Rac exchange factor Vav can be recruited directly to activated PVR and EGFR (Bianco et al., 2007), and reducing the expression of Vav showed similar effects as for Elmo, which is consistent with both Vav and Elmo being critical for early migration. In contrast, completely disrupting regulation of the MAPK pathway by expression of dominant-activated Raf strongly affected late migration, with a mild effect on early migration (Fig. S4). However, the selective effect of PVR versus EGFR is not altogether explained by this pathway analysis, as both receptors can signal through both pathways in somatic cells of the ovary (Jékely et al., 2005) as well as in other contexts. Other differences, such as in subcellular location of the receptors, may also contribute to PVR- versus EGFR-specific migration behaviors.
Effects of both guidance receptors: extensions and their productivity
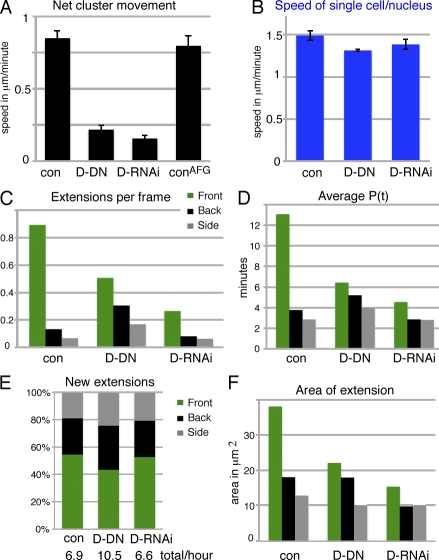
As PVR and EGFR have partially overlapping functions in border cells (Duchek et al., 2001), we needed to determine which features were affected by both receptors. For this, we first analyzed border cell clusters expressing DN versions of both RTKs. Such “double DN” (D-DN) clusters showed severe delays in initiating migration and strongly reduced forward-directed speed once migratory (Fig. 6 A), with normal apparent single-cell motility (Fig. 6 B). Consequentially, only the first half of the migration path was traversed during stage 9. Spatially, this corresponds to early migration, although temporally, the migration occurs late. As found in a previous study (Prasad and Montell, 2007), D-DN border cell clusters display a poorly polarized extension profile caused by a loss of forward extensions as well as an increase in other extensions (Fig. 6 C). This effect resulted from a substantial destabilization of front extensions and a slight stabilization of other extensions (Fig. 6 D) coupled with a small increase in the number of extensions formed in other directions (Fig. 6 E). Extension size grossly reflected the persistence time (Fig. 6 F). DN-PVR expression can produce effects different from simple loss of function (Rosin et al., 2004), and we had observed differences between the effects of DN-PVR and Pvr RNAi or the Pvf1 mutant background with regard to extensions (Fig. 4 E). We therefore also examined border cell clusters in which expression of both RTKs was strongly reduced by RNAi (Fig. 6, double RNAi [D-RNAi]). As expected, directional migration of the border cell cluster was severely perturbed (Fig. 6, A and B). There was a loss of front extensions but no gain of other extensions (Fig. 6 C). The difference from the wild type was solely at the level of persistence time with no change in the number of extensions formed (Fig. 6, D and E). In summary, the consistent effect of perturbing RTK guidance receptor signaling was a destabilization of front-directed extensions. The characteristics of extensions in other directions could also be affected, depending on how RTK perturbation was achieved.
Figure 6.
Effects of perturbing both guidance receptors. (A) Net movement of border cell clusters. Genotypes (for all panels): slboGal4, UAS-10×GFP/+ (control [con]) or UAS-DN-EGFR/UAS-DN-PVR;slboGal4, UAS-10×GFP/+ (D-DN) or hsFLP/+;UAS-EgfrRNAi/+;UAS-PvrRNAi/AFG, UAS-10×GFP (D-RNAi) and hsFLP/+;AFG, UAS-10×GFP/+ (conAFG; n = 37, 38, 18, and 22, respectively). SEM is indicated. AFG (actin-flipout-Gal4) was activated 2 d before imaging to give expression in all somatic cells, including border cells. Only migration at stage 9, up to 50% of path (early), was analyzed. The controls looked similar in all regards. (B) Apparent single-cell speed (tracked nuclei). n = 12–17. (C) Extensions per frame (n > 1,300); all differences are significant with P < 0.001, except the side versus back of D-RNAi (P < 0.05). (D) Mean lifetime of extensions (P(t)); there were significant differences (P < 0.02) for front extensions in different genotypes and compared with side and back extensions. (E) Direction of new extensions in percentages; indicated below is the total number of extensions per hour (n > 200). (F) Mean extension areas in micrometers squared based on all snapshots of extensions; all differences are significant (P < 0.01), except for side extensions and control versus D-RNAi. (D and F) n = 150–266.
The aforementioned results establish that one function of graded guidance receptor signaling is to selectively stabilize front-directed extensions or extensions from front cells. However, a front bias in extension formation remained in both double-receptor perturbation experiments (Fig. 6 E). This could be caused by an incomplete reduction in the RTK activity, as the migration defects were slightly milder than for double loss-of-function clones of null alleles (Jékely et al., 2005). Alternatively, there might be additional input reducing side and back extensions from border cell clusters. It is also clear that the reduced frequency and size of the front extensions do not fully explain the migration phenotype of guidance-deficient border cell clusters. When normalized for front extension size, there was still a two- to threefold reduction in net forward movement (Fig. S5). A similar tendency was observed with separate perturbations of PVR and EGFR (Fig. 4 and Fig. 5). This suggested that guidance input might also regulate other features of directional migration.
The analysis of wild-type border cell clusters indicated that the presence and size of forward extensions significantly affected forward-directed movement (Fig. 3). This was further supported by RTK manipulations producing parallel effects on forward extensions and net cluster movement. Extensions are also formed in other directions; do these affect net movement of the cluster as well? This was simplest to consider in the case of backward-directed extensions, which were well quantified in the analysis, and as for front extensions, effects would be expected along the axis of migration. As discussed initially, backward extensions are mainly actively formed extensions, growing outward from the back cell. If all outward extensions from cells of the cluster were equal, back cells might engage in a tug of war with front cells. To see whether this was occurring, we reconsidered the movement at each time point in our original dataset, now distinguishing between clusters with and without back extensions. In both early and late wild-type border cell clusters, the presence of a backward-directed extension had little or no effect on the net forward speed of the cluster (Fig. 7, A and B). This finding was particularly significant in cases in which clusters had a modest front extension (<30 µm2), and thus, front and back extensions were of similar size, and in cases without front extensions. In conclusion, back extensions appeared to be largely without effects, or nonproductive, with regard to the net cluster movement in the normal situation.
Figure 7.
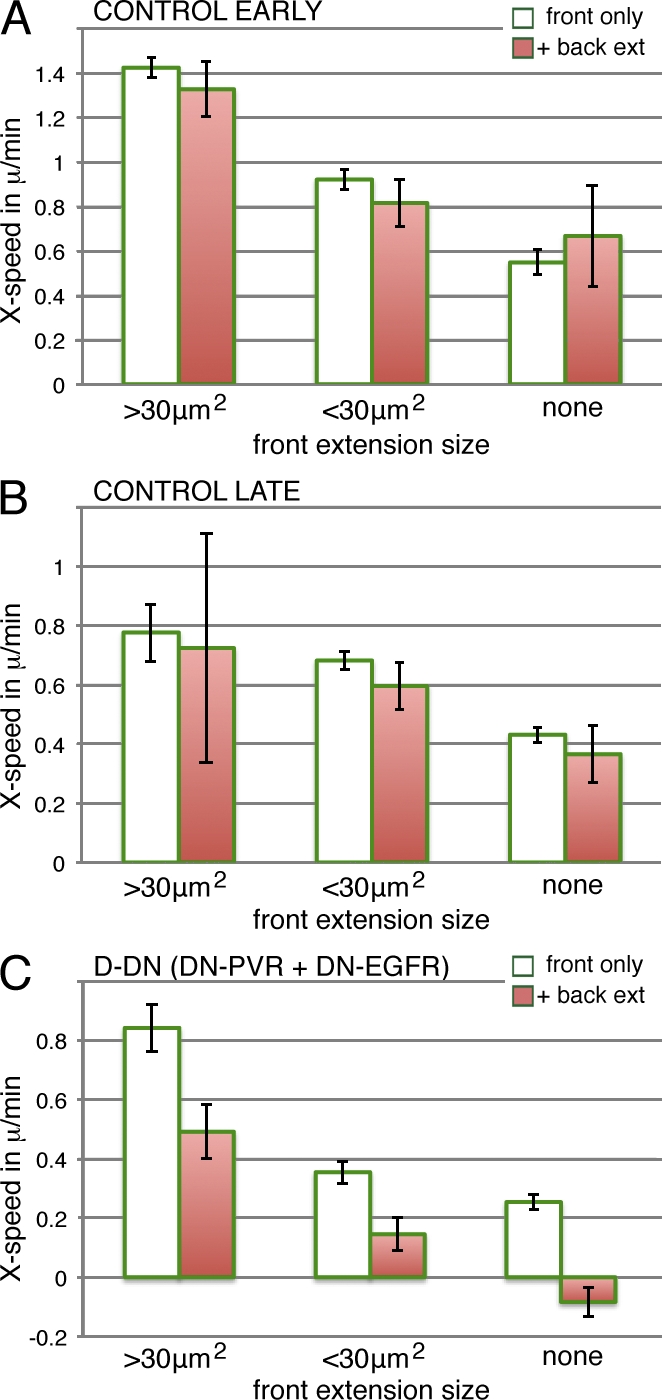
Effects of back extensions on cluster movement are guidance dependent. Mean instantaneous movement of the border cell cluster toward the oocyte binned by size of front extension as in Fig. 3 but divided into cases with only a front extension or both a front and a back extension. The rare events with side extensions as well were excluded. (A) Early control events; genotype, slboGal4, UAS-10×GFP/+. n = 1,336 (total) and 183 (with back extensions). (B) As in A, but with late events. n = 2,350 (total) and 142 (with back extensions). (C) As in A, but in the D-DN genotype, UAS-DN-EGFR/UAS-DN-PVR;slboGal4, UAS-10×GFP/+. The right-most category in A–C is without front extensions. n = 2,675 (total) and 664 (with back extensions). SEM is indicated. In C, the differences between front only and with back extensions are all significant (P < 0.01).
Extensions from the back of a cluster could be nonproductive for an external reason, for example, that the substrate has already been traversed, or it could be a feature of the migrating cells, specifically a guidance effect. To distinguish between these possibilities, we analyzed clusters in which guidance signaling was autonomously perturbed by expression of DN receptors (D-DN). The forward speed and size of the front extension were less correlated than in the control (Fig. S5), suggesting a decreased dominance of the front extension. More importantly, forward-directed migration was significantly slowed by the presence of a backward-directed extension in this genotype (Fig. 7 C). Reduction of a forward cluster movement by the presence of back extensions was observed for three size categories of front extensions (P < 0.01 for all three). Analysis of D-RNAi clusters showed the same tendency (Fig. S5), although it was not statistically significant because of limited numbers. Thus, perturbing guidance signaling allows backward-directed extensions to affect cluster movement, resulting in a tug of war between front and back cells. This indicates that the lack of an effect of the back extensions on cluster movement seen in the normal situation is caused by a guidance effect. The guidance input might provide a difference between front and back cells and, thereby, the behavior of front and back extensions, or the guidance input could directly and locally affect extensions depending on their direction. By either mechanism, guidance input could determine which extensions are productive and affect movement and which are not productive. In summary, directional guidance cues, acting through the RTKs PVR and EGFR, affect movement of the border cell by affecting the building and maintenance of cellular extensions as well as by regulating the productivity of such extensions.
Discussion
To better understand the directional movement of a cell group, we have examined migration-related features and how they are affected by input from guidance cues in the border cell system. Extensive quantitative analyses have been performed on single-cell migration in tissue culture. The collective migration studied here differs in that the migrating unit is a cluster of attached cells, meaning cells that affect one another physically, such that wild-type border cells can pull nonmigratory ones (Niewiadomska et al., 1999; Rørth et al., 2000) and can affect each others’ behavior (Wang et al., 2010). The migrating border cell cluster has a front and a back cell at any one point in time, but the cells change positions, and all the migratory cells are thought to be able to respond directly to guidance cues. The arrangement of the border cell cluster also means each migratory cell has an inherent polarity, abutting other border cells on the inside and contacting the substrate on the outside (Fig. 1 A′). Finally, the net movement of the cluster requires an invasion into a densely packed tissue consisting of large nurse cells, which constrains the movement spatially. We found little effect on apparent single-cell movement even with the most severe perturbations of both guidance receptors, indicating that border cells have a guidance-independent basic motility. Instead, we found that input from guidance cues rendered front and back cells and their outward-directed extensions different in two significant ways: the propensity to grow and maintain sizable cellular extensions and the ability of such extensions to influence actual movement of the cluster. Finally, we found a difference in how the two guidance receptors, both RTKs, affect cell behaviors.
How does guidance input steer the migration of border cells?
One set of characteristics that was affected by the guidance receptors and showed a positive correlation with cluster directionality was the size and persistence of the front extension. The main effect of productive RTK guidance signaling was to promote maintenance and growth of front extensions with limited effect on the directional bias in initiating new extensions. Although generally arising from one cell, the front extension could be controlled by many independent small events. Each of these events could stabilize or increase a small surface area of the extension as a direct readout of local guidance receptor signaling. In one model for single-cell chemotaxis, many such independent signaling events are argued to directly steer the cells (Arrieumerlou and Meyer, 2005). In other models, more emphasis is placed on the ability of guidance signaling to control an overall polarity in the cell using amplification and mutual inhibition (Xu et al., 2003; Van Haastert and Devreotes, 2004). PVR signaling has been monitored in border cells by the direct detection of an active receptor (Janssens et al., 2010). Active PVR was polarized, with the front of the front cell displaying a significantly higher signal than the side of the same cell. Interestingly, loss of the ligand Pvf1 changed the spatial distribution of PVR signaling but had little effect on the level of the signal. Given this, the observed failure to sustain forward extensions in the absence of a ligand is not easily explained by the first model of chemotaxis. In some directed cell migration, guidance input appears to act by allowing maintenance of the more favorable half of randomly split front protrusions (Andrew and Insall, 2007; Martini et al., 2009). Protrusion splitting is rare for border cells. However, understanding what allows a protrusion to be maintained or grow versus being retracted and whether retraction is a default may be the key to understanding the regulation of border cell extensions by guidance input.
The second feature affected by guidance input in border cells was the apparent productivity of cellular extensions. Specifically, backward extensions (outward extensions from the back cell) did not affect net movement of the cluster unless RTK guidance signaling was perturbed. This suggests that guidance signal differences affect the ability of an extension to adhere to the substratum or to supply the force required for the extension to exert pull on the cluster. One critical adhesion molecule in this context is E-cadherin, which is required in both border cells and the substrate nurse cells for productive movement (Niewiadomska et al., 1999), with a minor role in keeping the cluster intact (Pacquelet and Rørth, 2005). Both the strength of cadherin contacts and coupling to the cytoskeleton have been shown in different systems to be subject to regulation (Yap et al., 1997; Pokutta and Weis, 2007). This includes the regulation by RTK signaling and guidance cues (Hazan and Norton, 1998; Rhee et al., 2002). The dual effect of RTK guidance signaling on the presence of extensions and their “stickiness” or activity could reflect the use of different downstream pathways, but need not do so. For example, both extension stability (Ridley et al., 2003; Machacek et al., 2009) and the strength of cadherin adhesion (Chu et al., 2004) can be regulated via Rac, which has a key role in the guidance of border cells (Wang et al., 2010). The regulation of adhesion by guidance signaling has been observed in other contexts. For cells that migrate on the extracellular matrix, adhesion is mediated through integrins, including at focal adhesions, which display many opportunities for signal regulation and force transmission (Zaidel-Bar et al., 2007). Guidance receptors have been shown to affect integrins and focal adhesion kinase in neurons (Miao et al., 2000; Ren et al., 2004). One may assume that the adhesion monitored in these studies includes that which are required to make stable extensions as well as that which allows productive traction and force for movement. The contribution of both effects may be particularly critical for invasive migrations, such as that of border cells.
Finally, guidance signaling also appeared to affect migration in a manner independent of any extensions as defined in this study. There are several possible explanations for this: first, there are multiple ways of defining a cellular extension in complex 3D movement. The distinction between extensions and cell body used here leads us to disregard the base of broad extensions or very shallow extensions. Also, very small extensions and wrap-around extensions that do not project from the cluster were not quantified in this analysis. It is also possible that rolling or other forms of cell movement independent of extensions may contribute significantly to the directional movement. A more complete clarification of these issues will require a full 3D reconstruction of border cell clusters and individual cells during migration.
What are the roles of the two guidance receptors PVR and EGFR?
Genetically, it is established that EGFR is required for dorsal migration of border cells and reading the dorsal cue Gurken (Duchek and Rørth, 2001), whereas PVR and EGFR are redundantly involved in posterior migration (Duchek and Rørth, 2001). Only if both RTKs are perturbed are border cells prevented from finding their way to the oocyte. The present detailed analysis stresses the importance of PVR for the first half of migration to the oocyte, whereas EGFR becomes important in the second half. Why this spatial distinction, and why is EGFR needed for posterior migration at all given that the PVR ligand Pvf1 is expressed by the oocyte and should continue to function as an attractant? One key factor is the level of the receptor in the border cells: if border cells have excess PVR, they will show an efficient PVR/Pvf1-driven migration all the way to the oocyte. If they have half the normal PVR level, even early migration becomes EGFR dependent. So, the low endogenous PVR level is just sufficient for normal migration. We have not seen a change in total PVR level during migration that could explain its failure to function late. PVR signaling also does not seem to become saturated (Janssens et al., 2010). However, PVR may be sequestered, inactivated, or not able to properly interpret the high local Pvf1 concentrations close to the oocyte. Although it might seem inefficient to let EGFR take over, EGFR-driven guidance does work, and it must become dominant over time to allow the final dorsal migration. Preventing the switch by overexpression of PVR blocks dorsal border cell migration. Gurken itself, enriched at the dorsal side of the oocyte membrane, may also function as a posterior cue, as this membrane-tethered ligand appears to get cleaved (Ghiglione et al., 2002) and may diffuse anteriorly. Hampered by the limited extracellular space between the germline cells, the long-range extracellular distribution has not been determined for any of the guidance cues in this context.
Another difference between the two guidance receptors is which type of cell behavior is induced: sliding behavior induced by PVR or tumbling behavior induced by EGFR. In essence, PVR appears to be a better guidance receptor than EGFR. As might be expected from this, PVR guides migration of other cells, such as the single-cell migration of the macrophage-like hemocytes (Cho et al., 2002; Wood et al., 2006) and glia (Learte et al., 2008). EGFR is better known in Drosophila for its functions in cell differentiation, with both switchlike and graded responses (Schweitzer and Shilo, 1997). Both PVR and EGFR have been shown to affect cell survival and proliferation in different contexts (Schweitzer and Shilo, 1997; Brückner et al., 2004; Learte et al., 2008). Although able to activate the same or overlapping downstream pathways, EGFR may be optimized for cell-wide or nuclear responses, and PVR may be optimized for more local or polarized responses, that is, responses that scale to give a similar output over a large dynamic range as required for long-distance guidance. PVR could be a better substrate for inhibitory phosphatases or could be better recycled to the membrane given that endocytosis and recycling are important for polarized signaling (Jékely et al., 2005; Janssens et al., 2010). PVR could also link more effectively to Rac activation, which participates in a polarized output through feedback loops (Ridley et al., 2003). The best characterized Rac feedback loop involves lipid signaling through phosphatidylinositol 3′-kinase. Phosphatidylinositol 3′-kinase appears not to be essential for border cell guidance (Fulga and Rørth, 2002) but may participate. Also, Rac feedback loops and switches can be set up in multiple ways (Pankov et al., 2005).
As border cell migration is a collective migration by a strongly adherent cluster, guidance effects can, in principle, either be local and individual or cell based and collective (Rørth, 2007). Collective guidance occurs when there is a guidance-induced difference between cells of a group, and this, in turn, affects directional group movement. Local effects, which are essential for guidance in single-cell migration, do appear to occur in border cells. Some front extensions appear to not only project outward from the cluster but also turn slightly toward the oocyte. Also, local differences in PVR signaling have been directly observed (Janssens et al., 2010). However, the guidance effects characterized in this study are generally quantitative features of extensions from specific cells: first, stability and size of extensions and second, productivity of such extensions. For both features, guidance input may tune the cells relative to one another, or the effects may be local. Thus, the results are consistent with either cell-based or local guidance responses and do not distinguish between them. However, by showing which features are altered by guidance input, this analysis provides a better basis for more specifically testing what is under collective versus local control and what molecular mechanisms are responsible for each effect.
Materials and methods
Live imaging and processing
Egg chambers were dissected and cultured in imaging medium (Schneider’s medium + 2.5% fetal calf serum + 5 µg/ml insulin + 2 mg/ml trehalose + 5 µM methoprene + 1 µg/ml 20-hydroxyecdysone + 50 ng/ml adenosine deaminase + 9 µM FM 4–64) at ambient room temperature as described previously (Bianco et al., 2007). Multiposition confocal stacks were acquired using an inverted confocal microscope (SP5; Leica) with a 63×, 1.2 NA Plan Apochromat water immersion objective calibrated to the coverslip thickness of the imaging chambers and using Application Suite 2.2 (Leica). Fluorescence was excited with the 488-nm line of an argon ion laser, and the emitted fluorescence was acquired simultaneously for GFP (500–550 nm) and red (600–700 nm) for the membrane dye FM 4–64 in addition to the transmission image (differential interference contrast). Egg chambers were aligned by rotating the scan field with the anterior tip of the egg chamber aligned to the left and the image x axis going from this point through the middle of the oocyte (far right), and confocal sections 2 µm apart covering the migrating cluster were captured at 1–1.5-min intervals. Videos were subjected to quality control as described previously (Cliffe et al., 2007). Videos were broken into early (0–50% migration path) and late phases (50–100% migration path) to be used for later analysis; the minimum length used was 20 min. Frames in which clusters had not yet detached from the follicular epithelium or had made contact with the oocyte were not included.
For further analysis, maximum intensity projections of the GFP channel were created. These were then manually thresholded, and any GFP-positive follicle cells touching the border cell cluster were masked. All subsequent analysis was performed using a set of custom macros written for ImageJ (National Institutes of Health). Binary images were processed to remove outlying noisy pixels and subjected to one round of closing and hole filling. The cluster was then identified as the largest shape in a frame.
To identify the cluster body, we reasoned that an extension could be defined as a region too narrow to allow the entry of the cell nucleus. To define this region, we used a circle with a 30-pixel diameter (corresponding to 9.45 µm), which was determined by eye to give the most robust separation of body and extensions. The cluster body was defined by scanning this circular shaped element pixelwise over the image. Any region containing no empty pixels was marked as a cluster body by drawing a corresponding circle in a new image. The centroid of the cluster body was used to calculate cluster velocity. We defined the net cluster movement as the distance between the first and last time points divided by total time. See Videos 8 and 9 for examples of defining cluster body and extension (the videos are shown as nonsegmented).
Extensions were identified by subtracting the cell body image from the original binary image of the cluster for each frame. Manual inspection (as in Videos 8 and 9) confirmed that the extension did not overlap with border cell nuclei (>99% of cases). From this extension, only a set of images, any object larger than 30 pixels (>3 µm2), was counted. For each extension, a straight line was drawn from the body centroid to the furthest point of the extension tip. The length of the extension was measured from the edge of the cluster body to the extension tip. The angle of this line was taken relative to the x axis to give the extension angle. Extensions were classified as front (0–45° and 315–360°), side (>255 to <315° + >45 to >135°), and back (135–225°). To provide a temporal analysis of extensions, each frame was compared with the next time point, and objects that overlapped by ≥5 pixels were deemed to be the same extension. By identifying the first and last time points in which an extension was present, the persistence time of an extension was determined. This approach was validated by manually comparing several of the annotated videos against the original videos to ensure that minimal errors were produced. Side extensions are underestimated because the 2D projection as an extension in the z axis was not captured.
Generally, movement was tracked relative to the fixed xy imaging grid. Substrate (backward) sliding was observed in basically all videos but had to be an estimate, often based on fixed points beyond the actual migration path, and the effect was limited (Fig. 1). For these reasons, it was not included in the standard analysis. For instantaneous x-velocity analysis, relating forward speed to (front) extension size, the sum of the extension areas was used if more than one were present at one time point. Forward-directed speed was calculated using the distance between the centroid of the cluster body at this time point and the next in the x axis only.
Estimates of single-cell motility were taken as follows: for each cluster, the center of an individual outer border cell nucleus (marked by the absence of 10×GFP) was manually tracked in z stacks of GFP confocal sections time point by time point. This track was projected onto the 2D plane in which total cluster movement was followed, and the total path length was calculated in this plane. This procedure avoids errors from apparent jumps in the lower resolution z axis but also underestimates movement, as some movement also occurs in this axis. From a video (early and late phases) in which a single border cell was labeled, the projected body, centroid, and center of mass movement could be calculated. With tracked time points every 50 s, manual nuclear tracking appeared to systematically overestimate cell motility by 24 (early)–31% (late).
Analysis of fixed samples
For visualizing lacZ expression from slbo1310, ovaries were dissected in PBS, fixed with 0.5% glutaraldehyde, rinsed in PBS–0.1% Triton (PT), and stained in 10 mm NaH2PO4/Na2HPO4, pH 7.2, 150 mm NaCl, 1 mm MgCl2, 3.1 mm K4(FeII[CN]6), 3.1 mm K3(FeIII[CN]6), 0.3% Triton X-100, and 0.2% X-Gal at room temperature. Quantification of samples was performed blindly. For visualizing GFP, ovaries were fixed in 4% paraformaldehyde, rinsed in PT, and stained with Alexa Fluor 546–phalloidin (Invitrogen) and DAPI, and images were acquired by confocal microscopy.
Genetics
10×GFP was used as a marker for displayed experiments; an independent set of data for PVR and for EGFR manipulations using CD8-GFP as a marker is provided in Fig. S2. CD8-GFP extensions were not robust to the image analysis. A GFP fusion highlighting F-actin structures was used for preliminary experiments, but its expression showed a significant genetic interaction with guidance receptor perturbations.
All transgene expression (upstream activating sequence [UAS] or EPg) uses the binary Gal4 system and SlboGal4 to drive expression in border cells (plus additional cells) or actin-flipout-Gal4 (AFG), driving robust expression in all somatic cells after heat shock–induced expression of FLP recombinase. With AFG, RNAi expression was induced by heat shock to females 2 d before analysis. Transgenic constructs expressing full-length PVR, EGFR, or DN versions or ligands as well as the Pvf11624 mutant EPgPvf1 (drives Pvf1 expression) and Pvr1-null allele have been described previously (Duchek and Rørth, 2001; Duchek et al., 2001; Jékely et al., 2005). DNA encoding a 10×GFP fusion (gift from the Ellenberg laboratory, European Molecular Biology Laboratory, Heidelberg, Germany) was cloned into the UAS vector; this fusion is cytoplasmic and excluded from the nucleus.
The RNAi lines in this study used were GD13502 (KK105353 gave a similar, slightly weaker phenotype) for Pvr, GD43268 for Egfr, GD10455 for Elmo, and GD6243 and KK103820 for Vav. Fly stocks were obtained from the Bloomington or Vienna Drosophila RNAi Center stock centers. To analyze the statistical significance of difference between two datasets, a two-tailed Student’s t test was used.
Online supplemental material
Fig. S1 shows plots of rotational movement of tracked nuclei representing single cells. Fig. S2 shows the relationship between persistence time and maximum area for front extensions. Fig. S3 shows migration data for control and PVR or EGFR perturbation genotypes with UAS-CD8-GFP. Fig. S4 shows migration data of Elmo, Vav, and Raf perturbations. Fig. S5 shows the relationship between front extension size and forward movement for D-DN as well as the effect of back extensions on cluster movement and D-RNAi. Also available online are seven videos showing border cell migration in different phases (Videos 1–3 show early to late control) and genotypes (UAS-DN-PVR early [Video 4], UAS-PVR in both phases and late [Videos 5 and 6], and UAS-EGFR in both phases [Video 7]) as well as two videos showing examples of how the automatic processing defines body and extensions in early (Video 8) and late (Video 9) migration in border cell clusters. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010003/DC1.
Acknowledgments
We are grateful to the Temasek Life Sciences Laboratory for support in the early phase of this project. We thank Smitha Vishnu for contributing additional wild-type videos.
Funding from the Agency for Science, Technology, and Research to the Institute of Molecular and Cell Biology is acknowledged.
Footnotes
Abbreviations used in this paper:
- DN
- dominant negative
- EGFR
- EGF receptor
- PVR
- PDGF/VEGF-related receptor
- RTK
- receptor tyrosine kinase
- UAS
- upstream activating sequence
References
- Andrew N., Insall R.H. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 9:193–200 10.1038/ncb1536 [DOI] [PubMed] [Google Scholar]
- Arrieumerlou C., Meyer T. 2005. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev. Cell. 8:215–227 10.1016/j.devcel.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Berzat A., Hall A. 2010. Cellular responses to extracellular guidance cues. EMBO J. 29:2734–2745 10.1038/emboj.2010.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C.M., Fulga T.A., Rørth P. 2007. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 448:362–365 10.1038/nature05965 [DOI] [PubMed] [Google Scholar]
- Brückner K., Kockel L., Duchek P., Luque C.M., Rørth P., Perrimon N. 2004. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell. 7:73–84 10.1016/j.devcel.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Cho N.K., Keyes L., Johnson E., Heller J., Ryner L., Karim F., Krasnow M.A. 2002. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 108:865–876 10.1016/S0092-8674(02)00676-1 [DOI] [PubMed] [Google Scholar]
- Christiansen J.J., Rajasekaran A.K. 2006. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 66:8319–8326 10.1158/0008-5472.CAN-06-0410 [DOI] [PubMed] [Google Scholar]
- Chu Y.S., Thomas W.A., Eder O., Pincet F., Perez E., Thiery J.P., Dufour S. 2004. Force measurements in E-cadherin–mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 167:1183–1194 10.1083/jcb.200403043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe A., Poukkula M., Rørth P. 2007. Culturing Drosophila egg chambers and imaging border cell migration. Nature Protocol Exchange. http://www.nature.com/protocolexchange/protocols/25110.1038/nprot.2007.289
- deBakker C.D., Haney L.B., Kinchen J.M., Grimsley C., Lu M., Klingele D., Hsu P.K., Chou B.K., Cheng L.C., Blangy A., et al. 2004. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr. Biol. 14:2208–2216 10.1016/j.cub.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Duchek P., Rørth P. 2001. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 291:131–133 10.1126/science.291.5501.131 [DOI] [PubMed] [Google Scholar]
- Duchek P., Somogyi K., Jékely G., Beccari S., Rørth P. 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 107:17–26 10.1016/S0092-8674(01)00502-5 [DOI] [PubMed] [Google Scholar]
- Friedl P., Gilmour D. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., Hegerfeldt Y., Tusch M. 2004. Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 48:441–449 10.1387/ijdb.041821pf [DOI] [PubMed] [Google Scholar]
- Fulga T.A., Rørth P. 2002. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat. Cell Biol. 4:715–719 10.1038/ncb848 [DOI] [PubMed] [Google Scholar]
- Geisbrecht E.R., Montell D.J. 2004. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 118:111–125 10.1016/j.cell.2004.06.020 [DOI] [PubMed] [Google Scholar]
- Ghiglione C., Bach E.A., Paraiso Y., Carraway K.L., III, Noselli S., Perrimon N. 2002. Mechanism of activation of the Drosophila EGF Receptor by the TGFalpha ligand Gurken during oogenesis. Development. 129:175–186 [DOI] [PubMed] [Google Scholar]
- Hazan R.B., Norton L. 1998. The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton. J. Biol. Chem. 273:9078–9084 10.1074/jbc.273.15.9078 [DOI] [PubMed] [Google Scholar]
- Insall R.H., Machesky L.M. 2009. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell. 17:310–322 10.1016/j.devcel.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Janssens K., Sung H.H., Rørth P. 2010. Direct detection of guidance receptor activity during border cell migration. Proc. Natl. Acad. Sci. USA. 107:7323–7328 10.1073/pnas.0915075107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G., Sung H.H., Luque C.M., Rørth P. 2005. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev. Cell. 9:197–207 10.1016/j.devcel.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Katoh H., Negishi M. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 424:461–464 10.1038/nature01817 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D.A., Horwitz A.F. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Learte A.R., Forero M.G., Hidalgo A. 2008. Gliatrophic and gliatropic roles of PVF/PVR signaling during axon guidance. Glia. 56:164–176 10.1002/glia.20601 [DOI] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini F.J., Valiente M., López Bendito G., Szabó G., Moya F., Valdeolmillos M., Marín O. 2009. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 136:41–50 10.1242/dev.025502 [DOI] [PubMed] [Google Scholar]
- Miao H., Burnett E., Kinch M., Simon E., Wang B. 2000. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2:62–69 10.1038/35000008 [DOI] [PubMed] [Google Scholar]
- Montell D.J. 2003. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4:13–24 10.1038/nrm1006 [DOI] [PubMed] [Google Scholar]
- Murphy A.M., Montell D.J. 1996. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol. 133:617–630 10.1083/jcb.133.3.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P., Godt D., Tepass U. 1999. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J. Cell Biol. 144:533–547 10.1083/jcb.144.3.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A., Rørth P. 2005. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J. Cell Biol. 170:803–812 10.1083/jcb.200506131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. 2005. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170:793–802 10.1083/jcb.200503152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S., Weis W.I. 2007. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 23:237–261 10.1146/annurev.cellbio.22.010305.104241 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Prasad M., Montell D.J. 2007. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell. 12:997–1005 10.1016/j.devcel.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Ren X.R., Ming G.L., Xie Y., Hong Y., Sun D.M., Zhao Z.Q., Feng Z., Wang Q., Shim S., Chen Z.F., et al. 2004. Focal adhesion kinase in netrin-1 signaling. Nat. Neurosci. 7:1204–1212 10.1038/nn1330 [DOI] [PubMed] [Google Scholar]
- Rhee J., Mahfooz N.S., Arregui C., Lilien J., Balsamo J., VanBerkum M.F. 2002. Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat. Cell Biol. 4:798–805 10.1038/ncb858 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Paterson H.F., Johnston C.L., Diekmann D., Hall A. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 70:401–410 10.1016/0092-8674(92)90164-8 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rørth P. 2007. Collective guidance of collective cell migration. Trends Cell Biol. 17:575–579 10.1016/j.tcb.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Rørth P. 2009. Collective cell migration. Annu. Rev. Cell Dev. Biol. 25:407–429 10.1146/annurev.cellbio.042308.113231 [DOI] [PubMed] [Google Scholar]
- Rørth P., Szabo K., Texido G. 2000. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell. 6:23–30 10.1016/S1097-2765(00)00004-6 [DOI] [PubMed] [Google Scholar]
- Rosin D., Schejter E., Volk T., Shilo B.Z. 2004. Apical accumulation of the Drosophila PDGF/VEGF receptor ligands provides a mechanism for triggering localized actin polymerization. Development. 131:1939–1948 10.1242/dev.01101 [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Shilo B.-Z. 1997. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 13:191–196 10.1016/S0168-9525(97)01091-3 [DOI] [PubMed] [Google Scholar]
- Theveneau E., Marchant L., Kuriyama S., Gull M., Moepps B., Parsons M., Mayor R. 2010. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 19:39–53 10.1016/j.devcel.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert P.J., Devreotes P.N. 2004. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5:626–634 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y.I., Hahn K.M., Montell D.J. 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12:591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijer C.J. 2009. Collective cell migration in development. J. Cell Sci. 122:3215–3223 [DOI] [PubMed] [Google Scholar]
- Wood W., Faria C., Jacinto A. 2006. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. J. Cell Biol. 173:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Brock A.R., Wang Y., Fujitani K., Ueda R., Galko M.J. 2009. A blood-borne PDGF/VEGF-like ligand initiates wound-induced epidermal cell migration in Drosophila larvae. Curr. Biol. 19:1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H.R. 2003. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 114:201–214 [DOI] [PubMed] [Google Scholar]
- Yap A.S., Brieher W.M., Gumbiner B.M. 1997. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 13:119–146 10.1146/annurev.cellbio.13.1.119 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma’ayan A., Iyengar R., Geiger B. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]