Complexes containing tafazzin, which remodels newly synthesized cardiolipin, are destabilized by mutations associated with Barth syndrome.
Abstract
Deficits in mitochondrial function result in many human diseases. The X-linked disease Barth syndrome (BTHS) is caused by mutations in the tafazzin gene TAZ1. Its product, Taz1p, participates in the metabolism of cardiolipin, the signature phospholipid of mitochondria. In this paper, a yeast BTHS mutant tafazzin panel is established, and 18 of the 21 tested BTHS missense mutations cannot functionally replace endogenous tafazzin. Four BTHS mutant tafazzins expressed at low levels are degraded by the intermembrane space AAA (i-AAA) protease, suggesting misfolding of the mutant polypeptides. Paradoxically, each of these mutant tafazzins assembles in normal protein complexes. Furthermore, in the absence of the i-AAA protease, increased expression and assembly of two of the BTHS mutants improve their function. However, the BTHS mutant complexes are extremely unstable and accumulate as insoluble aggregates when disassembled in the absence of the i-AAA protease. Thus, the loss of function for these BTHS mutants results from the inherent instability of the mutant tafazzin complexes.
Introduction
The term mitochondrial medicine was first coined in 1994 by Luft, who 32 years earlier had described a patient suffering from hypermetabolism associated with profuse sweating and what were described as mitochondria defective in normal respiratory control (Ernster et al., 1959; Luft et al., 1962; Luft, 1994). Luft syndrome was the first documented mitochondrial disease, and amazingly, the causative gene has still not been identified. As a relatively recent medical subspecialty, mitochondrial medicine is in its nascency. A major complication in patient diagnosis is that mutations in two genomes can result in mitochondrial disease, a factor that contributes to the panoply of associated phenotypes and varied timing for the onset of disease symptoms (Thorburn, 2004; Haas et al., 2008). Still, as 99% of proteins resident to mitochondria are encoded in the nuclear genome (Sickmann et al., 2003; Pagliarini et al., 2008), the majority of pediatric cases of mitochondrial disease exhibit typical Mendelian inheritance (Thorburn, 2004). Given the mitochondrion’s central role in ATP production, not surprisingly, oxidative phosphorylation disorders represent a major class of mitochondrial disease (DiMauro and Schon, 2003; Thorburn et al., 2004). In fact, current estimates indicate that 1 in 5,000 children will develop an oxidative phosphorylation disorder, placing this disease category on par with lysosomal storage diseases as the most prevalent causes of inherited metabolic disease (Thorburn, 2004).
Of the 79 known nuclear mitochondrial disease–associated genes (Calvo et al., 2006; Calvo and Mootha, 2010), only one, TAZ1 (G4.5 or TAFAZZIN), the mutant gene associated with the X-linked disease Barth syndrome (BTHS; Online Mendelian Inheritance in Man [OMIM] accession no. 302060), is involved in the metabolism of the mitochondrial-specific phospholipid cardiolipin (CL; Barth et al., 1983; Bione et al., 1996; Schlame and Ren, 2006). BTHS patients suffer from skeletal and cardiomyopathy and cyclic neutropenia. Heart failure and opportunistic infection are major causes of patient mortality, and there is currently no documented universal therapeutic strategy. Mitochondria isolated from BTHS patients and models exhibit deficits in oxidative phosphorylation assembly and/or function (Barth et al., 1983, 1996; Ma et al., 2004; Brandner et al., 2005; McKenzie et al., 2006; Claypool et al., 2008b). CL, which is highly enriched in the mitochondrial inner membrane (IM), its site of synthesis, is an unusual dimeric phospholipid consisting of two phosphatidic acids bridged by a central glycerol moiety (Schlame et al., 2000). The four typically saturated acyl chains of newly synthesized CL undergo a remodeling process, the end result of which is the incorporation of more unsaturated fatty acyl chains that display a high degree of symmetry (Schlame et al., 2005). Tafazzin (Taz1p), the gene product of TAZ1, is a key player in the remodeling of newly synthesized CL, functioning as a monolysocardiolipin (MLCL) transacylase (Xu et al., 2006). Remodeling is initiated by a phospholipase, which is identified in yeast as CL-specific deacylase 1 (Cld1; Beranek et al., 2009). In the absence of tafazzin activity, MLCL (only three acyl chains) accumulates, as the remodeling pathway is initiated but cannot be completed. Thus, MLCL is a biochemical signature of BTHS (Valianpour et al., 2005; Houtkooper et al., 2009a).
Like many inherited disorders, there are a multitude of mutations in TAZ1 associated with BTHS (Fig. 1). In addition to frameshift mutations that cause tafazzin truncation and mutations affecting splice donor and acceptor sites, 28 missense mutations have been documented. Based on the wide distribution of the missense mutations, it is reasonable to predict that there will be multiple mechanisms for the loss of function of BTHS mutant tafazzins. Support for this notion was provided by our previous demonstration that four BTHS mutations in a unique membrane anchor of Taz1p, when individually modeled in the Saccharomyces cerevisiae Taz1p orthologue, resulted in two distinct biochemical fates: mislocalization to the mitochondrial matrix or altered assembly in macromolecular complexes (Claypool et al., 2006).
Figure 1.
ClustalW alignment of exon 5–deleted human and mouse Taz1p and full-length yeast Taz1p. BTHS mutations that occur at identical and conserved amino acids are indicated. Truncation mutations are indicated with an X above the last amino acid translated in the mutant protein. Gray boxes highlight the conserved acyltransferase motifs A–E; the black box reveals the determined location of the integral interfacial membrane anchor of Taz1p. Asterisks indicate identical residues. Single dots indicate lesser conserved residues, and double dots indicate highly conserved residues.
In the present study, we generate the complete collection of yeast BTHS mutant tafazzins by modeling the 21 missense mutations that occur at identical or conserved residues between the human and yeast orthologues. The power of the yeast model system to characterize the assorted BTHS mutant tafazzins is demonstrated by the fact that 18 of the 21 BTHS mutant tafazzins fail to complement the Δtaz1 yeast strain based on the accumulation of significant levels of the BTHS biomarker MLCL. Our further in-depth characterization of four BTHS mutant tafazzins (four mutations at three loci) expressed at low levels demonstrates that each is degraded by the intermembrane space (IMS) AAA (i-AAA) protease quality control apparatus of the mitochondrial IM, suggesting misfolding of the mutant polypeptides. Paradoxically, each of these mutant tafazzins assembles in normal protein complexes, arguing against a gross folding defect. Furthermore, in the absence of the i-AAA protease, the phospholipid profile of two of the BTHS mutants is improved, indicating recovered function. However, the BTHS mutant complexes are significantly more labile than wild-type (wt) Taz1p–containing complexes and, once disassembled, accumulate as insoluble protein aggregates in the absence of the i-AAA protease. Thus, we define a third mechanism explaining BTHS mutant tafazzin loss of function, tafazzin complex lability. To our knowledge, complex lability as a pathogenic mechanism has not been previously documented.
Results
Characterizing the yeast BTHS mutant tafazzin panel
The expression of human tafazzin is complicated by the presence of two potential start sites and alternative splicing (Bione et al., 1996; Houtkooper et al., 2009b). Of the multitude of detected human Taz1p splice variants, only the exon 5–deleted isoform complemented a yeast strain lacking endogenous tafazzin (Δtaz1; Vaz et al., 2003). Consistent with this functional homology, a comparison of the amino acid sequences of exon 5–deleted human and mouse Taz1p with the yeast orthologue revealed a substantial degree of conservation (∼18% identical and ∼41% conserved amino acids; Fig. 1). Furthermore, of the 28 missense mutations currently documented in the BTHS patient population, 21 reside at either identical (Fig. 1, highlighted in red) or conserved (Fig. 1, highlighted in green) residues between the human and yeast orthologues.
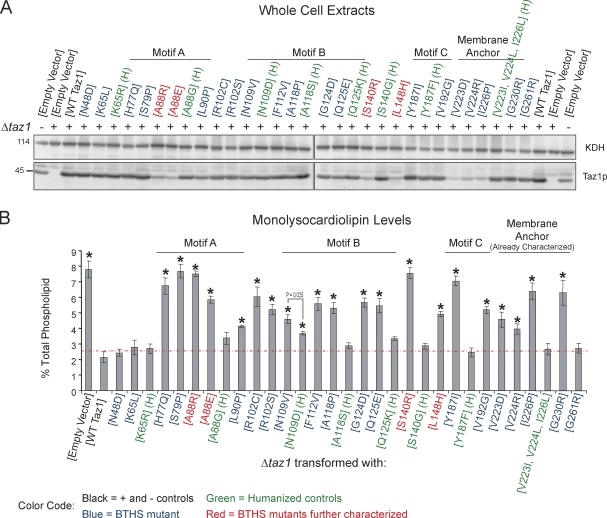
Each of the identical and conserved BTHS missense mutations was individually modeled in yeast tafazzin, generating a yeast BTHS mutant tafazzin panel (Fig. 2, highlighted in blue or red). Humanized yeast tafazzin constructs (Fig. 2, highlighted in green) were additionally generated for the BTHS mutations that occur at conserved residues to allow discrimination between a BTHS mutant–specific phenotype versus simple differences between the human and yeast polypeptides. The yeast BTHS mutant tafazzin panel was transformed into the Δtaz1 strain, and their expression was determined by immunoblotting (Fig. 2 A). Most of the mutant tafazzins were expressed at levels similar to wt Taz1p. However, some, including the previously characterized three membrane anchor mutants (V223D, V224R, and I226P), were expressed at drastically lower levels, even though all of the tafazzin variants are under control of the endogenous TAZ1 promoter. Next, the ability of each mutant tafazzin to restore the altered phospholipid profile of the Δtaz1 strain was assessed. Critically, 18 of the 21 BTHS mutant tafazzins failed to rescue the Δtaz1 strain based on the significant mitochondrial accumulation of MLCL relative to Δtaz1 yeast transformed with wt Taz1p (Fig. 2 B). Importantly, with only one exception, humanized tafazzin constructs functioned as well as wt Taz1p. Furthermore, the humanized construct that failed to phenocopy wt Taz1p (N109D) did accumulate significantly less MLCL than the corresponding BTHS mutant (N109V). Interestingly, greater variability was observed in the abundance of CL in the BTHS mutant tafazzin strains, with many containing significantly more CL than the empty vector–transformed Δtaz1 yeast strain (Fig. S1). From these analyses, clearly not every BTHS mutation is equivalent to a complete loss of function; otherwise, the MLCL and CL levels in each BTHS mutant strain would be expected to be the same as either the empty vector– or wt Taz1p–transformed Δtaz1 strain. Finally, these results validate using yeast to biochemically characterize each BTHS mutant tafazzin that fails to complement the Δtaz1 yeast strain (18/21).
Figure 2.
18/21 BTHS mutations result in dysfunctional yeast Taz1p. (A) The relative expression of each of the BTHS mutants was determined from whole-cell extracts by immunoblotting for Taz1p (bottom) with α-ketoglutarate dehydrogenase (KDH) serving as a loading control (top). (B) The relative abundance of MLCL was determined for each strain and is expressed as a percentage of the total phospholipid in each strain (means ± SEM; n = 3). The dashed red line indicates the highest level of MLCL detected in Δtaz1 (WT Taz1). Asterisks indicate significant accumulations of MLCL relative to Δtaz1 (WT Taz1; P < 0.001) as determined by one-way analysis of variance (ANOVA) with Holm–Sidak pairwise comparisons.
i-AAA protease–mediated degradation of four BTHS mutant tafazzins
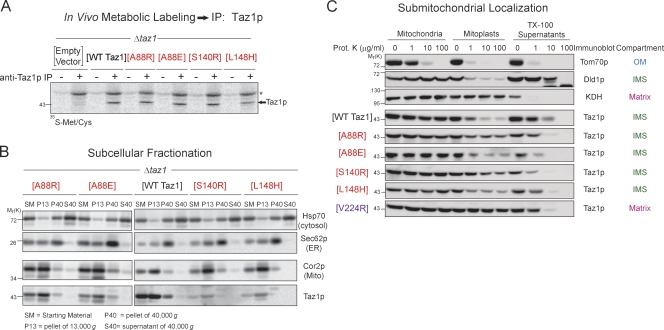
To begin to characterize the BTHS mutant panel, we focused on four mutants occurring at three BTHS loci that were expressed at significantly lower levels than wt Taz1p: A88R/E, S140R, and L148H (Fig. 2 A, highlighted in red). Immunoprecipitation of Taz1p from [35S]methionine/cysteine pulse-radiolabeled yeast extracts indicated that, as expected, all four mutant tafazzins are translated as robustly as wt Taz1p (Fig. 3 A). Unlike most mitochondrial proteins, Taz1p lacks a typical N-terminal targeting signal. Thus, mutations within the tafazzin polypeptide could inactivate a cryptic internal mitochondrial localization signal. However, each of the four mutant tafazzins cofractionated with mitochondria (Fig. 3 B) just like wt Taz1p. Thus, each BTHS mutant tafazzin retains normal mitochondrial targeting information. Yeast tafazzin normally associates with both the outer and inner mitochondrial membranes always facing the IMS (Claypool et al., 2006; Gebert et al., 2009). Previously, we demonstrated that three BTHS mutations that occur in the unusual membrane anchor of yeast Taz1p (V223D, V224R, and I226P) result in their mislocalization to the mitochondrial matrix (Claypool et al., 2006). Importantly, each of these membrane anchor mutants is expressed at drastically reduced levels similar to the A88R/E, S140R, and L148H BTHS mutant tafazzins (Fig. 2 A). To determine whether the A88R/E, S140R, and/or L148H BTHS mutant tafazzin is mislocalized within mitochondria, their submitochondrial localization was ascertained by a proteinase K accessibility assay (Fig. 3 C). As with wt Taz1p, each of these BTHS mutant tafazzins was degraded by added proteinase K upon disruption of the mitochondrial outer membrane (OM; mitoplast), demonstrating that they are resident to IMS-facing membranes. In contrast, the V224R BTHS mutant tafazzin, which is mislocalized to the mitochondrial matrix, is only degraded by proteinase K upon the addition of the detergent Triton X-100 (TX-100). It is worth mentioning that CL-deficient mitochondria exhibit a defective swelling response (Fig. S3; Ma et al., 2004); therefore, the failure to completely degrade Dld1p (positive control IMS protein) and the four BTHS mutant tafazzins upon mitoplasting simply reflects an incomplete rupturing of the OM. Thus, the A88R/E, S140R, and L148H BTHS mutant tafazzins, all of which are expressed at steady state at extremely low levels, are translated as well as wt Taz1p and still localize to and within mitochondria normally.
Figure 3.
The BTHS mutant tafazzins localize to and within mitochondria normally. (A) Immunoprecipitation (IP) of Taz1p from the indicated metabolically labeled yeast extracts. The asterisk highlights a nonspecific band. (B) Subcellular fractions were prepared from the indicated yeast strains through a series of differential centrifugations. 25 µg of each fraction was separated by SDS-PAGE and analyzed by immunoblotting using antisera specific for the indicated subcellular organelle. (C) Submitochondrial localization of wt and BTHS mutant tafazzins. Intact mitochondria, mitochondria subjected to osmotic shock (mitoplasts), or mitochondria solubilized with 0.1% TX-100 were incubated alone or in the presence of the indicated concentration of proteinase K. 50 µg/lane wt Taz1p and 100 µg/lane BTHS mutants were resolved by SDS-PAGE and immunoblotted as indicated. For simplicity, only one set of control immunoblots is shown. The controls for every source of mitochondria are provided in Fig. S2. The four BTHS mutants being characterized in the present study are shown in red. The previously characterized matrix-mislocalized BTHS mutant tafazzin is shown in purple. KDH, α-ketoglutarate dehydrogenase. Mito, mitochondria. (A–C) n = 3.
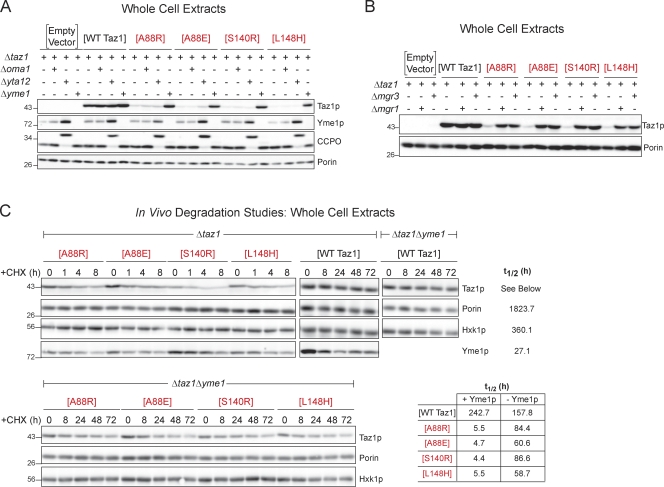
Next, the possibility that each of these BTHS mutant tafazzins was unable to fold subsequent to their proper sorting to and within mitochondria was addressed. The mitochondrial IM contains three resident proteases implicated in enforcing quality control in this compartment: Oma1p, the matrix AAA (m-AAA) protease, and the i-AAA protease (Tatsuta, 2009; Tatsuta and Langer, 2009). The m-AAA protease, in yeast composed of Yta10p and Yta12p, and the i-AAA protease, consisting of Yme1p and the adaptor subunits Mgr1p and Mgr3p, are both embedded in the IM but have their active sites facing opposite sides. Oma1p has recently been demonstrated to degrade presumably misfolded Cox1p in Δcoa2 yeast (Bestwick et al., 2010). The expression of the four BTHS mutant tafazzins was compared in Δtaz1 strains harboring additional deletions of Oma1p (Δoma1; deletion monitored by PCR), the m-AAA protease (Δyta12; deletion confirmed by aberrant processing of its substrate, cytochrome c peroxidase), or the i-AAA protease (Δyme1; Fig. 4 A). The expression level of wt Taz1p was invariant in the absence of any of the three IM proteases. In contrast, the expression level of the A88R/E, S140R, and L148H BTHS mutant tafazzins was restored to wt levels in the absence of the major i-AAA protease subunit Yme1p. There was no impact on their expression in the absence of either Oma1p or the m-AAA protease. Yme1p is the major building block of the oligomeric i-AAA protease and is responsible for its proteolytic capacity. Recently, two additional subunits of the i-AAA protease, Mgr1p and Mgr3p, have been identified (Dunn et al., 2006, 2008). Consistent with the conclusion that Mgr1p and Mgr3p function as i-AAA protease adaptors that increase the efficiency of Yme1p-mediated proteolysis, the expression of the A88R/E, S140R, and L148H BTHS mutant tafazzins was partially restored in the absence of either of these two proteins (Fig. 4 B). Next, in vivo degradation experiments were performed to determine the half-life of wt Taz1p and the four BTHS mutants, in the presence and absence of Yme1p, after the inhibition of new protein synthesis by the addition of cycloheximide (CHX; Fig. 4 C). Relative to wt Taz1p (t1/2 = ∼243 h), each of the four BTHS mutant tafazzins has a significantly shorter half-life (t1/2 of 4–6 h; Fig. 4 C). Although not restored to wt Taz1p values, the half-life of the A88R/E, S140R, and L148H BTHS mutant tafazzins was significantly increased in the absence of Yme1p. The increase in the half-lives of the four BTHS mutants in the absence of Yme1p is in contrast to the shorter half-life calculated for wt Taz1p in the absence of the i-AAA protease (t1/2 = ∼158 h). One possible explanation for this observation is that perhaps Yme1p is involved in the routine quality control of Taz1p, and in its absence, more suboptimal Taz1p exists that is intrinsically less stable and capable of being degraded by proteases in addition to Yme1p. Interestingly, compared with wt Taz1p (t1/2 = ∼243 h), another mitochondrial protein (porin, t1/2 = ∼1,824 h), and a cytosolic protein (hexokinase [Hxk1p], t1/2 = ∼360 h), Yme1p itself has a relatively short half-life (t1/2 = ∼27 h).
Figure 4.
The BTHS mutant tafazzins are degraded by the i-AAA protease. (A and B) Steady-state expression was determined from whole-cell extracts derived from the indicated strains by immunoblotting for Taz1p, Yme1p, cytochrome c peroxidase (CCPO), and the loading control, porin. n = 3. (C) Increased half-life of the BTHS mutants in the absence of Yme1p. Whole-cell extracts were harvested after incubation with cycloheximide (CHX) for the indicated times, and the Taz1p remaining was determined by immunoblotting. Yme1p and porin are mitochondrial controls, and hexokinase (Hxk1p) is a cytosolic control. The four BTHS mutants being characterized in the present study are shown in red. Relative molecular masses are shown on the left.
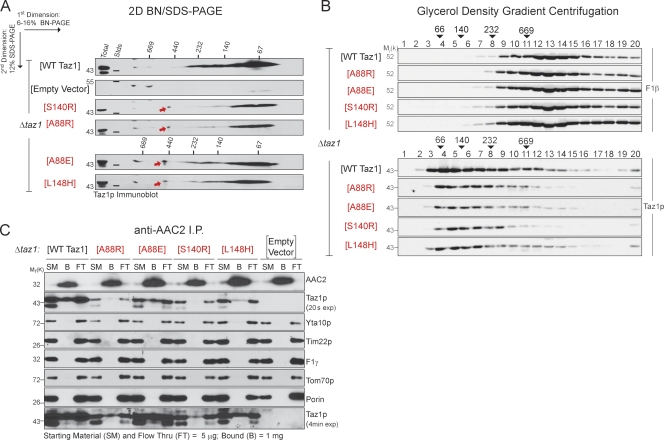
Collectively, these results indicate that the A88R/E, S140R, and L148H BTHS mutant tafazzins are targeted to and within mitochondria normally where they are rapidly degraded by the i-AAA protease. The simplest explanation for the rapid turnover of the four mutants is that they are unable to fold normally. However, when their assembly status was determined by 2D blue native (BN)/SDS-PAGE, each of the four BTHS mutant tafazzins formed, with the exception of a single aberrant complex of ∼480 kD (Fig. 5 A, red arrows), the normal Taz1p complexes (Fig. 5 A). The assembly status of the four BTHS mutants was additionally assessed by glycerol density gradient centrifugation (Fig. 5 B). Consistent with the results of the 2D BN/SDS-PAGE, the BTHS mutant tafazzins were detected in a range of complexes similar in size to those formed by wt Taz1p. Finally, as the formation of complexes presumes normal folding of a polypeptide, the ability of the four BTHS mutant tafazzins to associate with a known binding partner of yeast Taz1p, the major ADP/ATP carrier AAC2 (Claypool et al., 2008a), was directly assessed (Fig. 5 C). Indeed, each of the four BTHS mutant tafazzins was coimmunoprecipitated with endogenous AAC2. The relatively weak association between the A88R and S140R BTHS mutants and AAC2 compared with the A88E and L148H mutants could reflect a slightly lower expression level of the former and/or indicate that these two mutants are more severely impaired than the A88E and L148H mutants. Thus, the assembly of the A88R/E, S140R, and L148H mutants as determined by both 2D BN/SDS-PAGE and glycerol density gradient centrifugation reflects their ability to engage in normal interactions, indicating that the BTHS mutant tafazzins are not, as predicted, absolutely unable to fold. If these four BTHS mutant tafazzins are not grossly misfolded, why are they being degraded by the i-AAA protease?
Figure 5.
The BTHS mutant tafazzins assemble in normal complexes. (A) 1.5% (wt/vol) digitonin extracts from mitochondria derived from the indicated strains were resolved by 2D BN/SDS-PAGE, and Taz1p complexes were detected by immunoblotting. 150 µg Δtaz1 (WT Taz1) and 250 µg Δtaz1 yeast transformed with empty vector or BTHS mutant tafazzin were analyzed. The red arrows highlight a complex only detected in mutant tafazzin extracts. (B) Digitonin extracts from mitochondria derived from the indicated strains were separated by glycerol density gradient centrifugation, and collected fractions (1 = top and 20 = bottom) were immunoblotted for F1-β (top) and Taz1p (bottom). 100% of each BTHS mutant fraction was analyzed versus 40% of Δtaz1 (WT Taz1) fractions. The numbers above the arrowheads indicate the molecular weights of the high molecular weight standards (Stds). n = 3. (C) Endogenous AAC2 was immunoprecipitated from digitonin extracts from the indicated mitochondria. 0.5% of the starting material and final flow through versus 100% of the material remaining attached to the immunoprecipitation beads after washing were immunoblotted for the indicated mitochondrial proteins. exp, exposure. The four BTHS mutants being characterized in the present study are shown in red. (A and C) n = 5.
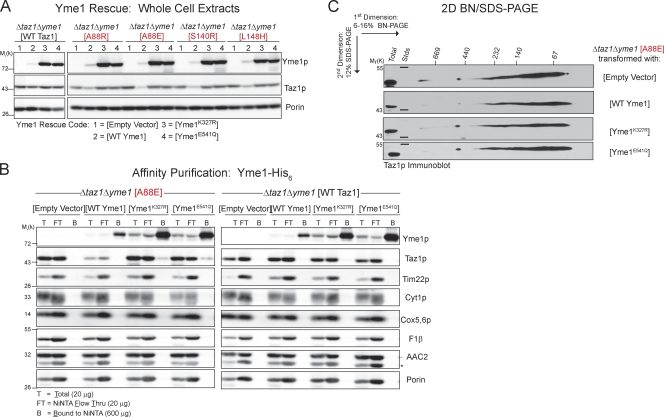
Reintroduction of wt Yme1p into the Δtaz1Δyme1 strain restored the expression of the four BTHS mutant tafazzins to low levels (Fig. 6 A). This prompted us to probe the ability of two defined Yme1p mutants to regulate the expression of the A88R/E, S140R, and L148H mutant tafazzins. The i-AAA protease contributes to mitochondrial quality control through its proteolytic activity and via its ability to provide, in some cases, chaperone-like properties (Leonhard et al., 1999). Mutation of glutamate 541 within the catalytic domain renders Yme1p proteolytically dead (Leonhard et al., 1996). Mutation of lysine 327 within the Walker A motif disrupts ATP binding. In the absence of ATP binding, both Yme1p proteolytic and chaperone-like activities are lost (Leonhard et al., 1999). Interestingly, the expression level of Yme1K327R and Yme1E541Q was higher than wt Yme1p (each construct under control of the YME1 promoter), suggesting that Yme1p regulates its own expression (Fig. 6 A). The expression level of the four BTHS mutants was the same in Δtaz1Δyme1 transformed with either Yme1K327R or Yme1E541Q as the empty vector control. Thus, a catalytically active Yme1p is required to degrade the A88R/E, S140R, and L148H BTHS mutant tafazzins.
Figure 6.
i-AAA protease mutants bind the A88E BTHS mutant, but not wt, tafazzin. (A) Steady-state expression was determined from whole-cell extracts derived from Δtaz1Δyme1 yeast transformed with the indicated Taz1p and Yme1p variants by immunoblotting for Taz1p, Yme1p, and the loading control, porin. (B) Digitonin extracts from mitochondria derived from the indicated strains were subjected to Ni2+ nitrilotriacetic acid (NiNTA) chromatography. The indicated amount of total, TCA-precipitated flow through, and bound material was resolved by SDS-PAGE and immunoblotted as indicated. The asterisk highlights the cross-reaction with porin of the AAC2 antiserum. (C) Digitonin extracts from mitochondria derived from the indicated strains were resolved by 2D BN/SDS-PAGE, and Taz1p complexes were detected by immunoblotting. 150 µg Δtaz1Δyme1 yeast transformed with empty vector or Yme1 mutants and 250 µg Δtaz1Δyme1 (Wt Yme1) were analyzed. The four BTHS mutants being characterized in the present study are shown in red. Stds, molecular weight standards. (A–C) n = 3.
Proteolysis is expected to occur subsequent to physical recognition. To test this, Yme1p was affinity purified by virtue of an appended C-terminal His6 tag from Δtaz1Δyme1 strains expressing either the A88E BTHS mutant or wt Taz1p and wt Yme1p, Yme1K327R, Yme1E541Q, or the empty vector (Fig. 6 B). wt Yme1p failed to copurify either the A88E or wt Taz1p. In contrast, Yme1K327R and Yme1E541Q copurified a small amount of the A88E mutant polypeptide. Importantly, wt Taz1p and the controls Tim22p, Cyt1p, Cox5,6p, F1-β, AAC2 (all resident to IM), and porin (OM) were not copurified with either Yme1K327R or Yme1E541Q. Consistent with the minor detected interaction, there was no discernable difference in the assembly of the A88E BTHS mutant tafazzin in the absence or presence of Yme1p, functional or not functional (Fig. 6 C). The failure to copurify the A88E mutant with wt Yme1p implies that substrate binding by Yme1p is quickly followed by substrate degradation.
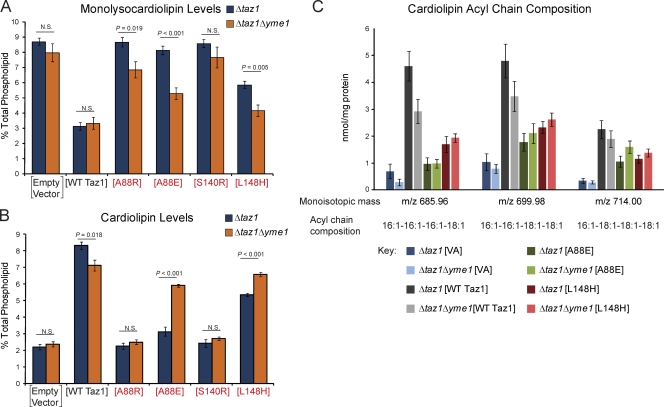
Absence of Yme1p restores activity of A88E and L148H BTHS mutant tafazzins
Although not expected given the fact that the A88R/E, S140R, and L148H BTHS mutants are degraded by the mitochondrial quality control executioner Yme1p, the fact that these mutants did assemble normally (Fig. 5, A and B) prompted us to determine whether their accumulation in the absence of Yme1p restored MLCL and/or CL levels to normal. Amazingly, there were both significant decreases in the abundance of MLCL (Fig. 7 A) and increases in the abundance of CL (Fig. 7 B) in strains expressing the A88E and L148H BTHS mutants in the absence versus the presence of Yme1p. These results suggest that, in the absence of i-AAA protease–mediated degradation, for two of these four mutant tafazzins, increased steady-state abundance correlates with the recovery of Taz1p function. If this is true, the CL that accumulates should resemble mature remodeled CL. Indeed, the abundance of the most common mature forms of CL in wt Taz1p–expressing yeast is increased in A88E and L148H BTHS mutant strains in the absence of the i-AAA protease (Fig. 7 C).
Figure 7.
Increased function of the A88E and L148H BTHS mutants in the absence of the i-AAA protease. (A and B) The relative abundance of MLCL (A) and CL (B) was determined for each strain and is expressed as a percentage of the total phospholipid in each strain (means ± SEM; n = 5). Student t tests were performed for each construct as expressed in Δtaz1 and Δtaz1Δyme1 yeast strains with significant differences indicated. (C) Comparison of CL in Δtaz1 and Δtaz1Δyme1 yeast strains transformed as indicated. The mass content of each molecular form of CL (acyl chain composition indicated) was determined by multidimensional mass spectrometric array analyses by comparison of the peak intensity of each individual ion to that of the internal standard (means ± SEM; n = 3). The four BTHS mutants being characterized in the present study are shown in red. m/z, mass to charge ratio.
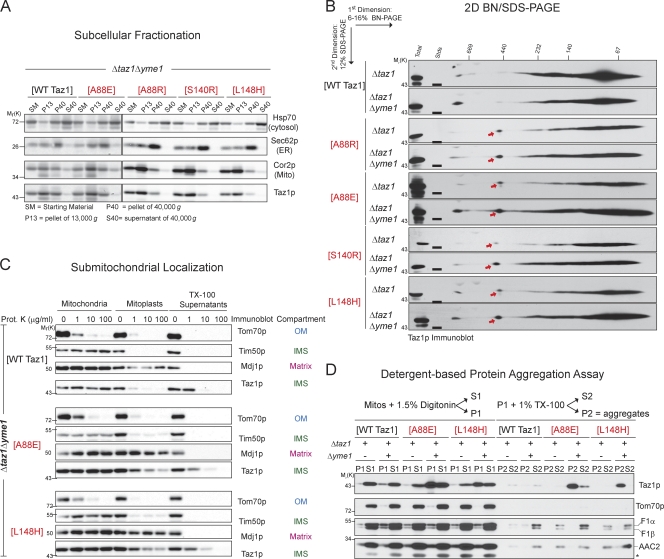
Do the BTHS mutants accumulate in a nonmitochondrial compartment in the absence of the i-AAA protease? Based on their continued cofractionation with mitochondria, the A88R/E, S140R, and L148H mutant tafazzins localize normally to mitochondria even in the absence of Yme1p (Fig. 8 A). Moreover, the assembly status of wt, A88R/E, S140R, and L148H tafazzins is unaffected by the absence of Yme1p (Fig. 8 B). The only discernable difference minus Yme1p is that the BTHS mutant complexes are more abundant. Finally, focusing on the A88E and L148H BTHS mutant tafazzins that had recovered function in the absence of the i-AAA protease, wt, A88E, and L148H tafazzins still associate with IMS-facing membranes in the absence of Yme1p (Fig. 8 C). Notably, the overall rupturing of the OM (tracked by a decrease in the IMS signal with the addition of proteinase K to mitoplasts) was consistently better for A88E and L148H mitochondria in the absence versus the presence of Yme1p, which is consistent with the restored function of both mutants in the absence of the i-AAA protease (Fig. 7).
Figure 8.
Increased assembly, but with protein aggregation, of the A88E and L148H BTHS mutants in the absence of the i-AAA protease. (A) Fractions were prepared from the indicated yeast strains through a series of differential centrifugations. 25 µg of each fraction was separated by SDS-PAGE and analyzed by immunoblotting for the indicated subcellular organelle. (B) Digitonin extracts from mitochondria derived from the indicated strains were resolved by 2D BN/SDS-PAGE, and Taz1p complexes were detected by immunoblotting. The red arrows highlight a complex only detected in mutant tafazzin extracts. 250 µg (Δtaz1 transformed with [A88R], [A88E], [S140R], or [L148H]) and 150 µg (all the rest) were analyzed. Stds, molecular weight standards. (A and B) n = 3. (C) Mitochondria derived from the indicated strains were treated exactly as described in Fig. 3 C. 50 µg/lane of each sample was resolved by SDS-PAGE and immunoblotted as indicated. (D) Solubility of Taz1p in detergents. Mitochondria isolated from the indicated strains were solubilized with digitonin and separated into a supernatant (S1) and pellet (P1) by centrifugation. Nonextracted material (P1) was solubilized with TX-100 and fractionated into a supernatant (S2) and pellet (P2) by centrifugation. Fractions were resolved by SDS-PAGE and immunoblotted as indicated. The asterisk highlights the cross-reaction with porin of the AAC2 antiserum. The four BTHS mutants being characterized in the present study are shown in red. Mito, mitochondria. (B and D) n = 4.
As a routine component of 2D BN/SDS-PAGE, the solubilization efficiency of every source of mitochondria is determined. Through these analyses, it was noticed that the solubilization efficiency of the mutant tafazzins was specifically impaired in the absence of Yme1p. Thus, to determine whether the mutant tafazzins are prone to aggregation in the absence of Yme1p, a sequential detergent solubilization assay was performed (Fig. 8 D). In this assay, protein aggregates accumulate in the TX-100 insoluble fraction (P2). In the presence of a functional i-AAA protease, both the A88E and L148H BTHS mutant tafazzins, similar to wt Taz1p, were readily solubilized by digitonin. However, in the absence of Yme1p, a sizeable proportion of the A88E and L148H mutant tafazzins was resistant to solubilization by digitonin (detected in P1). Further extraction with TX-100 demonstrated that these two BTHS mutants accumulated in TX-100–resistant aggregates (P2). wt Taz1p was equally extracted by digitonin in the presence or absence of Yme1p. Thus, whereas in the absence of Yme1p the abundance of A88E and L148H mutant tafazzin complexes is increased and the MLCL/CL levels are partially restored, both mutants additionally accumulate in presumably nonfunctional protein aggregates.
BTHS mutant tafazzin complex lability
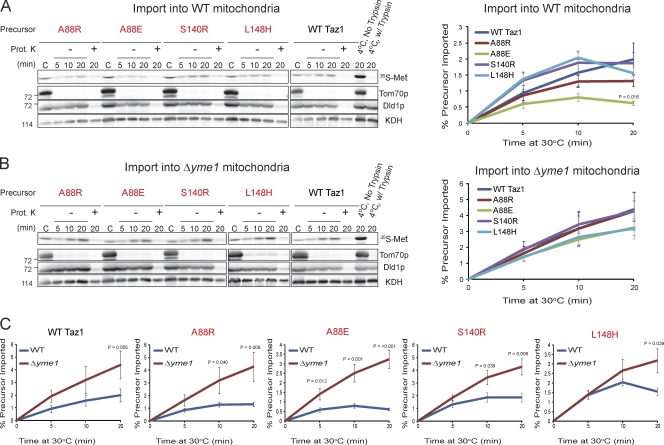
Still unclear is why the mutant tafazzins are even targeted for i-AAA protease–mediated degradation. One possibility is that the four mutant tafazzins are not imported into mitochondria as efficiently as wt Taz1p. To test this, wt mitochondria were incubated with 35S-labeled wt, A88R/E, S140R, or L148H tafazzins and treated with trypsin to remove nonimported proteins, and the OM was disrupted by osmotic shock in the absence or presence of proteinase K to degrade proteins in the IMS (Fig. 9 A). With only one exception, each of the BTHS mutants was imported into IMS-facing membranes to a similar extent as wt Taz1p. After a 20-min import, significantly less A88E mutant tafazzin was productively imported into wt mitochondria than wt Taz1p. However, compared with the steady increase in import of wt Taz1p over time, the amount of each BTHS mutant imported had plateaued or even decreased by the final import time point, suggesting that the mutants were already being recognized and degraded by the i-AAA protease. Therefore, in vitro import reactions were also performed using Δyme1 mitochondria (Fig. 9 B). Indeed, all four tested BTHS mutants were imported into Δyme1 mitochondria to a similar extent as wt Taz1p. Moreover, there was a steady increase in import into Δyme1 mitochondria for each of the BTHS mutants over time. Finally, the import of each of the four BTHS mutant tafazzins was significantly better in Δyme1 than wt mitochondria (Fig. 9 C). The improved import of wt Taz1p into Δyme1 mitochondria (approaching the level of significance by the final time point) also indicates that the i-AAA protease is intimately involved in the routine assessment of Taz1p biogenesis and either the folding/assembly of Taz1p is intrinsically inefficient or that chaperones required for Taz1p folding/assembly are saturated in the in vitro assay. Collectively, these results indicate that the A88R/E, S140R, and L148H BTHS mutants are imported into mitochondria as well as wt Taz1p.
Figure 9.
The A88R/E, S140R, and L148H BTHS mutants are recognized as defective by the i-AAA protease quickly after import. (A and B) In vitro import of 35S-radiolabeled wt Taz1 or the BTHS mutant Taz1 precursor into wt (A) or Δyme1 (B) mitochondria. The nonimported precursor was removed with trypsin, mitochondria were reisolated, and the OM was ruptured by osmotic shock in the absence or presence of proteinase K (Prot. K). C, control (2.5% of precursor proteins + 100 µg mitochondria). The samples were resolved by SDS-PAGE, the bottom half of the gel was analyzed by phosphoimaging, and the top half was immunoblotted for markers of the OM (Tom70p), membranes facing the IMS (Dld1p), and the matrix (α-ketoglutarate dehydrogenase [KDH]). The percentage of each precursor imported at each time point was determined after phosphoimaging. Significant differences were determined by one-way ANOVA with Holm–Sidak pairwise comparisons and are indicated. Relative molecular masses are shown on the left. (C) Comparison of import into wt versus Δyme1 mitochondria. Significant differences between the percentage of each precursor imported into wt versus Δyme1 mitochondria were determined at each time point by Student t tests, with significant differences indicated. The four BTHS mutants being characterized in the present study are shown in red. Values are means ± SEM (n = 5–7 for import into wt mitochondria, and n = 4 for import into Δyme1 mitochondria).
The results from the in vitro import experiments further imply that the A88R/E, S140R, and L148H BTHS mutant tafazzins are recognized as defective by the i-AAA protease soon after their import into IMS-facing membranes. Perhaps the subsequent folding and/or assembly of the mutant tafazzins is less efficient than wt Taz1p, and the stalled assembly intermediates are recognized and degraded by the i-AAA protease. In the absence of the i-AAA protease, some of the stalled assembly intermediates eventually mature into fully assembled Taz1p complexes. The detection of more abundant mutant Taz1p complexes in the absence of Yme1p is entirely consistent with this possibility. Unfortunately, conditions have not been defined as of yet that allow assembly of wt Taz1p into mature Taz1p complexes in organello (unpublished data). Thus, the kinetics of import and assembly of the mutant tafazzins cannot be compared with wt Taz1p at present.
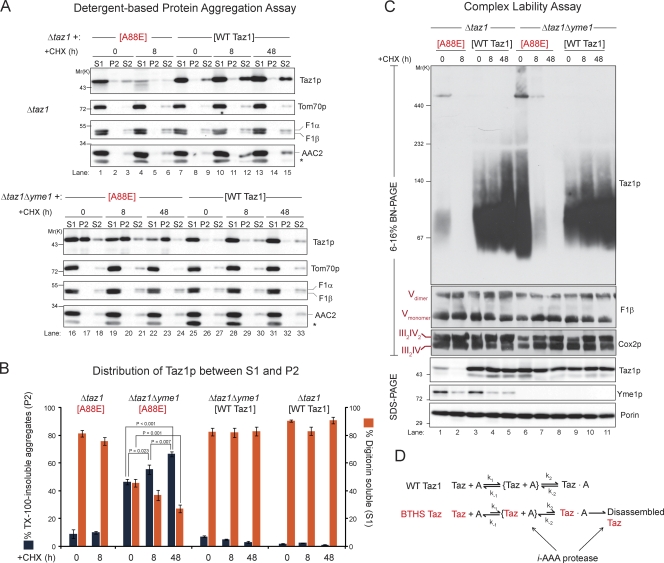
Another possible explanation as to why the A88R/E, S140R, and L148H BTHS mutant tafazzins are degraded by the i-AAA protease is that complexes containing the mutant tafazzins assemble but fall apart at significantly faster rates. Furthermore, perhaps the accumulation in TX-100–insoluble aggregates is a consequence of the unregulated disassembly of BTHS mutant–containing complexes. To test this hypothesis, mitochondria were isolated from yeast strains expressing wt Taz1p or the A88E BTHS mutant, in the presence or absence of Yme1p, after incubation with CHX for 8 or 48 h. wt Taz1p was readily solubilized by digitonin, regardless of the presence or absence of Yme1p, even when new protein synthesis was inhibited for 2 d (Fig. 10, A [lanes 7–15 and 25–33] and B). As expected, the absolute abundance of the A88E BTHS mutant in the presence of Yme1p was significantly decreased after 8-h CHX incubation (Fig. 10 C, bottom, lanes 1 and 2). Similar to wt Taz1p, the A88E mutant tafazzin remaining after 8-h CHX incubation was readily solubilized by digitonin when a functional i-AAA protease was present (Fig. 10, A [lanes 1–6] and B). In contrast, whereas the steady-state abundance of the A88E BTHS mutant in the absence of Yme1p remained stable throughout the CHX chase (Fig. 10 C, bottom, lanes 6–8), the fraction of A88E mutant tafazzin solubilized by digitonin decreased significantly after new protein synthesis was halted for 48 h (Fig. 10, A [lanes 16–24] and B). Moreover, in the absence of Yme1p, the abundance of the A88E BTHS mutant detectable in the TX-100–insoluble P2 fraction steadily and significantly increased throughout the CHX chase (Fig. 10, A and B). Similar to the solubilization data, wt Taz1p complexes were stable in the absence of new protein synthesis for up to 2 d regardless of the presence or absence of the i-AAA protease (Fig. 10 C, top, lanes 3–5 and 9–11; 2D BN/SDS-PAGE analyses are shown in Fig. S4). In the presence of Yme1p, after only an 8-h incubation with CHX, A88E BTHS mutant complexes, like the polypeptide, were virtually undetectable (Fig. 10 C, lanes 1 and 2). Strikingly, in mitochondria lacking Yme1p, inhibition of new protein synthesis for only 8 h significantly reduced A88E BTHS mutant tafazzin complexes compared with control mitochondria not incubated with CHX. After 2-d CHX incubation, in the absence of Yme1p, A88E BTHS mutant complexes were undetectable even though the A88E polypeptide remained stable throughout the CHX chase (Fig. 10 C, lanes 6–8, compare top BN-PAGE with bottom SDS-PAGE). In stark contrast to the A88E mutant tafazzin complexes in the absence of Yme1p, additional protein complexes of the mitochondrial IM (ATP synthase, respiratory supercomplexes, and AAC2 complexes) and OM (porin complexes) persisted even in the absence of new protein synthesis for 2 d (Fig. 10 C and Fig. S4). Thus, A88E BTHS mutant tafazzin complexes are specifically and selectively much more transient than wt Taz1p complexes. Moreover, the increased lability of the mutant complexes when expressed in the absence of the i-AAA protease correlates directly with the accumulation of BTHS mutant polypeptide in TX-100–insoluble aggregates.
Figure 10.
BTHS mutant tafazzin complexes are unstable, and the mutant polypeptide accumulates in aggregates. Mitochondria were harvested from the indicated strains after incubation with CHX for the indicated times. (A) Solubility of Taz1p in detergents was determined as described in Fig. 8 D. The asterisk highlights the cross-reaction with porin of the AAC2 antiserum. For Taz1p immunoblots, 50 µg (Δtaz1 [A88E]) and 20 µg (all the rest) were analyzed. For immunoblots of control proteins (Tom70p, F1-α/β, and AAC2), 20 µg was analyzed. (B) Versadoc-captured images were quantified using the affiliated Quantity One software, and the percentage of Taz1p in TX-100–insoluble aggregates and the percentage of Taz1p solubilized by digitonin were determined (means ± SEM; n = 5). Significant differences were determined by one-way ANOVA with Holm–Sidak pairwise comparisons and are indicated. (C) Digitonin extracts from mitochondria derived from the indicated strains were resolved by 1D BN-PAGE, and immunoblotting was performed for Taz1p, the ATP synthase/complex V (F1-β), and respiratory supercomplexes (Cox2p). The migrations of the Vdimer, Vmonomer, III2IV2, and III2IV supercomplexes are indicated where appropriate. 250 µg (Δtaz1[A88E]) and 150 µg (all the rest) were analyzed. 25 µg mitochondrial protein was additionally resolved by SDS-PAGE and immunoblotted as indicated. n = 3. (D) wt Taz1p assembles with partner proteins (generically denoted A) into final Taz1p complexes that are very stable. In contrast, the characterized BTHS mutant tafazzins have a retarded rate of assembly into final Taz1p complexes that quickly disassemble. The i-AAA protease recognizes both the delayed rate of BTHS mutant Taz1p assembly and the unregulated disassembly of Taz1p complexes. In the absence of the i-AAA protease, disassembled BTHS Taz1 forms aggregates. The BTHS mutants being characterized in the present study are shown in red.
Discussion
Currently there are 28 known missense mutations in tafazzin associated with the X-linked disease BTHS. With the exception of our previous characterization of four mutations present in the membrane anchor of Taz1p (Claypool et al., 2006), an explanation for the inability to properly function as an MLCL transacylase had not been provided for any of the other 24 BTHS mutant tafazzins. Here, we generate a panel of BTHS mutant yeast strains, focusing on the 21 missense mutations that occur at conserved positions in the human and yeast orthologues. Critically, 18 of the 21 tested BTHS mutant yeast strains accumulated significant quantities of the stalled CL-remodeling intermediate and BTHS biomarker MLCL, providing yet another illustration of the conservation of many basic mitochondrial processes from humans to fungi. Thus, the BTHS mutant yeast panel serves as another superb example of the power of the S. cerevisiae model system to characterize pathogenic mutations that cause human disease.
The detailed characterization of four BTHS mutations occurring at three loci uncovered a paradox. Whereas the A88R/E, S140R, and L148H BTHS mutants are degraded by an integral component of the mitochondrial quality control machinery, the i-AAA protease, suggestive of a severe folding defect of the mutants, each of the BTHS mutant tafazzins assembled in roughly normal Taz1p complexes, just of reduced abundance. Moreover, each BTHS mutant tafazzin retained the ability to associate with the ADP/ATP carrier. Yeast Taz1p associates with the ATP synthase and the ADP/ATP carrier in discrete complexes (Claypool et al., 2008a). Still, the composition of the majority of the Taz1p interactome remains a mystery. The ability to engage in a normal range of physical interactions is strong evidence that the BTHS mutants are in fact able to fold properly. In the absence of Yme1p, the major subunit of the i-AAA protease, more BTHS mutant tafazzin complexes are detected. This strongly suggests that the mutant tafazzins exhibit a retarded rate of assembly into final Taz1p-containing complexes (Fig. 10 D). This interpretation is based on the rationale that, if a BTHS mutant tafazzin had an increased frequency of forming dead-end assembly intermediates independent of Yme1p, the absence of the i-AAA protease should simply increase the abundance of such misassemblies. The fact that more BTHS mutant tafazzin complexes are detected in the absence of Yme1p argues that the mutants can fold and assemble normally; they just do so with a reduced efficiency. Based on the significantly increased in vitro import into mitochondria devoid of Yme1p, the folding and/or assembly of Taz1p is normally monitored by the i-AAA protease. Yme1p recognizes the presumed defect in the folding and/or assembly of the four BTHS mutant tafazzins and then quickly and actively proteolyzes the mutant polypeptides subsequent to a physical association. Thus, the delayed folding and assembly and consequent degradation by Yme1p undoubtedly contribute to the overall loss of function associated with these particular BTHS mutant tafazzins.
Strikingly, complexes containing the A88E BTHS mutant tafazzin fall apart much faster than wt Taz1p complexes (Fig. 10 D). Moreover, in the absence of i-AAA protease–mediated degradation, disassembled BTHS mutant tafazzin accumulates in TX-100–insoluble aggregates. Therefore, although these four BTHS mutant tafazzins undoubtedly display a reduced rate of complex formation, the inherent instability of the formed complexes ensures that they fail to perform their physiological tasks. To our knowledge, complex lability has never been described as an underlying pathogenic mechanism for a disease-associated mutation.
Why are the mutant tafazzin complexes more labile? One possibility is that perhaps while retaining the ability to fold, these four BTHS mutant tafazzins do so more loosely. Mutations that destabilize but not completely destroy tafazzin structures would be expected to both delay folding and complex assembly and weaken complexes that are formed during a loosening event of the mutant tafazzin. Furthermore, such destabilized mutants might be predicted to be detected during their folding and assembly by mitochondrial chaperones and/or Taz1p assembly factors, resulting in the degradation of the mutant polypeptides by the i-AAA protease. The dual observations that more mutant tafazzin complexes are detected in the absence of Yme1p but these complexes rapidly fall apart suggest that chaperones assisting in the folding and assembly of Taz1p are unable to efficiently promote the reassembly of mutant Taz1p complexes. Thus, for the four BTHS mutant tafazzins, the equilibrium between assembly and disassembly has been shifted significantly in the direction of disassembly. This strongly implies that the formation of stable Taz1p complexes is critical for normal tafazzin function. Therefore, the partial recovery in the MLCL and CL levels upon deletion of Yme1p was driven by the continual supply of newly translated A88E and L148H BTHS mutant tafazzins.
An obvious question is whether the results obtained with yeast accurately model the same mutations in human tafazzin as expressed either in patient samples or a mammalian model. Unfortunately, our efforts to raise antibodies against human Taz1p have yielded reagents that lack the required sensitivity and specificity to detect any of the endogenous tafazzin isoforms predicted at the mRNA level (Houtkooper et al., 2009b). This has significantly hampered the development of a mammalian cell culture model that is experimentally tractable and yet physiologically relevant. Similar to BTHS-derived cells, knockdown of tafazzin in HeLa cells results in a decrease in the abundance of CL (Gonzalvez et al., 2008). However, in contrast to BTHS-derived cells, tafazzin knockdown HeLa cells did not significantly accumulate MLCL, which is an established biomarker for BTHS (Houtkooper et al., 2009a). The Human Tafazzin (TAZ) Gene Mutation and Variation database available on the Barth Syndrome Foundation website is a wonderful resource that lists defined mutations, provides salient patient information, and, when available, indicates whether decreased CL and/or increased MLCL was detected in a patient with a specific tafazzin lesion. Unfortunately, information regarding the CL and MLCL profiles in patients harboring G80R/E, G131R, and L139H tafazzin mutations, corresponding to the yeast A88R/E, S140R, and L148H BTHS mutants characterized in detail in the present study, are not currently available. The absence of this data likely reflects the fact that awareness of this serious human disease is incomplete at best and that an affordable and reliable method to detect CL and MLCL from blood spots has only recently been established (Kulik et al., 2008). Thus, validation of the yeast BTHS mutant tafazzin library remains an important but challenging goal for our future investigations.
Materials and methods
Yeast strains
All strains were derived from the wt parental S. cerevisiae yeast strain GA74-1A (MATa, his3 –11,15, leu2, ura3, trp1, ade8, rho+, mit+). The Δtaz1His1.5 (MATa, leu2, ura3, trp1, ade8, Δtaz1::HISMX6) strain (Claypool et al., 2006) served as the parent for the BTHS tafazzin panel that contained all of the IM proteases. To generate the Δtaz1Δoma1 (MATa, leu2, trp1, ade8, Δtaz1::URAMX, Δoma1::HISMX6), Δtaz1Δyta12 (MATa, leu2, his3–11,15, ade8, Δtaz1::URAMX, Δyta12::TRP), Δtaz1Δyme1 (MATa, leu2, trp1, ade8, Δtaz1::URAMX, Δyme1::HISMX6), Δtaz1Δmgr1 (MATa, leu2, trp1, ade8, Δtaz1::URAMX, Δmgr1::HISMX6), and Δtaz1Δmgr3 (MATa, leu2, trp1, ade8, Δtaz1::URAMX, Δmgr3::HISMX6) strains, the entire open reading frame of each gene was replaced using the PCR-mediated one-step gene replacement strategy (Wach et al., 1994). The Δyme1His1.1 (MATa, leu2, ura3, trp1, ade8, Δyme1::HISMX6) strain has been described previously (Hwang et al., 2007).
Molecular biology and recombinant protein expression
pRS425wt Taz1 containing ∼300 bp of a 5′ untranslated region has been described previously (Claypool et al., 2006). The series of BTHS point mutations were generated by overlap extension (Ho et al., 1989) using pRS425wt Taz1 as a template. Each construct was subcloned into pRS425wt Taz1. For in vitro transcription/translation, the open reading frame of Taz1p was subcloned into pSP64. The A88R/E, S140R, and L148H BTHS mutants were shuttled from pRS425-based constructs into pSP64yTaz. Yme1, including ∼500 bp of a 5′ untranslated region and a C-terminal His6 tag, was amplified by PCR using genomic DNA isolated from the GA74-6A yeast strain as a template. Yme1K327R and Yme1E541Q were generated by overlap extension. All Yme1 constructs were cloned into the 2-µm vector pRS424. To generate yeast Dld1p (d-lactate dehydrogenase) containing an N-terminal His6 tag, the entire open reading frame was cloned into the pET28a vector (EMD) in frame and downstream of the His6 tag, and the thrombin cleavage site was provided by the vector. His6Dld1 was induced in BL21-CodonPlus(DE3)-RIL Escherichia coli (Agilent Technologies), and inclusion bodies were isolated from native protein extracts by centrifugation at 10,000 g for 20 min at 4°C. Inclusion bodies were solubilized with 3 ml of inclusion body solubilization buffer (1.67% [wt/vol] sarkosyl, 0.1-mM EDTA, 10-mM DTT, 10-mM Tris-CL, pH 7.4, and 0.05% polyethylene glycol 3350) by vigorous vortexing, incubated on ice for 20 min, and diluted with 6 ml of 10-mM Tris-CL, pH 7.4, and the solubilized proteins were recovered after centrifugation for 10 min at 12,000 g at 4°C. After dialysis against phosphate-buffered saline, His6Dld1 was purified under native conditions using Ni2+-agarose (QIAGEN) as per the manufacturer’s instructions. The sequence of every construct was verified by sequencing.
Antibodies
Most of the antibodies used in this work were generated in the Schatz or Koehler laboratories and have been described previously (Poyton and Schatz, 1975; Djavadi-Ohaniance et al., 1978; Maccecchini et al., 1979; Daum et al., 1982; Ohashi et al., 1982; Riezman et al., 1983; Scherer et al., 1990; Glick et al., 1992; Horst et al., 1997; Koehler et al., 1998; Claypool et al., 2006, 2008b; Hwang et al., 2007). Antibodies were raised in rabbits using His6Dld1 as an antigen. Other antibodies used were mouse anti-Sec62p (gift from D. Meyers, University of California, Los Angeles, Los Angeles, CA), mouse anti-yAAC2 (clone 6H8; Panneels et al., 2003), and horseradish peroxidase–conjugated secondary antibodies (Thermo Fisher Scientific).
Metabolic labeling and immunoprecipitation of Taz1p
Yeast were grown overnight at 30°C in synthetic complete (SC)–Leu medium. Each culture was diluted 1:10 in SC-Leu and grown at 30°C for 90 min to OD600s of 0.4–1.0. 2.5 OD600 units of cells were resuspended with 0.4 ml of fresh SC-Leu and incubated at 30°C while shaking at 200 rpm for 15 min. Proteins were labeled at 30°C for 10 min with 150 µCi (two experiments) or 500 µCi (one experiment) trans–35S-label (MP Biomedicals). Labeling was terminated by transferring the cells to an equal volume of ice-cold 2× azide stop mix (20-mM NaN3, 40-mM cysteine, 40-mM methionine, and 0.5 mg/ml BSA). Sedimented cells were washed with 0.5 ml of ice-cold distilled H2O, resuspended with 0.5 ml of 1% TX-100 (vol/vol) lysis buffer (20-mM Hepes-KOH, pH 7.4, 50-mM NaCl, 1-mM EDTA, 2.5-mM MgCl2, and 0.1% SDS [wt/vol]) supplemented with protease inhibitors (1-mM PMSF, 10 µM leupeptin, 2 µM pepstatin A, and 10 µM chymostatin), transferred to a microfuge tube containing 0.1 ml of glass beads, and disintegrated by vortexing on high for ∼30 min at 4°C. The lysate was transferred to a new tube, and the beads were washed with 0.5 ml of 1% TX-100 (vol/vol) lysis buffer to recover all of the extract. Lysates were precleared twice (rotating at 4°C for 1 h/preclear) with normal rabbit serum (Lampire Biological Laboratories, Inc.) prebound to protein A–Sepharose (GE Healthcare). Tafazzin was captured from precleared lysates with rabbit anti–yeast tafazzin antisera prebound to protein A–Sepharose rotating at 4°C overnight. Immune complexes were sequentially washed (1 ml and 10-min rotating at 4°C/wash) twice with lysis buffer containing 0.1% TX-100 (vol/vol), twice with high salt wash buffer (0.1% TX-100 [vol/vol], 20-mM Hepes-KOH, pH 7.4, and 500-mM NaCl), and once with low salt wash buffer (0.1% TX-100 [vol/vol] and 20-mM Hepes-KOH, pH 7.4). Bound material was resolved on 12% SDS-PAGE gels, and labeled proteins were detected using a K screen and an image scanner (Pharos FX Imager; Bio-Rad Laboratories).
In vivo degradation experiments
In vivo degradation experiments were performed essentially as described previously (Metzger et al., 2008). In brief, overnight cultures were resuspended to OD600 = 4.0 in SC media and incubated with shaking for 5 min at 30°C, and new protein synthesis was inhibited by the addition of CHX ([200 µg/ml]final). At each time point, 0.5 ml of cells was transferred to a tube containing an equal volume of ice-cold 2× azide stop mix (20-mM NaN3 and 0.5 mg/ml BSA). Cells were pelleted by centrifugation (845 g for 10 min at 4°C), the supernatant was aspirated, and pellets were stored at −80°C until all time points were collected. The preparation of yeast cell extracts was performed otherwise as described previously (Claypool et al., 2006). For quantitation of immunoblots, images captured with either a Versadoc (Bio-Rad Laboratories) or Fluorchem Q (Cell Biosciences, Inc.) quantitative digital imaging system were quantitated using Quantity One software (Bio-Rad Laboratories). Half-lives were determined from exponential curve fits using Excel (Microsoft). wt and BTHS mutant tafazzin half-lives were calculated using the mean of seven to eight individual repetitions. The half-life of Yme1p was calculated from experiments using Δtaz1 (wt Taz1; n = 3). Data from the longer CHX chase studies were pooled together to calculate the half-life of porin (n = 42) and Hxk1p (n = 18).
Glycerol density gradient centrifugation
Glycerol density gradient centrifugation was performed essentially as described previously (Tamura et al., 2009) with minor alterations. In brief, 5 mg/ml mitochondria was solubilized with lysis buffer base (20-mM Hepes-KOH, pH 7.4, 50-mM NaCl, 0.1-mM EDTA, and 10% glycerol) supplemented with 1.5% (wt/vol) digitonin (Biosynth International, Inc.) and 1-mM PMSF for 30 min on ice; insoluble material was removed by centrifugation for 30 min at 21,000 g at 4°C. The 100-µl supernatant was placed onto a 5-ml glycerol step gradient (20–40%) in 20-mM Hepes-KOH, pH 7.4, 50-mM NaCl, 50-mM 6-aminohexanoic acid, 0.1-mM EDTA, and 0.1% (wt/vol) digitonin and then centrifuged at 45,000 rpm for 15 h in a rotor (SW55Ti; Beckman Coulter) at 4°C. 270 µl of fractions was collected from the top. Proteins were precipitated with 20% TCA and then resolved by SDS-PAGE followed by immunoblotting.
Ni2+ nitrilotriacetic acid purification
Detergent solubilization of mitochondria (5 mg/ml) was performed for 30 min on ice with lysis buffer base (20-mM Hepes-KOH, pH 7.4, 100-mM NaCl, 20-mM imidazole, 1-mM CaCl2, and 10% glycerol) supplemented with 1.5% (wt/vol) digitonin and protease inhibitors; insoluble material was removed by centrifugation for 30 min at 21,000 g at 4°C. 0.6 mg digitonin extract was added to a tube containing 25 µl Ni2+ nitrilotriacetic acid beads (QIAGEN) and 0.88 ml lysis buffer base with added protease inhibitors and incubated by rotating at 4°C for 1 h. Nonbinding material was TCA precipitated. Beads were washed twice (1 ml and 10-min rotating at 4°C wash) with 0.1% digitonin wash buffer (20-mM Hepes-KOH, pH 7.4, 100-mM NaCl, 20-mM imidazole, 1-mM CaCl2, and 10% glycerol), and bound material was eluted with sample buffer supplemented with 300-mM imidazole.
Multidimensional mass spectrometry–based shotgun lipidomic analysis of mitochondrial lipids
The yeast mitochondrial lipidome was determined using multidimensional mass spectrometry–based shotgun lipidomics as previously described (Han and Gross, 2005; Han et al., 2006; Yang et al., 2009). In brief, a 400-µg aliquot of mitochondrial protein was transferred to a disposable borosilicate glass tube. Internal standards as a premixture were added based on protein concentration. A modified Bligh and Dyer procedure was used to extract lipids from each mitochondrial preparation. Each lipid extract was reconstituted with a volume of 200 µl/mg mitochondrial protein in chloroform/methanol (1:1; vol/vol). The lipid extracts were flushed with nitrogen, capped, and stored at −20°C for electrospray ionization mass spectrometric analysis.
Sequential solubilization assay
Protein aggregation was determined as previously described (Rabl et al., 2009). In brief, mitochondria were solubilized with 1.5% (wt/vol) digitonin lysis buffer supplemented with protease inhibitors as previously described in the Ni2+ nitrilotriacetic acid purification section. After centrifugation at 4°C for 10 min at 21,000 g, solubilized material was transferred to a new tube, and nonsolubilized material (P1) was either directly analyzed or resolubilized for 30 min on ice with 1% (vol/vol) TX-100 lysis buffer (20-mM Hepes-KOH, pH 7.4, 20-mM imidazole, 10% glycerol, 100-mM NaCl, and 1-mM CaCl2) supplemented with protease inhibitors. TX-100–insoluble material (P2) was separated from solubilized extract (S2) by centrifugation for 10 min at 21,000 g at 4°C. The percentage of Taz1p in TX-100–insoluble aggregates and the percentage of Taz1p solubilized by digitonin were calculated as follows: P2/(S1 + S2 + P2) × 100 and S1/(S1 + S2 + P2) × 100, in which S1, S2, and P2 are the volumes of Taz1p detected in fractions S1, S2, and P2, respectively.
In organello import into mitochondria
Radiolabeled precursors were generated using an SP6 Quick Coupled Transcription/Translation system and guidelines (Promega) and a 35S EasyTag (PerkinElmer). Radiolabeled precursors were incubated for 5, 10, or 20 min at 30°C with wt GA74-1A or Δyme1 mitochondria in import buffer (0.6-M sorbitol, 2-mM KH2PO4, 60-mM KCl, 50-mM Hepes, 10-mM MgCl2, 2.5-mM EDTA, pH 8.0, 5-mM l-methionine, 10 mg/ml BSA, 2-mM ADP, and 2-mM NADH). Import was stopped, and the nonimported precursor was degraded with an equal volume of ice-cold BB7.4 (0.6-M sorbitol and 20-mM Hepes-KOH, pH 7.4) containing 40 µg/ml trypsin. Trypsin was inhibited with 100 µg/ml of soybean trypsin inhibitor, and mitochondria were reisolated by spinning at 8,000 g for 5 min at 4°C. To rupture the OM, mitoplasting was performed in the absence or presence of 100 µg/ml proteinase K as previously described (Claypool et al., 2006). As a control, the wt Taz1 precursor was incubated with mitochondria on ice for 20 min and treated with trypsin, or not treated, to remove a nonimported precursor, and reisolated mitochondria were subjected to osmotic shock in the absence of proteinase K. This control demonstrates that import does not occur at 4°C and that trypsin removes all of the nonimported precursor. 100% of each time point and 2.5% of imported precursors were resolved on 12% SDS-PAGE gels and analyzed by phosphoimaging. The percentage of precursor imported was calculated as follows: (T/P) × 2.5, in which T is the volume of Taz1p at a given import time point, and P is the volume of the Taz1p precursor.
Miscellaneous
The performed experiments used mitochondria harvested from yeast grown at 30°C to an OD600 of ∼3 in synthetic lactate−Leu (0.17% yeast nitrogen base minus amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.2% Drop-out Mix Synthetic minus Leu, 0.05% dextrose, 2% lactic acid, 3.4-mM CaCl2 2H2O, 8.5-mM NaCl, 2.95-mM MgCl2 6H2O, 7.35-mM KH2PO4, and 18.7-mM NH4Cl) in Figs. 3 (B and C), 5, 7 C, 8, 10, S2, and S4; rich lactate medium (1% yeast extract, 2% tryptone, 0.05% dextrose, 2% lactic acid, 3.4-mM CaCl2 2H2O, 8.5-mM NaCl, 2.95-mM MgCl2 6H2O, 7.35-mM KH2PO4, and 18.7-mM NH4Cl) in Figs. 9 and S3; and synthetic lactate−Leu–His–Trp–Ura (0.17% yeast nitrogen base minus amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.2% Dropout Mix Synthetic minus Leu, His, Trp, and Ura, 0.05% dextrose, 2% lactic acid, 3.4-mM CaCl2 2H2O, 8.5-mM NaCl, 2.95-mM MgCl2 6H2O, 7.35-mM KH2PO4, and 18.7-mM NH4Cl) in Fig. 6 (B and C). The preparation of yeast cell extracts (SC media), subcellular fractionation, isolation of mitochondria, submitochondrial localization, 1D BN-PAGE, immunoblotting, and immunoblot quantitation were performed as previously described (Claypool et al., 2006); 2D BN/SDS-PAGE and AAC2 immunoprecipitation (Claypool et al., 2008b) and phospholipid labeling (synthetic lactate−Leu), extraction, and data collection (Claypool et al., 2006, 2008a) were performed as previously described. Statistical comparisons were performed using SigmaPlot 11 software (Systat Software, Inc.).
Online supplemental material
Fig. S1 shows the CL levels in the BTHS mutant tafazzin panel. Fig. S2 shows the submitochondrial localization of BTHS mutant tafazzins with additional controls. Fig. S3 shows the defective swelling response of CL-deficient mitochondria. Fig. S4 shows 2D BN/SDS-PAGE immunoblots of wt and A88E tafazzin, porin, and AAC2 in the presence and absence of Yme1p after incubation of yeast with CHX. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008177/DC1.
Acknowledgments
We would like to thank Dr. Jeff Schatz for antibodies, Yasushi Tamura and Hiromi Sesaki for help with glycerol density gradient centrifugation, and Matthew J. Maurer and Susan Michaelis for help with the CHX chase experiments.
This work was supported by grants from the American Heart Association (0640076N), Muscular Dystrophy Association (4033), and National Institutes of Health (R00HL089185 to S.M. Claypool, P01HL57278 to X. Han, and R01GM61721 and R01GM073981 to C.M. Koehler). X. Han serves as a consultant for LipoSpectrum, LLC. C.M. Koehler is an Established Investigator of the American Heart Association.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- BN
- blue native
- BTHS
- Barth syndrome
- CHX
- cycloheximide
- CL
- cardiolipin
- i-AAA
- IMS AAA
- IM
- inner membrane
- IMS
- intermembrane space
- m-AAA
- matrix AAA
- MLCL
- monolysocardiolipin
- OM
- outer membrane
- SC
- synthetic complete
- TX-100
- Triton X-100
- wt
- wild type
References
- Barth P.G., Scholte H.R., Berden J.A., Van der Klei-Van Moorsel J.M., Luyt-Houwen I.E., Van’t Veer-Korthof E.T., Van der Harten J.J., Sobotka-Plojhar M.A. 1983. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 62:327–355 10.1016/0022-510X(83)90209-5 [DOI] [PubMed] [Google Scholar]
- Barth P.G., Van den Bogert C., Bolhuis P.A., Scholte H.R., van Gennip A.H., Schutgens R.B., Ketel A.G. 1996. X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): respiratory-chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 19:157–160 10.1007/BF01799418 [DOI] [PubMed] [Google Scholar]
- Beranek A., Rechberger G., Knauer H., Wolinski H., Kohlwein S.D., Leber R. 2009. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J. Biol. Chem. 284:11572–11578 10.1074/jbc.M805511200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick M., Khalimonchuk O., Pierrel F., Winge D.R. 2010. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30:172–185 10.1128/MCB.00869-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S., D’Adamo P., Maestrini E., Gedeon A.K., Bolhuis P.A., Toniolo D. 1996. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12:385–389 10.1038/ng0496-385 [DOI] [PubMed] [Google Scholar]
- Brandner K., Mick D.U., Frazier A.E., Taylor R.D., Meisinger C., Rehling P. 2005. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Mol. Biol. Cell. 16:5202–5214 10.1091/mbc.E05-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Mootha V.K. 2010. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 11:25–44 10.1146/annurev-genom-082509-141720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S., Jain M., Xie X., Sheth S.A., Chang B., Goldberger O.A., Spinazzola A., Zeviani M., Carr S.A., Mootha V.K. 2006. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 38:576–582 10.1038/ng1776 [DOI] [PubMed] [Google Scholar]
- Claypool S.M., McCaffery J.M., Koehler C.M. 2006. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174:379–390 10.1083/jcb.200605043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., Boontheung P., McCaffery J.M., Loo J.A., Koehler C.M. 2008a. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell. 19:5143–5155 10.1091/mbc.E08-09-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool S.M., Oktay Y., Boontheung P., Loo J.A., Koehler C.M. 2008b. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182:937–950 10.1083/jcb.200801152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Böhni P.C., Schatz G. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028–13033 [PubMed] [Google Scholar]
- DiMauro S., Schon E.A. 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348:2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- Djavadi-Ohaniance L., Rudin Y., Schatz G. 1978. Identification of enzymically inactive apocytochrome c peroxidase in anaerobically grown Saccharomyces cerevisiae. J. Biol. Chem. 253:4402–4407 [PubMed] [Google Scholar]
- Dunn C.D., Lee M.S., Spencer F.A., Jensen R.E. 2006. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell. 17:213–226 10.1091/mbc.E05-06-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C.D., Tamura Y., Sesaki H., Jensen R.E. 2008. Mgr3p and Mgr1p are adaptors for the mitochondrial i-AAA protease complex. Mol. Biol. Cell. 19:5387–5397 10.1091/mbc.E08-01-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernster L., Ikkos D., Luft R. 1959. Enzymic activities of human skeletal muscle mitochondria: a tool in clinical metabolic research. Nature. 184:1851–1854 10.1038/1841851a0 [DOI] [PubMed] [Google Scholar]
- Gebert N., Joshi A.S., Kutik S., Becker T., McKenzie M., Guan X.L., Mooga V.P., Stroud D.A., Kulkarni G., Wenk M.R., et al. 2009. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr. Biol. 19:2133–2139 10.1016/j.cub.2009.10.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S., Brandt A., Cunningham K., Müller S., Hallberg R.L., Schatz G. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 69:809–822 10.1016/0092-8674(92)90292-K [DOI] [PubMed] [Google Scholar]
- Gonzalvez F., Schug Z.T., Houtkooper R.H., MacKenzie E.D., Brooks D.G., Wanders R.J., Petit P.X., Vaz F.M., Gottlieb E. 2008. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 183:681–696 10.1083/jcb.200803129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R.H., Parikh S., Falk M.J., Saneto R.P., Wolf N.I., Darin N., Wong L.J., Cohen B.H., Naviaux R.K.; Mitochondrial Medicine Society’s Committee on Diagnosis 2008. The in-depth evaluation of suspected mitochondrial disease. Mol. Genet. Metab. 94:16–37 10.1016/j.ymgme.2007.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Gross R.W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24:367–412 10.1002/mas.20023 [DOI] [PubMed] [Google Scholar]
- Han X., Yang K., Yang J., Cheng H., Gross R.W. 2006. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J. Lipid Res. 47:864–879 10.1194/jlr.D500044-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 77:51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- Horst M., Oppliger W., Rospert S., Schönfeld H.J., Schatz G., Azem A. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16:1842–1849 10.1093/emboj/16.8.1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Rodenburg R.J., Thiels C., van Lenthe H., Stet F., Poll-The B.T., Stone J.E., Steward C.G., Wanders R.J., Smeitink J., et al. 2009a. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Anal. Biochem. 387:230–237 10.1016/j.ab.2009.01.032 [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Turkenburg M., Poll-The B.T., Karall D., Pérez-Cerdá C., Morrone A., Malvagia S., Wanders R.J., Kulik W., Vaz F.M. 2009b. The enigmatic role of tafazzin in cardiolipin metabolism. Biochim. Biophys. Acta. 1788:2003–2014 10.1016/j.bbamem.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Hwang D.K., Claypool S.M., Leuenberger D., Tienson H.L., Koehler C.M. 2007. Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J. Cell Biol. 178:1161–1175 10.1083/jcb.200706195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R.J., Schatz G. 1998. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 279:369–373 10.1126/science.279.5349.369 [DOI] [PubMed] [Google Scholar]
- Kulik W., van Lenthe H., Stet F.S., Houtkooper R.H., Kemp H., Stone J.E., Steward C.G., Wanders R.J., Vaz F.M. 2008. Bloodspot assay using HPLC-tandem mass spectrometry for detection of Barth syndrome. Clin. Chem. 54:371–378 10.1373/clinchem.2007.095711 [DOI] [PubMed] [Google Scholar]
- Leonhard K., Herrmann J.M., Stuart R.A., Mannhaupt G., Neupert W., Langer T. 1996. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15:4218–4229 [PMC free article] [PubMed] [Google Scholar]
- Leonhard K., Stiegler A., Neupert W., Langer T. 1999. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 398:348–351 10.1038/18704 [DOI] [PubMed] [Google Scholar]
- Luft R. 1994. The development of mitochondrial medicine. Proc. Natl. Acad. Sci. USA. 91:8731–8738 10.1073/pnas.91.19.8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft R., Ikkos D., Palmieri G., Ernster L., Afzelius B. 1962. A case of severe hypermetabolism of nonthyroid origin with a defect in the maintenance of mitochondrial respiratory control: a correlated clinical, biochemical, and morphological study. J. Clin. Invest. 41:1776–1804 10.1172/JCI104637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Vaz F.M., Gu Z., Wanders R.J., Greenberg M.L. 2004. The human TAZ gene complements mitochondrial dysfunction in the yeast taz1Delta mutant. Implications for Barth syndrome. J. Biol. Chem. 279:44394–44399 10.1074/jbc.M405479200 [DOI] [PubMed] [Google Scholar]
- Maccecchini M.L., Rudin Y., Blobel G., Schatz G. 1979. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc. Natl. Acad. Sci. USA. 76:343–347 10.1073/pnas.76.1.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M., Lazarou M., Thorburn D.R., Ryan M.T. 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361:462–469 10.1016/j.jmb.2006.06.057 [DOI] [PubMed] [Google Scholar]
- Metzger M.B., Maurer M.J., Dancy B.M., Michaelis S. 2008. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 283:32302–32316 10.1074/jbc.M806424200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A., Gibson J., Gregor I., Schatz G. 1982. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J. Biol. Chem. 257:13042–13047 [PubMed] [Google Scholar]
- Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K., et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 134:112–123 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneels V., Schüssler U., Costagliola S., Sinning I. 2003. Choline head groups stabilize the matrix loop regions of the ATP/ADP carrier ScAAC2. Biochem. Biophys. Res. Commun. 300:65–74 10.1016/S0006-291X(02)02795-X [DOI] [PubMed] [Google Scholar]
- Poyton R.O., Schatz G. 1975. Cytochrome c oxidase from bakers’ yeast. IV. Immunological evidence for the participation of a mitochondrially synthesized subunit in enzymatic activity. J. Biol. Chem. 250:762–766 [PubMed] [Google Scholar]
- Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., et al. 2009. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185:1047–1063 10.1083/jcb.200811099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H., Hay R., Gasser S., Daum G., Schneider G., Witte C., Schatz G. 1983. The outer membrane of yeast mitochondria: isolation of outside-out sealed vesicles. EMBO J. 2:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P.E., Krieg U.C., Hwang S.T., Vestweber D., Schatz G. 1990. A precursor protein partly translocated into yeast mitochondria is bound to a 70 kd mitochondrial stress protein. EMBO J. 9:4315–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M., Ren M. 2006. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580:5450–5455 10.1016/j.febslet.2006.07.022 [DOI] [PubMed] [Google Scholar]
- Schlame M., Rua D., Greenberg M.L. 2000. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39:257–288 10.1016/S0163-7827(00)00005-9 [DOI] [PubMed] [Google Scholar]
- Schlame M., Ren M., Xu Y., Greenberg M.L., Haller I. 2005. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids. 138:38–49 10.1016/j.chemphyslip.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H.E., Schönfisch B., Perschil I., Chacinska A., Guiard B., et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 100:13207–13212 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Endo T., Iijima M., Sesaki H. 2009. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185:1029–1045 10.1083/jcb.200812018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T. 2009. Protein quality control in mitochondria. J. Biochem. 146:455–461 10.1093/jb/mvp122 [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Langer T. 2009. AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 160:711–717 10.1016/j.resmic.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Thorburn D.R. 2004. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 27:349–362 10.1023/B:BOLI.0000031098.41409.55 [DOI] [PubMed] [Google Scholar]
- Thorburn D.R., Sugiana C., Salemi R., Kirby D.M., Worgan L., Ohtake A., Ryan M.T. 2004. Biochemical and molecular diagnosis of mitochondrial respiratory chain disorders. Biochim. Biophys. Acta. 1659:121–128 10.1016/j.bbabio.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Valianpour F., Mitsakos V., Schlemmer D., Towbin J.A., Taylor J.M., Ekert P.G., Thorburn D.R., Munnich A., Wanders R.J., Barth P.G., Vaz F.M. 2005. Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J. Lipid Res. 46:1182–1195 10.1194/jlr.M500056-JLR200 [DOI] [PubMed] [Google Scholar]
- Vaz F.M., Houtkooper R.H., Valianpour F., Barth P.G., Wanders R.J. 2003. Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J. Biol. Chem. 278:43089–43094 10.1074/jbc.M305956200 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- Xu Y., Malhotra A., Ren M., Schlame M. 2006. The enzymatic function of tafazzin. J. Biol. Chem. 281:39217–39224 10.1074/jbc.M606100200 [DOI] [PubMed] [Google Scholar]
- Yang K., Cheng H., Gross R.W., Han X. 2009. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 81:4356–4368 10.1021/ac900241u [DOI] [PMC free article] [PubMed] [Google Scholar]