Arrays of Sir3 binding sites at telomeres are sufficient to promote long-range trans-telomere interactions.
Abstract
A general feature of the nucleus is the organization of repetitive deoxyribonucleic acid sequences in clusters concentrating silencing factors. In budding yeast, we investigated how telomeres cluster in perinuclear foci associated with the silencing complex Sir2–Sir3–Sir4 and found that Sir3 is limiting for telomere clustering. Sir3 overexpression triggers the grouping of telomeric foci into larger foci that relocalize to the nuclear interior and correlate with more stable silencing in subtelomeric regions. Furthermore, we show that Sir3′s ability to mediate telomere clustering can be separated from its role in silencing. Indeed, nonacetylable Sir3, which is unable to spread into subtelomeric regions, can mediate telomere clustering independently of Sir2–Sir4 as long as it is targeted to telomeres by the Rap1 protein. Thus, arrays of Sir3 binding sites at telomeres appeared as the sole requirement to promote trans-interactions between telomeres. We propose that similar mechanisms involving proteins able to oligomerize account for long-range interactions that impact genomic functions in many organisms.
Introduction
The spatial and temporal behavior of genomes and their regulatory proteins has emerged as an important, yet still poorly understood, control mechanism in genomic functions (Akhtar and Gasser, 2007; Heard and Bickmore, 2007; Misteli, 2007; Zhao et al., 2009). One key feature of nuclear organization is the existence of subcompartments in which specific DNA sequences and proteins associate, thereby creating microenvironments that can favor or impede particular enzymatic activities. A well-characterized example of these microenvironments results from the clustering of certain genes or repetitive DNAs (de Laat, 2007), such as telomeric repeats (in budding yeast) or centromeric heterochromatin (in fission yeast, Drosophila melanogaster, and mammals). These repetitive sequences generally nucleate patterns of histone modifications that are recognized by histone-binding repressors, and their clustering results in the sequestration of these general repressors into subcompartments. This phenomenon, conserved from yeast to man, in addition to its role in concentrating silencing factors, has a dominant impact on chromosome folding and positioning.
Telomeric foci of budding yeast represent one of the best-studied examples of subnuclear compartments. Indeed, the 32 telomeres of haploid cells cluster into three to eight foci that sequester silent information regulators (SIRs; Sir factors), such as the clustering of HP1-associated centromeric repeats, which concentrates silencing factors in metazoans (Guenatri et al., 2004). Intriguingly, a similar spatial juxtaposition of telomeres can be observed in the parasite Plasmodium falciparum, in which the clustering appears to favor the monoallelic expression of subtelomeric virulence factor loci, which is essential for the parasite to escape the immune system response (Scherf et al., 2008). In yeast, sequestration of SIRs into telomeric foci both favors subtelomeric repression and prevents promiscuous effects on a distinct subset of promoters (Taddei et al., 2009).
At the molecular level, budding yeast telomeres consist of 250–300 bp of irregular tandem repeats with the consensus sequence TG1–3 (Shampay et al., 1984). Bound to these sequences is the repressor activator protein 1 (Rap1; Shore and Nasmyth, 1987), in which the C terminus is a binding site for Sir3 and Sir4 (Moretti et al., 1994; Jeppesen, 1997; Wotton and Shore, 1997). The yKu70/yKu80 heterodimer, which binds the very end of telomeres, also recruits Sir4 (Tsukamoto et al., 1997; Bertuch and Lundblad, 2003; Roy et al., 2004), and both Sir4 and yKu70/yKu80 contribute to telomere anchoring to the nuclear periphery (Taddei et al., 2004; Bupp et al., 2007; Schober et al., 2008). Sir3 and Sir4 form a stoichometric complex with the NAD+-dependent histone deacetylase Sir2 that deacetylates H3 and H4 histone tails from neighboring nucleosomes, generating histone binding sites for Sir3 and Sir4. This leads to the spreading of the Sir2–3–4 stoichometric complex from sites of nucleation over a 2–3-kb subtelomeric domain (Rusche et al., 2003; Moazed et al., 2004) and to the transcriptional repression of subtelomeric regions (Aparicio et al., 1991; Johnson et al., 2009; Martino et al., 2009).
The SIR complex is also found at the cryptic mating-type loci HML and HMR, where it represses both endonucleolytic cleavage and transcription (Haber, 1998; Rusche et al., 2003). In addition, Sir2 is also enriched in the nucleolus (Gotta et al., 1997), where it protects ribosomal DNA from recombination and silences ectopically inserted RNA–polymerase II genes (Gottlieb and Esposito, 1989). Importantly, cellular amounts of Sir proteins, particularly Sir3, are limiting for the spread of silent chromatin from nucleation sites (Renauld et al., 1993; Hecht et al., 1996), and loci associated with Sir proteins compete for these limiting pools (Buck and Shore, 1995; Smith et al., 1998; Cockell et al., 2000; Michel et al., 2005).
The mechanism and the proteins that mediate telomere clustering remained elusive. Interactions between subtelomeres have been proposed to be governed only by some physical constraints, including chromosome arm length, centromere attachment to the spindle pole body, and nuclear crowding (Therizols et al., 2010). On the other hand, interaction between HM loci depends on correctly assembled heterochromatin at these loci (Miele et al., 2009). Although mutations in YKU70/YKU80 or SIR3–4 do affect clustering (Gotta et al., 1996; Laroche et al., 1998), these loss-of-function experiments are difficult to interpret because removing any of these proteins from telomeres impacts the recruitment of the others (Hoppe et al., 2002; Luo et al., 2002). However, components of the SIR complex are strong candidates for promoting trans-interactions between telomeres because they all have the ability to interact with each other and among themselves (Rusche et al., 2003; Norris and Boeke, 2010).
To decipher the mechanism underlying the clustering of telomeres, we have investigated the individual contribution of Sir2, Sir3, and Sir4 by monitoring the effect of their overexpression. We show that overexpressing Sir3 leads specifically to the hyperclustering of telomeres and silencing factors in foci mainly localized away from the nuclear periphery. By modulating Sir3 expression, we further show that the cellular amount of Sir3 is a determinant of telomere organization. In addition, we found that nonacetylable Sir3, which is deficient for silencing, is yet efficient for telomere clustering. Moreover, we show that Rap1-mediated recruitment of nonacetylable Sir3 to telomeres can promote telomere clustering in the absence of Sir2 and Sir4. These data lead us to propose a model in which arrays of binding sites for Sir3 are sufficient to promote trans-interactions between telomeres independently of silencing or anchoring to the nuclear periphery.
Results
Sir3 overexpression leads to the grouping of Rap1 foci
To test the individual contribution of the Sir2, Sir3, and Sir4 protein to telomere clustering, we overexpressed each individually by replacing the endogenous promoters of the SIR genes. We constructed a set of strains in which the strong inducible promoter of GAL1 (GAL1p) replaces the endogenous promoters of the SIR genes. Importantly, although overexpression of Sir3 from a multicopy plasmid was reported to be toxic (Holmes et al., 1997), the overexpression from the unique genomic copy of SIR2, SIR3, or SIR4 is not toxic (Fig. 1 A). Upon inducing conditions, these strains are competent for silencing at the cryptic mating-type loci (Fig. S1 A) and at telomere VIIL except for the strain overexpressing Sir4 (Fig. 1 A) as previously reported (Marshall et al., 1987; Cockell et al., 1995).
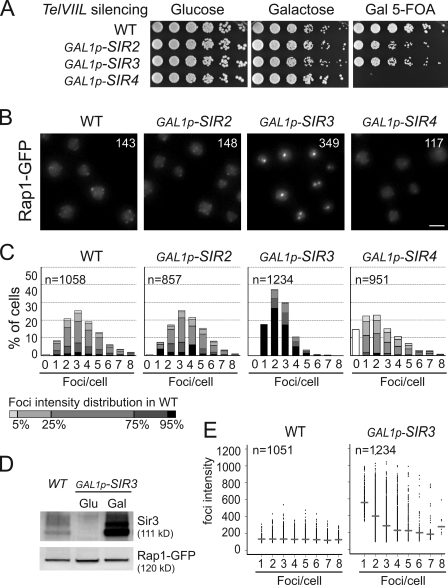
Figure 1.
Sir3 overexpression specifically leads to Rap1-GFP foci clustering. (A) Growth assay and telomeric silencing assay at telVIIL::URA3. To assess the toxicity of SIR overexpression, wild-type (WT; yAT232), GAL1p-SIR2 (yAT200), GAL1p-SIR3 (yAT208), and GAL1p-SIR4 (yAT202) cells were grown in glucose, and fivefold serial dilutions were plated either onto YPD or YPGal plates. To monitor telomeric silencing at telVIIL, strains were grown in galactose synthetic medium for 48 h and then plated onto 5-FOA galactose plates. Decreased growth on YPGal plates indicates SIR overexpression toxicity, and decreased growth on 5-FOA plates reflects a disruption of telomeric silencing. (B) Rap1 foci grouping upon Sir3 overexpression. Representative fluorescent images of the telomere-associated protein Rap1 tagged with GFP of the strains used in A. Cells were grown in galactose overnight, diluted to OD600nm = 0.2, and imaged at OD600nm = 1. Numbers represent the mean intensity of foci. Bar, 2 µm. (C) Quantification of images from B using an application developed in house (Q-Foci; see Materials and methods). Gray levels are set to represent the distribution of foci intensity in wild-type cells. (D) Levels of Rap1 are stable upon Sir3 overexpression. Immunoblots with anti-GFP and anti-Sir3 on crude extracts from wild type (yAT7; first lane), GAL1p-SIR3 (yAT370) in repressive conditions (second lane), and after 3 h of Sir3 induction (third lane). (E) Graphical representation of the foci intensity as a function of the number of foci per cells (data from C) in a wild type and in a strain overexpressing Sir3. Horizontal bars represent the median for each category of cells.
We studied telomere foci organization in those strains by following the telomere-bound protein Rap1 fused to GFP in living cells (Hayashi et al., 1998). As previously reported for cells grown in glucose-containing medium (Gotta et al., 1996), wild-type cells grown in galactose medium show a diffuse distribution of Rap1-GFP throughout the nucleoplasm with a limited number of bright spots or foci (Fig. 1 B).
Strikingly, whereas Sir2 overexpression had almost no effect, overexpressing either Sir3 or Sir4 profoundly affects Rap1-GFP foci but in opposite ways (Fig. 1, B and C). Indeed, these foci decreased in intensity upon Sir4 overexpression, coinciding with the absence of telomeric silencing (Fig. 1 A). In contrast, in a strain overexpressing Sir3, Rap1 foci are brighter and fewer in number. Importantly, the brightness of the Rap1-GFP clusters observed in strains with high Sir3 levels is not caused by increased levels of Rap1-GFP (Fig. 1 D). Furthermore, Sir3 overexpression does not have a major effect on the overall nuclear organization, as the nuclear diameter, the nucleolus, and centromere localization appeared normal under these conditions (Fig. S1, B–D). Thus, Sir3 overexpression appears to affect specifically the distribution of telomeres.
To quantify these observations, we developed a numerical method (see Materials and methods) that allowed the automatic detection of Rap1-GFP foci in interphase cells. Fig. 1 C illustrates the distribution of cells sorted according to the number of foci in different genetic contexts, with gray levels representing foci intensities. The wild-type population shows an expected distribution centered around 3.5 foci per cell (mean = 3.47 ± 0.03 foci). Importantly, foci intensities show a narrow distribution (Fig. 1 E), indicating that the variation of the number of telomeres per wild-type focus is limited. Furthermore, foci intensities are independent of the number of foci per cell from cells with one to eight foci, suggesting that most of the telomeres are not detected as Rap1-GFP foci in cells with few visible foci. Indeed, our simulation suggests that single telomeres or pairs of telomeres are hidden by the diffuse part of Rap1-GFP fluorescence and, thus, are not detectable as Rap1-GFP foci in this assay (Fig. S1 E).
In contrast to the wild-type situation, cells overexpressing Sir3 show fewer foci (mean = 2.48 ± 0.03 foci per cell) with a higher mean intensity (143 in wild-type cells vs. 349 in the GAL1p-SIR3 strain; Kolmogorov–Smirnov test adapted for continuous set of values, P < 2.2E-16; Fig. 1, B and C). Moreover and contrary to the wild-type situation, when the number of foci per cell increases, the intensity of the foci decreases (Fig. 1 E), suggesting that bright foci correspond to the grouping of smaller foci. Indeed, a time course experiment following Rap1 distribution upon Sir3 induction revealed that Rap1 foci appeared as small foci resembling wild-type cell foci at 45 min after induction (Fig. S2). These foci then diminished in number and increased in intensity to reach a maximum level of clustering at 8 h. Moreover, time-lapse acquisitions performed every 4 min over 9 h allowed us to observe some fusion events, although the time resolution was probably not sufficient to observe all these events (Video 1). Importantly, although the decrease in foci number is statistically significant in our quantitative experiments, it is probably underestimated because grouping telomeres simultaneously increases the number of detected telomeres and decreases the number of total clusters.
Finally, Sir3 overexpression induces this hyperclustering even in the presence of high levels of Sir2 and/or Sir4 as shown by co-overexpressing Sir3 with Sir2 and/or Sir4 (Fig. S3 A). Thus, high levels of the other Sir proteins cannot counteract the hyperclustering caused by high Sir3 levels. In conclusion, we propose that Sir3 overexpression specifically induces the hyperclustering of Rap1 foci.
Sir3 overexpression induces telomere hyperclustering in foci containing Sir2, Sir3, and Sir4
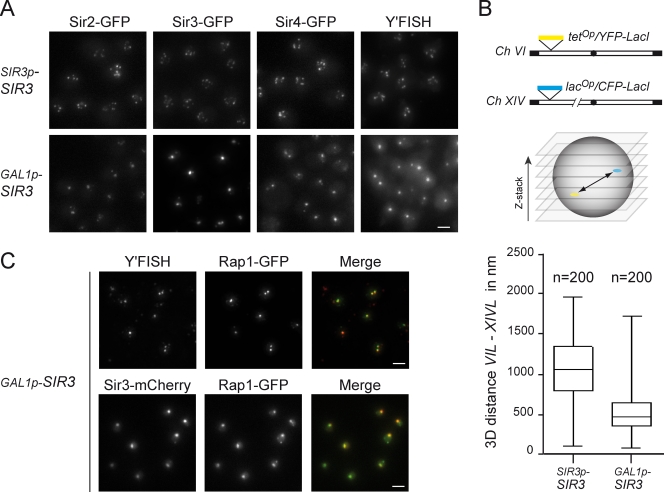
To test whether the grouping of Rap1 foci observed upon Sir3 overexpression coincides with the hyperclustering of silent chromatin, we studied the localization of Sir2, Sir3, Sir4, and telomeres in strains with either endogenous or high levels of Sir3. As expected, in wild-type cells grown in galactose, GFP-tagged Sir2, Sir3, and Sir4 are found in several foci, whereas upon Sir3 overexpression, most of the cells show one bright nuclear dot (Fig. 2 A). Thus, the overexpression of Sir3 leads to the grouping of Rap1-, Sir2-, Sir3-, and Sir4-containing foci. To rule out the possibility that the hyperclustering of these proteins was independent of the telomeres themselves, we evaluated the status of the telomeric chromatin by FISH experiments. In situ hybridizations were performed with a probe derived from the Y′ telomere–associated DNA sequences (Gotta et al., 1996). This analysis revealed a staining pattern very similar to the one observed with Rap1-GFP, Sir2-GFP, Sir3-GFP, and Sir4-GFP in a strain expressing endogenous levels of Sir3. Strikingly, when Sir3 was overexpressed, only one large bright focus was observed in most of the nuclei, indicating that the majority of the Y′-bearing telomeres localize in the same cluster (Fig. 2 A, right). In addition, we measured in vivo the distance between telomeres VIL and XIVL tagged with the tetracycline operator/tetracycline repressor and lactose operator/lactose repressor systems (Belmont, 2001), respectively. We found that these two telomeres were closer in cells overexpressing Sir3 than in wild-type cells, with a median distance decreasing from 1 µm in wild-type cells (as previously reported by Bystricky et al., 2005) to 550 nm in cells overexpressing Sir3 (Fig. 2 B). The distribution of these distances in a cell population indicated that telomeres remain dynamic and are probably associated only transiently, which is consistent with the dynamics of Rap1-GFP foci (Videos 1 and 2). Importantly, Rap1-GFP foci coincided with Y′ clusters and Sir3 foci in strains overexpressing Sir3, as shown by immuno-FISH and in vivo imaging, respectively (Fig. 2 C). Thus, Sir3 overexpression leads to telomere hyperclustering in foci containing Rap1 and the Sir2–3–4 complex.
Figure 2.
Sir3 overexpression leads to hyperclustering of telomeres and is associated with high levels of Sir2, Sir3, and Sir4. (A) Fluorescent images of strains expressing endogenous levels of Sir3 (top) or high levels of Sir3 (bottom). SIR2-GFP (yAT405), GAL1p-SIR3 SIR2-GFP (yAT718), SIR3-GFP (yAT779), GAL1p-SIR3 SIR3-GFP (yAT780), SIR4-GFP (yAT431), and GAL1p-SIR3 SIR4-GFP (yAT720) were grown in galactose synthetic medium before imaging. Immuno-FISH was performed with a Y′-repeat telomeric probe on wild-type (yAT126) and GAL1p-SIR3 (yAT960) strains grown in YPGal. (B) 3D position of telomeres VIL and XIVL relative to each other in living cells expressing endogenous levels of Sir3 (wild type, yAT56) and in strains overexpressing Sir3 (GAL1p-SIR3, yAT690). YFP–tetracycline repressor and CFP–lactose repressor fusions allowed the visualization of tetop and lacop arrays inserted at telomeres VIL and XIVL, respectively, as previously described (Bystricky et al., 2005). Cells were grown in galactose before imaging. Shown on the bottom are box plots for distances between telomeres VIL and XIVL. The line in the middle of the box represents the median of the values; the bottom and the top of the box are the 25th and 75th percentiles. The whiskers indicate the minimum and maximum data values. (C) Colocalization of telomeres with Rap1-GFP foci (top): GAL1p-SIR3 RAP1-GFP (yAT208) cells were grown in YPGal for immuno-FISH experiments. Colocalization of Sir3-mCherry foci with Rap1-GFP foci (bottom): GAL1p-SIR3-mCherry RAP1-GFP (yAT330) cells were grown in galactose synthetic medium for live-cell imaging. Bars, 2 µm.
The cellular amount of Sir3 is a determinant of telomere organization
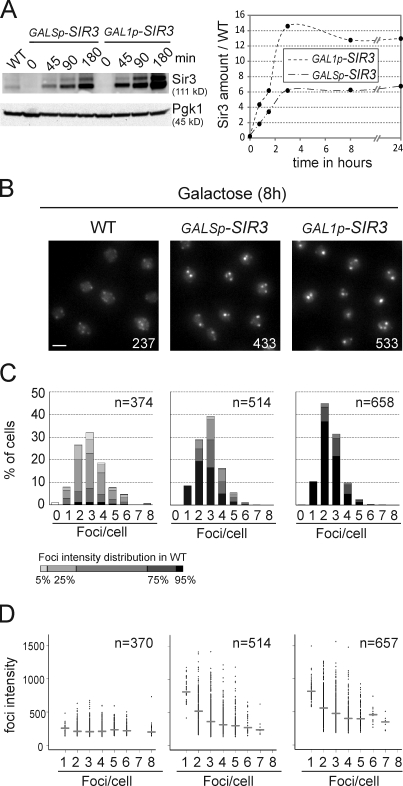
Next, we investigated the effect of inducing SIR3 at different levels by the strong GAL1 promoter or its weaker derivative the GALS promoter (Mumberg et al., 1994). Quantification by Western blot analysis showed that GAL1p and GALSp lead, respectively, to Sir3 amounts 15-fold and 6-fold higher than endogenous levels (Fig. 3 A). The sixfold increase of Sir3 amount obtained when GALSp drove SIR3 expression leads to some grouping of Rap1 foci, which are more intense and lower in number than those observed in wild-type cells (Fig. 3, B–D). However, the hyperclustering was more pronounced when Sir3 was overexpressed using the GAL1 promoter, indicating that the degree of telomere clustering is a reflection of the cellular amount of Sir3.
Figure 3.
Telomere hypercluster formation and maintenance depends on Sir3 levels. (A) Expression levels of Sir3 upon induction. Immunoblots with anti-Sir3 and anti-Pgk1 (loading control) on crude extracts from wild type (WT; yAT7), GALSp-SIR3 (yAT369), and GAL1p-SIR3 (yAT370). Graphical representation of the quantification of Sir3 levels during the time course in the GALSp-SIR3 and GAL1p-SIR3 strains. Sir3 amounts are normalized to endogenous levels. (B) Fluorescent images of wild-type (yAT7), GALSp-SIR3 (yAT369), and GAL1p-SIR3 (yAT370) cells after 8 h in galactose medium. Numbers correspond to the mean intensity of foci. Bar, 2 µm. (C) Quantification of images from B using Q-foci. (D) Graphical representation of the foci intensity as a function of the number of foci per cells. Horizontal bars represent the medians for each category of cells.
The behavior of the bright Rap1-GFP foci was tracked after Sir3 shutoff and revealed that only 31% of the cells still showed a bright focus after 6 h of repression and that the disappearance of the Rap1-GFP foci correlated with the dilution of Sir3 upon cell divisions (Fig. S3 B and Videos 2 and 3). Therefore, Sir3 levels correlate with the degree of telomere clustering, and high levels of Sir3 are necessary to maintain telomere hyperclustering.
Telomere hyperclusters are internal and correlate with more stable silencing
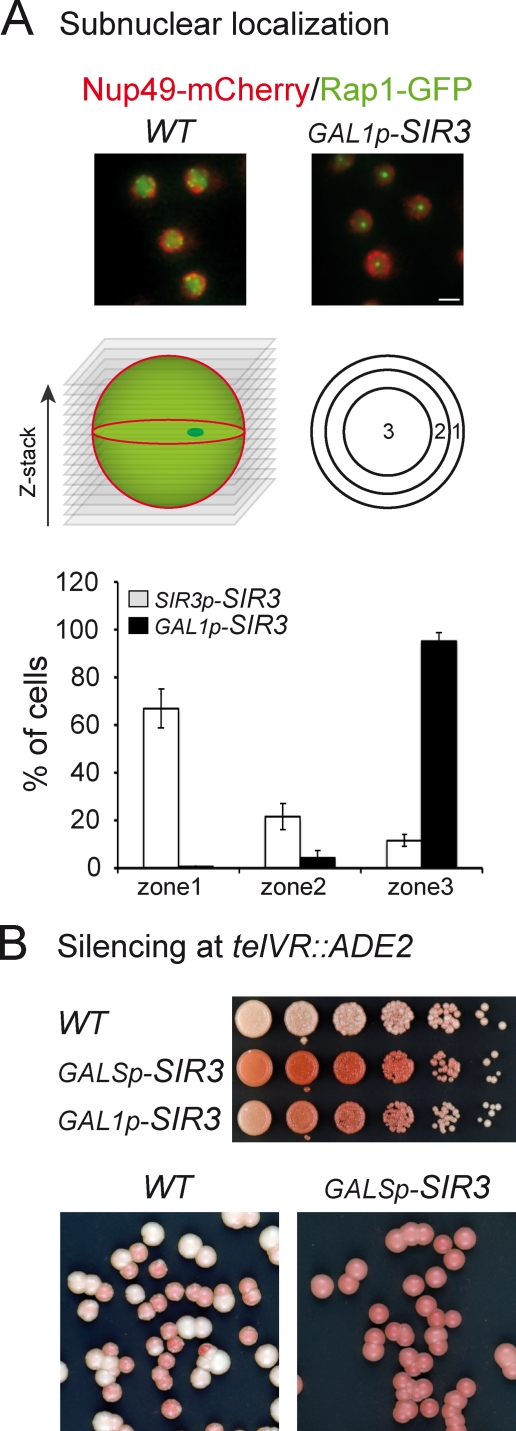
Because Sir3 overexpression modified the grouping of telomeres, we considered the possibility that the subnuclear position of telomeres was altered under these conditions. We thus monitored the position of the brightest telomere cluster relative to the nuclear envelope as previously described (Hediger et al., 2004) in cells overexpressing Sir3 or in wild-type cells (Fig. 4 A). Strikingly, although the brightest Rap1-GFP focus was mainly found adjacent to the nuclear envelope in wild-type cells (75% in zone 1), the telomere hypercluster was found in the innermost zone in most of the cells overexpressing Sir3 (>90% in zone 3; Fig. 4 A). Thus, Sir3 overexpression leads to the relocalization of telomeres from the nuclear envelope to the nuclear interior.
Figure 4.
The telomere hypercluster is internal and correlates with more stable silencing. (A) Rap1-GFP hypercluster localization relative to the nuclear pore. Two-color z-stack images were acquired on strains expressing Rap1-GFP, Nup49-mCherry, and either endogenous levels of Sir3 or high levels of Sir3 (yAT222 and yAT223 transformed with the NUP49-mCherry plasmid). The localization of the brightest Rap1-GFP spot in one of the three equal concentric zones was scored on the corresponding focal plane. This experiment was repeated twice: for experiment 1, nyAT222 = 69 and nyAT223 = 98, and for experiment 2, nyAT222 = 77 and nyAT223 = 173 (n is the number of nuclei analyzed). Error bars represent means ± SEM. Bar, 2 µm. (B) Telomeric silencing at the telVR::ADE2 (YPH499 background) in wild-type (WT; yAT7), GALSp-SIR3 (yAT369), and GAL1p-SIR3 (yAT370) strains. Cells were grown in YPGal medium and plated onto YPGal plates. The color of the colonies is indicative of the state of silencing of the ADE2 reporter gene at telVR: the ADE2 gene is expressed in white colonies and repressed in pink colonies. Fivefold dilution assay (top). Single-colony plating (bottom).
We then addressed the functional consequence of overexpressing Sir3 on the telomeric position effect. Using either the GAL1p or GALSp promoter to induce Sir3, we monitored silencing at the ADE2 reporter gene inserted at telomere VR by performing a colony color assay (Gottschling et al., 1990). Under inducing conditions, both inducible strains showed stronger ADE2 silencing than the wild-type strain (Fig. 4 B). We noticed that colonies from a wild-type strain showed pink and white sectors reflecting the variegated expression of the reporter gene (Gottschling et al., 1990), whereas strains overexpressing Sir3 through either GAL1p or GALSp showed uniformly pink colonies indicative of a more stable silencing (Fig. 4 B, bottom). This is consistent with a previous study showing that Sir3 overexpression increases the silencing of native telomeres up to 9 kb from the TG repeats at telomere XVR (Pryde and Louis, 1999). Thus, despite the internal localization of the telomere hypercluster, the transcriptional repression that characterizes usually peripheral subtelomeric sequences (Gottschling et al., 1990; Aparicio et al., 1991; Ottaviani et al., 2008) is not impaired upon Sir3 overexpression. This demonstrates that, as previously shown for silencing at HM loci (Gartenberg et al., 2004), efficient and stable silencing of telomeres can also be achieved internally when Sir3 is not limiting.
Separation-of-function mutants uncouple Sir3 silencing function from clustering
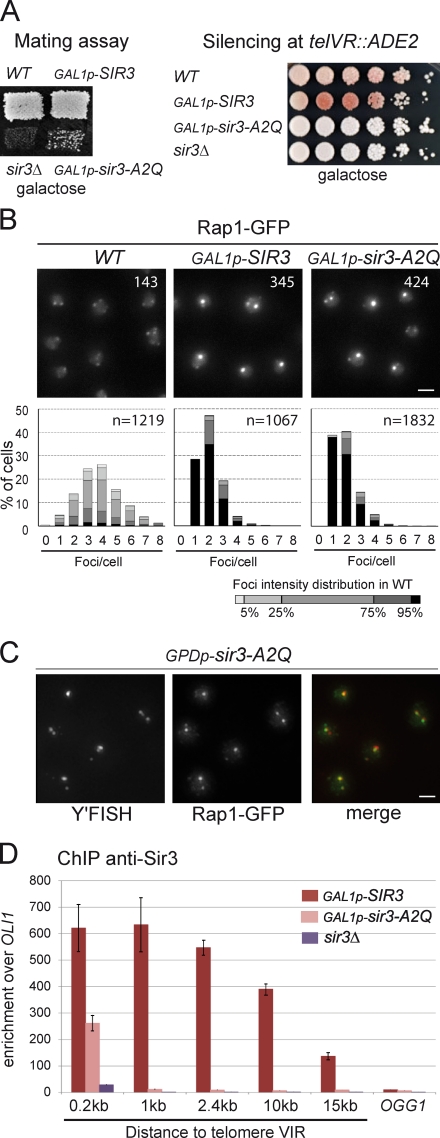
Because Sir3 overexpression increases both the clustering and the stability of telomeric silencing, we wondered whether the formation of a more stable heterochromatin structure could be the cause of the hyperclustering. To test this hypothesis, we assessed the ability of silencing-defective mutants to promote telomere clustering, and interestingly, we identified several alleles of SIR3 that are efficient for telomere clustering but not for telomere silencing. All of them were modified in their N terminus (unpublished data). As Sir3 is acetylated on Ala2 by the Nat1–Ard1 complex and this modification is essential for its function in silencing at telomeres (Geissenhöner et al., 2004; Wang et al., 2004), we tested whether the A2Q substitution could recapitulate this phenotype. As shown in Fig. 5 A, a strain overexpressing Sir3-A2Q had a severe defect for silencing at HML and was completely deficient for silencing at telomeres. However, this mutant protein was very efficient to promote telomere hyperclustering, as shown by following Rap1-GFP foci in vivo (Fig. 5 B) and telomeres by immuno-FISH (Figs. 5 C and S4 A). Importantly, when expressed on a centromeric plasmid under the control of the SIR3 promoter, Sir3-A2Q was also able to promote telomere clustering in the absence of wild-type Sir3 (Fig. S4 B). These results demonstrate that Sir3 has two distinct functions that can be separated: a function in telomere silencing and a function in telomere clustering.
Figure 5.
A silencing-defective allele of SIR3 is functional for telomere clustering. (A) Mating assays with wild type (WT; yAT7), GAL1p-SIR3 (yAT370), sir3Δ (yAT1196), and GAL1p-Sir3-A2Q (yAT1197). Cells were grown in YPGal plates and replica plated on a lawn of α-mating testers. Telomeric silencing at telVR::ADE2 in wild-type (yAT7), GAL1p-SIR3 (yAT370), GAL1p-Sir3-A2Q (yAT1197), and sir3Δ (yAT1196) strains. Cells were grown in YPGal liquid medium, and dilutions were plated onto YPGal plates. (B) Sir3-A2Q mediates telomere clustering. Fluorescent images of Rap1-GFP in wild-type (yAT7), GAL1p-SIR3 (yAT370), and GAL1p-Sir3-A2Q (yAT1197) cells. Cells were grown in synthetic complete galactose medium overnight, diluted to OD600nm = 0.2, and imaged at OD600nm = 1. Numbers correspond to the mean intensity of foci. Quantification of the images was performed using Q-foci. (C) Fluorescent images of an immuno-FISH experiment performed with a Y’-repeat telomeric probe and an anti-GFP on a strain expressing high levels of Sir3-A2Q (yAT1256). (D) ChIP analysis was carried out using an anti-Sir3 to study the spreading of Sir3 on telomere VIR: GAL1p-SIR3 (yAT208), GAL1p-Sir3-A2Q (yAT1205), and sir3Δ (yAT360) strains were grown in YPGal for 48 h. The bar graph represents the enrichment over the mitochondrial locus OLI1 for amplicons at different distances from the TG repeats as indicated. Enrichment at the internal chromosomal locus OGG1 is shown for comparison. The experiment was repeated three times. Error bars represent means ± SEM. Bars, 2 µm.
The second residue of Sir3 has been shown to be important for binding to nucleosomes (Sampath et al., 2009). We thus tested the ability of Sir3-A2Q to spread in subtelomeric regions when overexpressed by performing chromatin immunoprecipitation (ChIP) against Sir3. As was previously described (Hecht et al., 1996; Strahl-Bolsinger et al., 1997; Katan-Khaykovich and Struhl, 2005), overexpressed Sir3 spreads over 15 kb away from telomere VIR. In contrast, overexpressed Sir3-A2Q showed a twofold decrease in recruitment 200 bp away from the TG repeats and background levels of recruitment in subtelomeric regions (Fig. 5 D), which was consistent with its telomeric silencing defect (Fig. 5 A). Thus, Sir3-A2Q is found only at the very end of telomeres, where it promotes telomere clustering without detectable spreading in the subtelomeric regions, demonstrating that stable binding of the SIR complex in subtelomeric regions is not necessary for telomere clustering. In conclusion, Sir3’s ability to mediate telomere clustering can be separated from its role in silencing and spreading into subtelomeric regions.
The nonacetylable Sir3 promotes telomere clustering independently of Sir2 and Sir4 but requires Rap1 C terminus
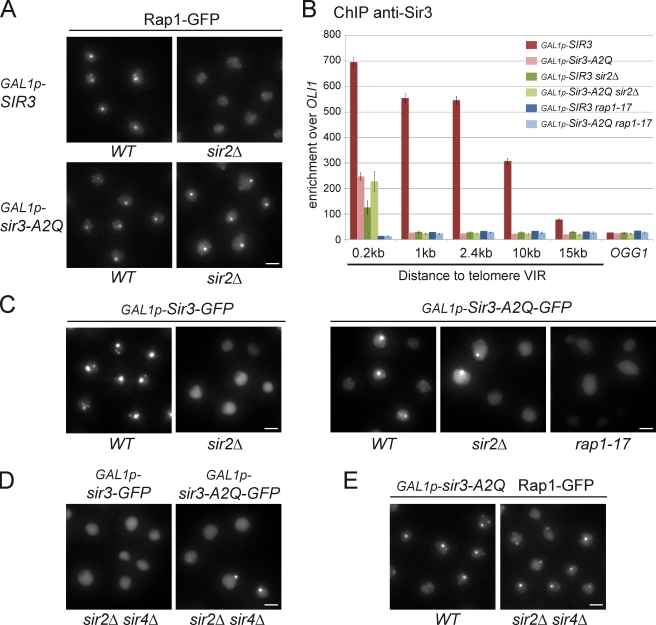
Having shown that nonacetylable Sir3 can promote telomere clustering independently of stable spreading in subtelomeric regions, we tested whether this activity requires an intact SIR complex. Intriguingly, overexpression of Sir3-A2Q, but not wild-type Sir3, leads to the grouping of Rap1-GFP foci in the absence of Sir2 (Fig. 6 A). Western blot analysis showed that this difference was not caused by a difference of the cellular amount of Sir3 versus Sir3-A2Q in a sir2 mutant (Fig. S4 C). An alternative explanation could be that the unacetylable Sir3 is recruited better to telomeres than wild-type Sir3 in the absence of Sir2. To test this model, we monitored the recruitment of Sir3 or Sir3-A2Q at telomere VIR by ChIP in a sir2Δ strain. As expected, Sir3 recruitment was severely decreased 200 bp away from the TG repeats (5.5-fold) and did not spread in subtelomeric regions in the absence of Sir2 (Fig. 6 B). In contrast, Sir3-A2Q recruitment was unaffected at the very end of telomeres in the absence of Sir2. Importantly, neither Sir3-A2Q nor Sir3 was recruited at telomeres in strains bearing the rap1–17 mutation (Liu et al., 1994), which results in the truncation of the Rap1 C-terminal part thought to contain sites for Sir3p and Sir4p association (Moretti et al., 1994). Thus, Sir3-A2Q is recruited to telomeres through interaction with the C terminus of Rap1, independently of Sir2.
Figure 6.
Nonacetylable Sir3 promotes telomere clustering in the absence of Sir2 and Sir4 but requires Rap1 C terminus. (A) Fluorescent images of Rap1-GFP in GAL1p-SIR3 (yAT208), GAL1p-SIR3 sir2Δ (yAT772), GAL1p-Sir3-A2Q (yAT1205), and GAL1p-Sir3-A2Q sir2Δ (yAT1334) cells grown in synthetic complete galactose medium overnight, diluted to OD600nm = 0.2, and imaged at OD600nm = 1. (B) ChIP analysis with an anti-Sir3 to study the spreading of Sir3 on telomere VIR: GAL1p-SIR3 (yAT208), GAL1p-Sir3-A2Q (yAT1205), GAL1p-SIR3 sir2Δ (yAT772), GAL1p-Sir3-A2Q sir2Δ (yAT1334), GAL1p-SIR3-GFP rap1–17 (yAT1357), and GAL1p-Sir3-A2Q-GFP rap1–17 (yAT1358) strains were grown in YPGal for 48 h. The bar graph represents the enrichment over the mitochondrial locus OLI1 for amplicons at different distances from the TG repeats as indicated. Enrichment at the internal chromosomal locus OGG1 is shown for comparison. The experiment was repeated three times. Error bars represent means ± SEM. (C) Fluorescent images of the overexpressed Sir3 or Sir3-A2Q proteins tagged with GFP in their C terminus: the GAL1p-SIR3-GFP (yAT780), GAL1p-SIR3-GFP sir2Δ (yAT782), GAL1p-Sir3-A2Q-GFP (yAT1337), GAL1p-Sir3-A2Q-GFP sir2Δ (yAT1338), and GAL1p-Sir3-A2Q-GFP rap1–17 (yAT1358) strains were grown as in A. (D) Fluorescent images of the GAL1p-SIR3-GFP sir2Δ sir4Δ (yAT1342) and GAL1p-Sir3-A2Q-GFP sir2Δ sir4Δ (yAT1340) strains grown as in A. (E) Fluorescent images of Rap1-GFP in GAL1p-Sir3-A2Q (yAT1205) and in GAL1p-Sir3-A2Q sir2Δ sir4Δ (yAT1336) cells grown as in A. WT, wild type. Bars, 2 µm.
To determine the nuclear distribution of Sir3-A2Q, we introduced a GFP tag in the C terminus of this mutant and found that, similar to Sir3-GFP, Sir3-A2Q-GFP formed bright foci when overexpressed (Fig. 6 C). However, contrary to Sir3-GFP, Sir3-A2Q-GFP formed bright foci when overexpressed in the absence of Sir2 (Fig. 6 C), which is consistent with the formation of Rap1-GFP hyperclusters independent of Sir2 upon Sir3-A2Q overexpression (Fig. 6 A). Importantly, Sir3-A2Q-GFP, as Sir3-GFP, did not form any detectable foci in rap1–17 strains (Fig. 6 C). Thus, Sir3-A2Q formed foci only when recruited to telomeres via its interaction with the C terminus of Rap1, ruling out the possibility that this protein forms aggregates when overexpressed. Furthermore, both overexpressed Sir3-A2Q-GFP and Rap1-GFP upon Sir3-A2Q overexpression formed bright foci in a sir2Δ sir4Δ strain, showing that Sir2 and Sir4 are not required for nonacetylable Sir3-promoted telomere clustering (Fig. 6, D and E). Together, these data strongly suggest that recruiting Sir3 to telomeres is the only requirement to promote trans-interactions between telomeres.
Discussion
Sir3 is a determinant of telomere clustering
Sir3 was previously shown to be limiting for silencing adjacent to telomeres (Renauld et al., 1993). When overexpressed, Sir3 extends silenced regions by spreading over 15 kb in subtelomeric regions (Hecht et al., 1996; Strahl-Bolsinger et al., 1997; Katan-Khaykovich and Struhl, 2005). Here, we show, in two different genetic backgrounds (W303 and YPH499), that Sir3 is also limiting for telomere clustering. Indeed, overexpressing Sir3 with the strong GAL1 promoter leads to a 15-fold increase in Sir3 levels and to the hyperclustering of telomeres. This hyperclustering corresponds to the grouping of wild-type telomere foci into one or two hyperclusters per cell as shown by DNA FISH and localization of telomere-associated proteins. Mild overexpression of Sir3 (sixfold above the endogenous level) through an attenuated version of the GAL1 promoter leads to an intermediate effect. Thus, the cellular amount of Sir3 is a determinant of the extent of telomere clustering.
Silencing occurs away from the nuclear periphery when Sir3 is overexpressed
In wild-type cells, telomeric foci are mainly found at the nuclear periphery (Gotta et al., 1996), where telomeres and silent chromatin are tethered through redundant pathways (Andrulis et al., 2002; Taddei et al., 2004; Bupp et al., 2007; Schober et al., 2009). Unexpectedly, telomere hyperclusters observed upon Sir3 overexpression are internally located, suggesting that an excess of Sir3 counteracts telomere-anchoring pathways. One of these pathways involves Sir4 through its binding with both the inner nuclear envelope–associated protein Esc1 (Andrulis et al., 2002; Taddei et al., 2004) and the transmembrane protein Mps3 (Bupp et al., 2007). It is possible that Sir3 competes with Mps3 and Esc1 for the binding of Sir4 because the Sir4 domains reported to interact with Esc1, Mps3, and Sir3 are all located in the C-terminal half of Sir4 (Moazed and Johnson, 1996; Andrulis et al., 2002; Bupp et al., 2007). Although the mechanism leading to the internal localization of these hyperclusters remains to be elucidated, this observation shows that telomere clustering can occur away from the nuclear periphery. Consistent with this, none of the proteins involved in telomere anchoring (Yku70, Sir4, Esc1, or Mps3) are essential for telomere clustering (Figs. 6 and S4, D–F).
Telomere clustering has been shown to promote and restrict silencing to specific regions by concentrating silencing factors (Maillet et al., 1996; Marcand et al., 1996; Andrulis et al., 1998; Taddei et al., 2009). Consistent with this notion, we show that the hyperclustering of telomeres in the nuclear interior leads to a strong enrichment of silencing factors in this subnuclear region and correlates with a more stable silencing at telomeres. Thus, stable silencing at telomeres occurs away from the nuclear envelope, possibly thanks to the hyperclustering of telomeres allowing the internal concentration of silencing factors.
Silencing and clustering functions of Sir3 can be separated
Sir3 has been the focus of numerous studies addressing its possible mode of action in transcriptional silencing (Norris and Boeke, 2010). However, the possible role of Sir3 in promoting trans-interaction between telomeres has not been specifically addressed in vivo. In this study, we demonstrate that Sir3 promotes telomere clustering and that this function is independent of its activity in silencing, as illustrated by the Sir3-A2Q substitution. This substitution impairs the N-terminal acetylation of Sir3 on Ala2 by the Nat1–Ard1 complex, which is essential for Sir3 function in telomeric and HML silencing (Geissenhöner et al., 2004; Wang et al., 2004). Here, we show that, although Sir3-A2Q is unable to spread in subtelomeric regions, it is efficient for telomere clustering when expressed at endogenous levels and leads to hyperclustering when overexpressed. Sir3 thus appears to have a dual function in silencing and clustering, which could be mediated by distinct domains.
Interestingly, the N and C termini of Sir3p have been shown to perform different and independent functions within the silencing complex, and expression of the two halves of Sir3 in trans partially complements a SIR3 deletion for silencing at HML (Gotta et al., 1998). On the one hand, the N-terminal part of Sir3 contains the conserved bromo-adjacent homology domain that is also found in Orc1 (Zhang et al., 2002) and shows histone tail– and nucleosome-binding activity (Onishi et al., 2007; Sampath et al., 2009). The bromo-adjacent homology domain has been proposed to play an essential role in Sir spreading and can silence HML and HMR in the absence of full-length Sir3 (Connelly et al., 2006; Buchberger et al., 2008). On the other hand, the 144–amino acid C-terminal domain of Sir3 represents the minimum domain for Sir3 homodimerization, a function that is conserved in related yeasts (Liaw and Lustig, 2006). In addition, the C-terminal domain can interact with a more internal part of Sir3 (King et al., 2006). These Sir3–Sir3 interactions could promote trans-interactions between Sir3-bound regions as suggested by in vitro studies (Georgel et al., 2001; McBryant et al., 2008; Adkins et al., 2009). Consistent with this hypothesis, overexpressing the N terminal part of Sir3 in the presence of endogenous Sir3 improves telomere silencing with almost the same efficiency as full-length Sir3 (Gotta et al., 1998). However, we observed no improvement of telomere clustering in this case (Fig. S5), demonstrating that the C-terminal part of Sir3 is necessary for telomere clustering.
Together, these data show that Sir3 function in clustering can be separated from silencing. Therefore, telomere clustering is not a consequence of silencing but can rather favor silencing by concentrating silencing factors.
Mechanism of telomere clustering
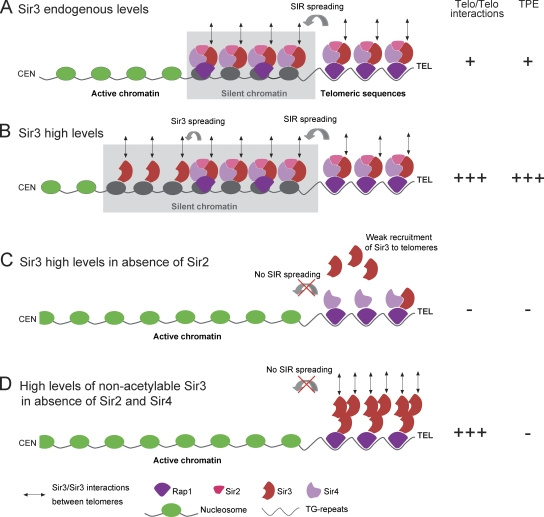
Together, these results led us to propose a model (Fig. 7) in which Sir3–Sir3 interactions (Liou et al., 2005; King et al., 2006; Liaw and Lustig, 2006; McBryant et al., 2008) promote telomere clustering in a dose-dependent manner (Fig. 7, A and B). In the absence of Sir2, wild-type Sir3 does not spread into subtelomeres and is poorly recruited to the TG repeats; as a consequence, it cannot mediate telomere clustering (Fig. 7 C). However, we showed that Sir3-A2Q, whose recruitment is not affected by Sir2 deletion, is able to mediate telomere clustering (Fig. 7 D). This gain of function of the N-terminally altered Sir3 could stem from its inability to bind to nucleosomes as indicated by its incapacity to spread into subtelomeres and consistent with previous work (Sampath et al., 2009). The lack of affinity of this Sir3 mutant for nucleosomes could favor homotypic interactions and/or its binding to Rap1 by eliminating competitive interactions. Furthermore, unacetylable Sir3 can mediate telomere clustering in the absence of Sir4 and Sir2 but requires the C terminus of Rap1, which is necessary to recruit Sir3 to telomeres (Moretti et al., 1994). Thus, recruiting Sir3 to telomeres appears to be the only requirement to promote trans-interactions between telomeres. In the future, it will be interesting to explore how the cell regulates telomere clustering in response to various stresses, knowing that the degree of Sir sequestration in telomeric foci varies in response to nutrient- and damage-induced stress, responding in part to a phosphorylation cascade that targets Sir3 (Stone and Pillus, 1996; Martin et al., 1999; Mills et al., 1999; Ai et al., 2002).
Figure 7.
Model for Sir3 as a determinant of telomere clustering. Schematic representation of yeast telomeres in different conditions (endogenous/overexpressed Sir3 and wild-type/mutant strains). (A) When Sir3 is expressed at endogenous levels, the Sir2–3–4 trimeric complex is recruited to the TG repeats by Rap1 and spread over 2–3 kb away from the telomere (TEL) ends (Rusche et al., 2003; Moazed et al., 2004). We propose that Sir3–Sir3 interactions promote trans-interaction between Sir3-bound telomeres leading to the grouping of three to five telomeres. (B) Upon Sir3 overexpression, Sir3 (but not Sir2 and Sir4) spreads further away from telomere ends (15 kb; Figs. 5 D and 6 B; Hecht et al., 1996; Strahl-Bolsinger et al., 1997; Katan-Khaykovich and Struhl, 2005), generating extended silent chromatin and long arrays of Sir3 proteins in subtelomeric regions. The additional pool of Sir3 bound to telomere ends and subtelomeric regions increases telomere–telomere (Telo/Telo) interactions leading to the formation of telomere hyperclusters. (C) In the absence of Sir2, Sir3 is not correctly recruited to the TG repeats and, thus, cannot mediate telomere–telomere interactions. (D) However, nonacetylable Sir3 is well recruited at the TG repeats independently of Sir2 and mediates telomere hyperclustering when overexpressed through Sir3–Sir3 interactions occurring only at the very end of telomeres. CEN, centromere. TPE, telomeric position effect.
In conclusion, we propose that arrays of chromatin-bound proteins with the ability to oligomerize are sufficient to promote trans-interactions between chromatin regions, which in turn favors the concentration of factors associated with these regions, such as silencing factors. Such a mechanism could account for the clustering of heterochromatin in other species given that many heterochromatin proteins involved in long-range chromatin interactions, including HP1 and Polycomb group proteins, have the ability to self-interact.
Materials and methods
Media and growth conditions
Yeast cells were grown either in rich medium (YPD [yeast extract–peptone-dextrose]) or in synthetic medium (yeast nitrogen base; MP Biomedicals) supplemented with 2% glucose, raffinose, or galactose (wt/vol) and the appropriate supplement mixture (complete or lacking a nutrient; BIO 101). Liquid synthetic media were enriched for complete synthetic medium (2× complete synthetic medium as final concentration; Gomes et al., 2007). All the strains were grown at 30°C.
For galactose induction in rich medium, cells were precultured in YPD and switched to YPGal medium (yeast extract– peptone–2% galactose [wt/vol]) for induction of the GAL1 promoter. For time course experiments, cells were precultured in synthetic medium containing 2% raffinose (wt/vol), and galactose was added to a final concentration of 2% (wt/vol) to start the induction. For telomeric silencing assays, 5-fluoroorotic acid (5-FOA; Zymo Research) plates were prepared by adding 5-FOA to a final concentration of 0.1% to supplemented synthetic medium.
Strains
The strains used in this study are listed in Tables S1 and S2. They are derivatives of W303 (Thomas and Rothstein, 1989) and YPH499 (Sikorski and Hieter, 1989) strains. Gene deletions, gene tagging, and insertions of alternative promoters were performed by PCR-based gene targeting (Longtine et al., 1998; Janke et al., 2004).
Plasmids
pAT146 is a centromeric vector expressing Sir3 under the control of its endogenous promoter. A SalI–SalI fragment containing the SIR3 gene from pRS6.3 was inserted into pRS314 digested with SalI. pAT334 is a centromeric vector expressing Sir3-A2Q under the control of the SIR3 promoter. This plasmid was obtained by PCR-mediated mutagenesis on pAT146 using primer pair am877-CTAACAATTGGATTAGCTAAAATGCAGAAAACATTGAAAGATTTGGACGG/am883-CCGTCCAAATCTTTCAATGTTTTCTGCATTTTAGCTAATCCAATTGTTAG. The pUN100-Nup49-mCherry plasmid was provided by B. Palancade and V. Doye (L’Institut Jacques Monod, Paris, France).
Silencing assays
For mating assays, strains were patched onto YPD or YPGal plates for 18 h and then replica plated onto minimal plates covered by a lawn of mating-type testers (GA-857 MATa and GA-858 MATα; gifts from S. Gasser, Friedrich Miescher Institute, Basel, Switzerland). To assay for successful diploid formation, plates were grown for 72 h. For telomeric silencing assays, cultures were grown in liquid medium and plated in fivefold serial dilutions starting at OD600nm = 1 (corresponding to 107 cells/ml) onto appropriate plates.
Protein immunoblotting
Crude extracts were prepared by postalkaline extraction: 2 × 107 cells/ml were harvested and resuspended in 100 µl of water. Then, 100 µl of 0.2-M NaOH was added to the cell suspension. After 5 min at RT, cells were pelleted and resuspended in 100 µl of sample loading buffer. Extracts were denatured by heating at 95°C for 3 min. For immunoblotting, we used monoclonal antibodies raised against GFP at 1:1,000 (clones 7.1 and 13.1; Roche), 3-phosphoglycerate kinase (Pgk1) at 1:20,000 (clone 22C5; Invitrogen), and polyclonal antibodies against Sir3 at 1:5,000 (a gift from L. Pillus, University of California, San Diego, San Diego, CA). Loading was normalized according to Pgk1 levels, and quantification was performed after normalization using Quantity One software (Bio-Rad Laboratories).
ChIP and quantitative PCR analyses
ChIP was adapted from Borde et al. (2008). Cells were grown on a YPGal plate for 24 h, seeded in liquid YPGal at OD600nm = 0.005, and grown overnight to OD600nm = 1. Cells were cross-linked with 1% paraformaldehyde for 15 min at 30°C (Sigma-Aldrich), quenched at 30°C with 125 mM glycin for 5 min (Invitrogen), and washed twice in TBS. Pellets were resuspended in 500 µl of lysis buffer (20 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and 2.5 µl protease inhibitors [P-1860; Sigma-Aldrich]) and lysed with 0.5 mm zirconium/silica beads (Biospec Products) for three times for 30 s in the Fastprep instrument (MP Biomedicals). The chromatin was fragmented to a mean size of 500 bp by sonication in the Bioruptor sample processor (Diagenode) for 14 min at high power with 30 s on/30 s off. For Sir3 ChIP, cleared lysate was added to 50 µl of magnetic beads (Dynabeads Protein A; Invitrogen) preincubated for 4 h at 4°C with 4 µg polyclonal antibody anti-Sir3 (raised against the full-length untagged protein expressed in bacculovirus; a gift from F. Martino, Medical Research Council, Cambridge, England, UK). Precipitates were washed, and reversal cross-linking was performed by heating overnight at 65°C. Proteins were digested with proteinase K in the presence of glycogen, and the remaining DNA was purified on columns (QIAquick PCR Purification kit; QIAGEN). Finally, samples were treated with RNase.
ChIP quantification by quantitative PCR was performed on 1/20 of the immunoprecipitated DNA or 1/1,800 of the DNA from the whole-cell extract. Primers were designed with the Primer Express software (Applied Biosystems); primer sequences used in this study are the following: for OLI1, am648-GAGCAGGTATTGGTATTGCTATCG/am649-TTGATGGGTTTCTTGATACACCAT; for OGG1, am643-CAATGGTGTAGGCCCCAAAG/am644-ACGATGCCATCCATGTGAAGT; for 0.2-kb telVIR, am615-TGAGGCCATTTCCGTGTGTA/am616-CCCAGTCCTCATTTCCATCAA; for 1-kb telVIR, am617-TGATGAATTACAAGGGAACAATGAG/am618-CATCAAACAAGTAGGAATGCGAAA; for 2.4-kb telVIR, am619-TCTCCTTGTCGTCATGTGAAAGTC/am620-AGAGGAGAGTTGCTGCTTCATCA; for 10-kb telVIR, am645-ATTTCCCATTTTCTTGAAGGTTTCT/am646-GGGTTTGTAAAGGAACACCGTTT; and for 15-kb telVIR, am625-GGTCTCGCTGTCAACTGTAAACA/am626-TGCCCAAGGAATTGATGGAT. PCR reactions were conducted at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 30 s on a real-time quantitative PCR system (7900HT Fast Real-Time PCR; Applied Biosystems). Sequences of interest were amplified using the SYBR Green PCR Master Mix (Applied Biosystems). Each real-time PCR reaction was performed in triplicate. Triplicates giving cycle threshold (Ct) values differing >0.2 from two other triplicates were eliminated. Each experiment was conducted at least three times.
A dilution series of genomic DNA from 1 to 10−4 ng was used to generate a standard curve. The log (concentration of template) was plotted against the Ct for each dilution. The curve was then used to calculate the efficiency for each primer pair (10[−1/slope]). The Ct values of the diluted genomic DNA were then used to normalize the experimental samples. The signal from a given region was normalized to the one from the OLI1 (Q0130) control locus in immunoprecipitated and input DNA samples. Plots represent the mean value obtained for at least three independent experiments; error bars correspond to SEM.
Immuno-FISH
Immuno-FISH was performed according to Gotta et al. (1999) with a few variations. The probe was obtained by PCR on a plasmid containing 4.8 kb of Y′ element and TG repeats (pEL42H10; Louis and Borts, 1995) with primer pair am151-GAAGAATTGGCCTGCTCTTG/am152-CCGTAAGCTCGTCAATTATT. The PCR purification was followed by a nick translation labeling reaction using the Nick Translation kit from Vysis (Abbott Molecular, Inc.). The fluorophore used in the reaction was SpectrumRed (Vysis). The probe was denatured for 5 min at 98°C, purified by ethanol precipitation, and resuspended in the hybridization mix (50% formamide, 10% dextran sulfate, and 2× SSC). 30 OD (1 OD corresponding to 107 cells) of cells was grown overnight to mid–logarithmic phase (∼1–2 × 107 cells/ml) in 30 ml YPD or YPGal and harvested at 1,200 g for 5 min at RT. Cells were resuspended in 25 ml of 4% paraformaldehyde for 20 min at RT, washed twice with 20 ml H20, and resuspended in 2 ml of 0.1-M EDTA-KOH, pH 8.0, and 10 mM DTT for 10 min at 30°C with gentle agitation. Cells were then collected at 800 g at RT, and the pellet was carefully resuspended in 2 ml YPD and 1.2-M sorbitol. Next, cells were spheroplasted at 30°C with Zymolyase (8–16 µl Zymolyase 100T at 5 mg/ml to 1 ml YPD-sorbitol cell suspension). Spheroplasting was stopped by the addition of 40 ml YPD and 1.2-M sorbitol. Cells were washed twice in YPD and 1.2-M sorbitol, and the pellet was resuspended in 1 ml YPD. Cells were dropped on diagnostic microscope slides and superficially air dried for 2 min. The slides were put in methanol at −20°C for 6 min, transferred to acetone at −20°C for 30 s, and air dried for 3 min. For immunofluorescence, the slides were incubated in PBS, 1% BSA, and 0.1% Triton X-100 for 20 min and overlayed with anti-GFP at 1:500 (rabbit, fraction A11122; Invitrogen) overnight at 4°C. The slides were covered with a coverslip to avoid drying of the antibody solution. After the primary antibody incubation, the slides were washed three times in PBS and 0.1% Triton X-100 for 5 min, and an anti–rabbit FITC was added at 1:100 for 1 h at 37°C. The secondary antibody was then washed three times in PBS and 0.1% Triton X-100 for 5 min before proceeding to the FISH. The cells were fixed afterward in 4× SSC and 4% paraformaldehyde during 20 min at RT and rinsed three times for 3 min in 4× SSC. After an overnight incubation at RT in 4× SSC, 0.1% Tween, and 20 µg/ml RNase, the slides were washed in H20 and dehydrated in ethanol 70, 80, 90, and 100% consecutively at −20°C for 1 min in each bath. Slides were air dried, and a solution of 2× SSC and 70% formamide was added for 5 min at 72°C. After a second step of dehydration, the denatured probe was added to the slides for 10 min at 72°C followed by a 37°C incubation for 24–60 h at 37°C in a humid chamber. The slides were then washed twice in 0.05× SSC at 40°C for 5 min and incubated twice in BT buffer (0.15-M NaHCO3 for 30 min, 0.1% Tween, and 0.05% BSA) at 37°C. 15 µl/spot of antifading compound in glycerol, pH 7.5 (DABCO), was added before imaging.
Microscopy
Sets of images from any given figure panel were acquired the same day using identical acquisition parameters on cells grown in the same culture conditions. The live-cell images were acquired using a wide-field microscopy system based on an inverted microscope (TE2000; Nikon) equipped with a 100×/1.4 NA oil immersion objective, a charge-coupled device (CCD) camera (Coolsnap HQ2; Photometrics), and a xenon arc lamp for fluorescence (Lambda LS; Sutter Instrument Co.), a collimated white light-emitting diode for the transmission, and a UV filter on the two illumination paths (LP 400 and GG400; Nikon). A Dual-View microimager (Optical Insights) was positioned in the optical path. When used, this device spatially split emitted light and allowed the simultaneous measurement of two-color information on the same sensor. Single-color images were acquired using either a GFP filter block (excitation: band pass (BP), 465–500 nm and dichroic, 506 nm; emission: BP, 516–556 nm; Semrock) for green fluorescence or a G2-A filter block for red fluorescence (excitation: BP, 510–560 nm and dichroic 565 nm; emission: long pass, 590 nm; Chroma Technology Corp.).
GFP-mCherry two-color images were acquired simultaneously on two halves of the same sensor using a GFP-mCherry filter block (excitation: double BP, 460–490/550–590 nm and dichroic double BP 500–550/600–665 nm) and the Dual View. The Dual View was equipped with adapted filter sets to observe green fluorescence (GFP, dichroic 565 nm and emission BP 499–529 nm; Semrock) on the left channel and red fluorescence (mCherry, dichroic 565 nm and emission BP 604–656 nm; Semrock) on the right channel. A home ImageJ macro (National Institutes of Health) was used to align and recombine channels. The position shift was estimated using the correlation function peak in transmitted light data (which is the same in the two channels) and used for fluorescent image alignment.
Immuno-FISH images were acquired with a wide-field microscope (Deltavision RT; Applied Precision) using a 100×/1.4 NA oil immersion objective (Olympus), a CCD camera (Coolsnap HQ2), and the softWoRx software (Applied Precision). The filters comprised the standard filter set suitable for FITC and RD-TR-PE (rhodamine, Texas red, and phycoerythrin). Images were deconvolved with softWoRx (additive method; eight iterations).
CFP–YFP two-color images were acquired on a spinning-disk confocal microscope (Revolution XD Confocal System; ANDOR) equipped with a spinning-disk unit (CSU-X1; Yokogawa), a microscope (Ti 2000; Nikon) with a 100×/1.4 NA oil immersion objective, and an EM CCD camera (iXON DU-885; ANDOR). CFP and YFP signals were acquired sequentially for each z section using solid-state 445- and 514-nm diodes and appropriate filters (confocal scanner unit triple dichroic mirror for 445, 514, and 640 nm and a double BP 464/547 emission filter from Semrock).
For fluorescent images, the axial (z) step is 200 nm, except for 4D movies, which have an axial (z) step of 300 nm. All fluorescent images are a z projection of z-stack images.
Microscopy data processing
Deconvolution was made using the Meinel algorithm in Metamorph (eight iterations; Σ = 0.8; frequency 3; MDS Analytical Technologies). Videos were denoised using the Safir-nD algorithm (Institut National de Recherche en Informatique et en Automatique Vista).
Telomere cluster quantification
Analyses have been performed using a home-made Matlab (MathWorks) application (Q-foci). A smoothing of data using a double Gaussian model, whose parameters were determined according to Zhang et al. (2007), was applied on deconvolved images. For segmentation and labeling of individual nuclei in 3D images, the diffuse Rap1-GFP fluorescence signal was considered as a nucleoplasm staining. Otsu thresholding was used for nuclei segmentation (Otsu, 1979). Additional filters were used to discard nonvalid objects. First, a morphological opening (disk kernel, radius of 4 pixels) was used to suppress segmentation artifacts. Incomplete objects touching the border of the 3D data stack and adjacent nuclei were also discarded. Local intensity maxima detected in segmented nuclei were considered as telomere cluster candidates. They were then attributed a score according to local curvature and mean intensity (Thomann et al., 2002). Because Rap1-GFP foci brightness is highly variable (depending on the number of telomeres in the cluster), results did not show a clear cutoff in scores between small clusters and false positives, as in other studies (Thomann et al., 2002; Berger et al., 2008). Consequently, the threshold for classification of a candidate as a telomere cluster was set manually based on the control (wild type) of the experiment and then applied on data corresponding to other conditions. The resulting data file lists all nuclei present in a series of 3D data stacks, each representing tenths of cells, along with the number of telomere clusters each cell contains and the intensity corresponding to these clusters, which were measured as the intensity component in the scoring method. 3D distances between telomeres VIL and XIVL were quantified using the SpotDistance ImageJ plugin with a visual inspection.
Simulations for the detection of telomere clusters in synthetic nuclei
Parameters required for these simulations were fitted experimentally based on microscopy images presented in Fig. 1, including noise, nucleus size, intensity, and microscope characteristics. Here, noise was considered as following a normal distribution; nuclei are considered as a sphere of radius 800 nm; single telomeres and clusters are considered as subresolution particles; total intensity of nuclei presented in Fig. 1 is equivalent to 4,390 Rap1 molecules per cell as previously described (Ghaemmaghami et al., 2003). The number of Rap1 molecules bound to each telomere was set to 40, assuming that 15–20 Rap1 molecules bind the TG repeats (Shore and Nasmyth, 1987) and 10–15 are spreading on neighboring nucleosomes (Hecht et al., 1996). The remaining pool of Rap1 was considered as diffusing freely in the nucleus. These simulations were then convolved using the measured point spread function of the microscope to reproduce as accurately as possible the experimental conditions.
Online supplemental material
Fig. S1 shows characterization of Sir3 overexpression and simulations of Rap1-GFP clusters. Fig. S2 shows the dynamics of telomere foci formation upon Sir3 induction (fluorescent images and quantifications using Q-foci). Fig. S3 shows the effect of Sir2–3–4 co-overexpression and Sir3 cellular amount on telomere clustering. Fig. S4 shows the requirements for Sir3 acetylation, Sir1, Esc1, yku70, and mps3 for telomere clustering. Fig. S5 shows that the overexpression of the N-terminal domain of Sir3 strengthens telomeric silencing without improving telomere clustering. Video 1 shows the appearance of Rap1-GFP hyperclusters upon Sir3 induction. Video 2 shows the disappearance of Rap1-GFP hyperclusters upon Sir3 shutoff. Video 3 shows the dynamics of Rap1-GFP hyperclusters during cell growth. Table S1 contains a list of the strains used in the main figures. Table S2 contains a list of the strains used in the supplemental data. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008007/DC1.
Acknowledgments
We thank Patricia Le Baccon for assistance with microscopy, Sebastien Huart for the ImageJ macro, Clémentine Brocas for strain constructions, Philippe Hupé and Gaël Millot for advice in statistical analysis, L. Pillus for strains and Sir3 antibody, Fabrizio Martino for providing recombinant Sir3, and Susan Gasser, Valérie Doye, and Benoît Palancade for reagents. We also thank Geneviève Almouzni, Tricia Laurenson, Sébastien Léon, Emmanuelle Martini, and Lorraine Pillus for critical reading of the manuscript and the members of the Taddei laboratory for helpful discussions.
M. Ruault was supported by a postdoctoral fellowship from the Association pour la Recherche sur le Cancer. The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Program (FP7/2007–2013/European Research Council grant agreement 210508) and the Agence Nationale de la Recherche Jeune chercheur grant.
Footnotes
Abbreviations used in this paper:
- 5-FOA
- 5-fluoroorotic acid
- BP
- band pass
- CCD
- charge-coupled device
- ChIP
- chromatin immunoprecipitation
- mRFP
- monomeric RFP
- SIR
- silent information regulator
References
- Adkins N.L., McBryant S.J., Johnson C.N., Leidy J.M., Woodcock C.L., Robert C.H., Hansen J.C., Georgel P.T. 2009. Role of nucleic acid binding in Sir3p-dependent interactions with chromatin fibers. Biochemistry. 48:276–288 10.1021/bi801705g [DOI] [PubMed] [Google Scholar]
- Ai W., Bertram P.G., Tsang C.K., Chan T.F., Zheng X.F. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell. 10:1295–1305 10.1016/S1097-2765(02)00695-0 [DOI] [PubMed] [Google Scholar]
- Akhtar A., Gasser S.M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 394:592–595 10.1038/29100 [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Zappulla D.C., Ansari A., Perrod S., Laiosa C.V., Gartenberg M.R., Sternglanz R. 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22:8292–8301 10.1128/MCB.22.23.8292-8301.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Billington B.L., Gottschling D.E. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 66:1279–1287 10.1016/0092-8674(91)90049-5 [DOI] [PubMed] [Google Scholar]
- Belmont A.S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250–257 10.1016/S0962-8924(01)02000-1 [DOI] [PubMed] [Google Scholar]
- Berger A.B., Cabal G.G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J.C., Gadal O., Zimmer C. 2008. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 5:1031–1037 10.1038/nmeth.1266 [DOI] [PubMed] [Google Scholar]
- Bertuch A.A., Lundblad V. 2003. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol. Cell. Biol. 23:8202–8215 10.1128/MCB.23.22.8202-8215.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V., Robine N., Lin W., Bonfils S., Géli V., Nicolas A. 2008. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28:99–111 10.1038/emboj.2008.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger J.R., Onishi M., Li G., Seebacher J., Rudner A.D., Gygi S.P., Moazed D. 2008. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:6903–6918 10.1128/MCB.01210-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S.W., Shore D. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9:370–384 10.1101/gad.9.3.370 [DOI] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Laroche T., van Houwe G., Blaszczyk M., Gasser S.M. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168:375–387 10.1083/jcb.200409091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Palladino F., Laroche T., Kyrion G., Liu C., Lustig A.J., Gasser S.M. 1995. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 129:909–924 10.1083/jcb.129.4.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M.M., Perrod S., Gasser S.M. 2000. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics. 154:1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.J., Yuan P., Hsu H.C., Li Z., Xu R.M., Sternglanz R. 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 26:3256–3265 10.1128/MCB.26.8.3256-3265.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W. 2007. Long-range DNA contacts: romance in the nucleus? Curr. Opin. Cell Biol. 19:317–320 10.1016/j.ceb.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Gartenberg M.R., Neumann F.R., Laroche T., Blaszczyk M., Gasser S.M. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967 10.1016/j.cell.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Geissenhöner A., Weise C., Ehrenhofer-Murray A.E. 2004. Dependence of ORC silencing function on NatA-mediated Nalpha acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:10300–10312 10.1128/MCB.24.23.10300-10312.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P.T., Palacios DeBeer M.A., Pietz G., Fox C.A., Hansen J.C. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA. 98:8584–8589 10.1073/pnas.151258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gomes P., Sampaio-Marques B., Ludovico P., Rodrigues F., Leão C. 2007. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech. Ageing Dev. 128:383–391 10.1016/j.mad.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B.K., Grunstein M., Gasser S.M. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243–3255 10.1093/emboj/16.11.3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Palladino F., Gasser S.M. 1998. Functional characterization of the N terminus of Sir3p. Mol. Cell. Biol. 18:6110–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Gasser S.M. 1999. Analysis of nuclear organization in Saccharomyces cerevisiae. Methods Enzymol. 304:663–672 10.1016/S0076-6879(99)04040-9 [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R.E. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 56:771–776 10.1016/0092-8674(89)90681-8 [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 63:751–762 10.1016/0092-8674(90)90141-Z [DOI] [PubMed] [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561–599 10.1146/annurev.genet.32.1.561 [DOI] [PubMed] [Google Scholar]
- Hayashi A., Ogawa H., Kohno K., Gasser S.M., Hiraoka Y. 1998. Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells. 3:587–601 10.1046/j.1365-2443.1998.00215.x [DOI] [PubMed] [Google Scholar]
- Heard E., Bickmore W. 2007. The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 19:311–316 10.1016/j.ceb.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger S., Grunstein M. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 383:92–96 10.1038/383092a0 [DOI] [PubMed] [Google Scholar]
- Hediger F., Taddei A., Neumann F.R., Gasser S.M. 2004. Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol. 375:345–365 10.1016/S0076-6879(03)75022-8 [DOI] [PubMed] [Google Scholar]
- Holmes S.G., Rose A.B., Steuerle K., Saez E., Sayegh S., Lee Y.M., Broach J.R. 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 145:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G.J., Tanny J.C., Rudner A.D., Gerber S.A., Danaie S., Gygi S.P., Moazed D. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167–4180 10.1128/MCB.22.12.4167-4180.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jeppesen P. 1997. Histone acetylation: a possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 19:67–74 10.1002/bies.950190111 [DOI] [PubMed] [Google Scholar]
- Johnson A., Li G., Sikorski T.W., Buratowski S., Woodcock C.L., Moazed D. 2009. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol. Cell. 35:769–781 10.1016/j.molcel.2009.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K. 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24:2138–2149 10.1038/sj.emboj.7600692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.A., Hall B.E., Iwamoto M.A., Win K.Z., Chang J.F., Ellenberger T. 2006. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J. Biol. Chem. 281:20107–20119 10.1074/jbc.M512588200 [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653–656 10.1016/S0960-9822(98)70252-0 [DOI] [PubMed] [Google Scholar]
- Liaw H., Lustig A.J. 2006. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol. Cell. Biol. 26:7616–7631 10.1128/MCB.01082-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G.G., Tanny J.C., Kruger R.G., Walz T., Moazed D. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 121:515–527 10.1016/j.cell.2005.03.035 [DOI] [PubMed] [Google Scholar]
- Liu C., Mao X., Lustig A.J. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for Telomeric silencing in Saccharomyces cerevisiae. Genetics. 138:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Louis E.J., Borts R.H. 1995. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics. 139:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Vega-Palas M.A., Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528–1539 10.1101/gad.988802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10:1796–1811 10.1101/gad.10.14.1796 [DOI] [PubMed] [Google Scholar]
- Marcand S., Buck S.W., Moretti P., Gilson E., Shore D. 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 10:1297–1309 10.1101/gad.10.11.1297 [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney D., Rose A., Hicks J.B., Broach J.R. 1987. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Laroche T., Suka N., Grunstein M., Gasser S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 97:621–633 10.1016/S0092-8674(00)80773-4 [DOI] [PubMed] [Google Scholar]
- Martino F., Kueng S., Robinson P., Tsai-Pflugfelder M., van Leeuwen F., Ziegler M., Cubizolles F., Cockell M.M., Rhodes D., Gasser S.M. 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell. 33:323–334 10.1016/j.molcel.2009.01.009 [DOI] [PubMed] [Google Scholar]
- McBryant S.J., Krause C., Woodcock C.L., Hansen J.C. 2008. The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol. Cell. Biol. 28:3563–3572 10.1128/MCB.01389-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A.H., Kornmann B., Dubrana K., Shore D. 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19:1199–1210 10.1101/gad.340205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Bystricky K., Dekker J. 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5:e1000478 10.1371/journal.pgen.1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.D., Sinclair D.A., Guarente L. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 97:609–620 10.1016/S0092-8674(00)80772-2 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Moazed D., Johnson D. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 86:667–677 10.1016/S0092-8674(00)80139-7 [DOI] [PubMed] [Google Scholar]
- Moazed D., Rudner A.D., Huang J., Hoppe G.J., Tanny J.C. 2004. A model for step-wise assembly of heterochromatin in yeast. Novartis Found. Symp. 259:48–56; discussion 56–62, 163–169 10.1002/0470862637.ch4 [DOI] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D. 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8:2257–2269 10.1101/gad.8.19.2257 [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller R., Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767–5768 10.1093/nar/22.25.5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A., Boeke J.D. 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 24:115–122 10.1101/gad.1865510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Liou G.G., Buchberger J.R., Walz T., Moazed D. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell. 28:1015–1028 10.1016/j.molcel.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Otsu N. 1979. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9:62–66 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Ottaviani A., Gilson E., Magdinier F. 2008. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 90:93–107 10.1016/j.biochi.2007.07.022 [DOI] [PubMed] [Google Scholar]
- Pryde F.E., Louis E.J. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18:2538–2550 10.1093/emboj/18.9.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio O.M., Zierath P.D., Billington B.L., Chhablani S.K., Gottschling D.E. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133–1145 10.1101/gad.7.7a.1133 [DOI] [PubMed] [Google Scholar]
- Roy R., Meier B., McAinsh A.D., Feldmann H.M., Jackson S.P. 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279:86–94 10.1074/jbc.M306841200 [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier A.L., Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 10.1146/annurev.biochem.72.121801.161547 [DOI] [PubMed] [Google Scholar]
- Sampath V., Yuan P., Wang I.X., Prugar E., van Leeuwen F., Sternglanz R. 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29:2532–2545 10.1128/MCB.01682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Lopez-Rubio J.J., Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 62:445–470 10.1146/annurev.micro.61.080706.093134 [DOI] [PubMed] [Google Scholar]
- Schober H., Kalck V., Vega-Palas M.A., Van Houwe G., Sage D., Unser M., Gartenberg M.R., Gasser S.M. 2008. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 18:261–271 10.1101/gr.6687808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23:928–938 10.1101/gad.1787509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J.W., Blackburn E.H. 1984. DNA sequences of telomeres maintained in yeast. Nature. 310:154–157 10.1038/310154a0 [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 51:721–732 10.1016/0092-8674(87)90095-X [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Brachmann C.B., Pillus L., Boeke J.D. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 149:1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.M., Pillus L. 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135:571–583 10.1083/jcb.135.3.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83–93 10.1101/gad.11.1.83 [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312 10.1038/sj.emboj.7600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E., Gasser S.M. 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 19:611–625 10.1101/gr.083881.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Duong T., Dujon B., Zimmer C., Fabre E. 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. USA. 107:2025–2030 10.1073/pnas.0914187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann D., Rines D.R., Sorger P.K., Danuser G. 2002. Automatic fluorescent tag detection in 3D with super-resolution: application to the analysis of chromosome movement. J. Microsc. 208:49–64 10.1046/j.1365-2818.2002.01066.x [DOI] [PubMed] [Google Scholar]
- Thomas B.J., Rothstein R. 1989. Elevated recombination rates in transcriptionally active DNA. Cell. 56:619–630 10.1016/0092-8674(89)90584-9 [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato J., Ikeda H. 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 388:900–903 10.1038/42288 [DOI] [PubMed] [Google Scholar]
- Wang X., Connelly J.J., Wang C.L., Sternglanz R. 2004. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics. 168:547–551 10.1534/genetics.104.028803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748–760 10.1101/gad.11.6.748 [DOI] [PubMed] [Google Scholar]
- Zhang B., Zerubia J., Olivo-Marin J.C. 2007. Gaussian approximations of fluorescence microscope point-spread function models. Appl. Opt. 46:1819–1829 10.1364/AO.46.001819 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hayashi M.K., Merkel O., Stillman B., Xu R.M. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21:4600–4611 10.1093/emboj/cdf468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Bodnar M.S., Spector D.L. 2009. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 19:172–179 10.1016/j.gde.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]