Abstract
Circulating tumor cells (CTCs) shed from primary and metastatic cancers are admixed with blood components and are thus rare, making their isolation and characterization a major technological challenge. CTCs hold the key to understanding the biology of metastasis and provide a biomarker to noninvasively measure the evolution of tumor genotypes during treatment and disease progression. Improvements in technologies to yield purer CTC populations amenable to better cellular and molecular characterization will enable a broad range of clinical applications, including early detection of disease and the discovery of biomarkers to predict treatment responses and disease progression.
Introduction
Blood-borne metastasis is initiated by cancer cells that are transported through the circulation from the primary tumor to vital distant organs, and it is directly responsible for most cancer-related deaths. Addressing this challenge, however, is confounded by our limited understanding of the process by which tumor cells exit from their primary site, intravasate into the circulation, and then establish distant lesions in the lung, brain, liver, or bone. Tumor cells that are identified in transit within the blood stream are referred to as circulating tumor cells (CTCs). Although their exact composition is unknown, a fraction of these are thought to be viable metastatic precursors capable of initiating a clonal metastatic lesion. However, CTCs are extraordinarily rare (estimated at one CTC per billion normal blood cells in the circulation of patients with advanced cancer); our understanding of their biological properties has thus been limited by the availability of technologies capable of isolating them in sufficient numbers and under conditions that are compatible with detailed molecular and functional experiments. Despite the limitations of current CTC-isolating methods, circulating cancer cells have been detected in a majority of epithelial cancers, including those from breast, prostate, lung, and colon. Patients with metastatic lesions are more likely to have CTCs detected in their blood; however, these have also been reported in some localized cancers. A better understanding of the identity of CTCs and the factors underlying their shedding into the vasculature is critical to identifying the key drivers of human cancer metastasis and devising rational therapeutic approaches.
Much of our current understanding of processes involved in cancer metastasis has been derived from mouse models of metastasis. Recent studies in these models have raised interesting mechanistic insights. For example, CTCs captured in xenograft prostate cancer models have highlighted the importance of pathways conferring resistance to apoptosis in these cells (Berezovskaya et al., 2005; Howard et al., 2008; Helzer et al., 2009). In a mouse model of breast cancer, disseminated tumor cells (DTCs) in the bone marrow can be detected in the premalignant phases of breast cancer, suggesting an early spread to distant organs (Hüsemann et al., 2008). Studies of the effects of epithelial–mesenchymal transition (EMT) in the generation of CTCs and distal metastases have suggested that this mesenchymal transformation may enhance the ability of cells to intravasate but may reduce their competence to initiate overt metastases (Tsuji et al., 2008, 2009). Mouse studies have also identified bone marrow–derived hematopoietic progenitor cells that express VEGF receptor 1 (VEGFR1) and may form a premetastatic niche that precedes the arrival of tumor cells (Kaplan et al., 2005). Moreover, Kim et al., (2009) have recently proposed a new concept of tumor self-seeding, in which injected tagged human cancer cell lines may colonize an existing tumor deposit, with the newly recruited tumor cells conferring increased aggressiveness to the existing tumor. Finally, the possibility of intravascular proliferation of CTCs adherent to vascular endothelium has been proposed based on in vivo imaging of tagged cells (Al-Mehdi et al., 2000).
Although these mouse studies offer fascinating insights into potential mechanisms of metastasis, certain limitations apply: xenograft models using established human cancer cell lines do not recapitulate the complex evolving vasculature and microenvironment of endogenous cancers, nor, of course, does direct intravascular inoculation of cancer cell lines into the tail vein. On the other hand, most endogenous mouse tumor models metastasize late, if at all, often in the setting of a massive primary tumor. Thus, although instructive, mechanistic insights from experimental mouse models must be validated by observational studies in human cancer.
The emergence of increasingly advanced and sensitive technologies to isolate human CTCs provides the opportunity to extend studies of cancer metastasis directly to human cancer. The full range of potential applications for CTC analyses include real-time noninvasive monitoring of CTCs as biomarkers of either sensitivity or acquired resistance to new cancer therapies, identifying new potential therapeutic targets to directly suppress cancer metastasis, and as technologies for CTC detection become increasingly sensitive and reliable, applying these at earlier stages of cancer progression with the goal of early cancer detection.
Although the potential applications of CTC analyses appear extraordinarily promising, the development of appropriate, high throughput, and reliable technological platforms for rare tumor cell detection within blood specimens remains the critical impediment. In fact, appropriate interpretation of many reported molecular analyses of CTCs requires an understanding of the technical limitations of the assays used to make these observations. In the following sections, we will review the technologies currently available for CTC isolation and the utility of CTCs as a diagnostic and prognostic marker in various cancers and focus on the molecular properties of these rare cells, which may help define their biology.
CTC detection technologies
The presence of CTCs in an autopsy of a patient who succumbed to advanced metastatic cancer was first reported in 1869 (Ashworth, 1869). To date, however, only limited information is available about the numbers of these cells in the blood stream at different stages of cancer and in different types of cancer, their molecular and biological heterogeneity, and their significance in the natural history of the disease. In the absence of any gold standard with which to measure various technologies, defining their absolute accuracy, sensitivity, and specificity in detecting CTCs remains a challenge. The ultimate goal, i.e., to efficiently isolate this rare population of cells in a viable and intact state and with high purity from the vast number of surrounding blood cells, presents a formidable technological challenge. A variety of currently used approaches relying on either the physical properties, expression of biomarkers, or functional characteristics of CTCs are reviewed in the following sections. Although none of these current approaches constitute the optimal platform for CTC isolation, they each represent significant advances and provide a basis from which to anticipate ongoing technological developments.
Nucleic acid–based detection of CTCs.
Free DNA and, to a lesser extent, RNA circulating in plasma from patients with cancer have been described previously (Papadopoulou et al., 2006; Gormally et al., 2007; Zanetti-Dällenbach et al., 2008; Yoon et al., 2009). Schwarzenbach et al. (2007, 2009) have suggested a link between the presence of CTCs (and DTCs) and the detection of free tumor-derived DNA in serum/plasma of prostate cancer patients. However, the origin of these nucleic acids may also include direct shedding from necrotic cells in tumor deposits, tumor-derived exosomes, or cellular fragments or lysis of CTCs in the bloodstream. Some approaches have focused on isolating nucleic acids directly from cell-free plasma (Papadopoulou et al., 2006; Yoon et al., 2009), whereas others have first purified nucleated cells followed by lysis and nucleic acid extraction. Molecular analysis of DNA mutations have been described previously (Igetei et al., 2008), and individual tumor-specific translocation breakpoints have recently been sequenced by next-generation approaches (Leary et al., 2010). Overall, the relatively low sensitivity of nucleic acid analyses from free plasma or unpurified blood cell components has been addressed by sampling relatively large blood volumes as well as application of such next-generation sequencing technologies with extraordinary sensitivity. Nonetheless, interpretation of a negative test remains a concern because absence of a defined molecular abnormality within a blood specimen cannot be readily distinguished from insufficient amounts of tumor-derived DNA in the circulation from an individual patient. At the RNA level, RT-PCR analyses have also been applied, either to unpurified plasma nucleic acids (Papadopoulou et al., 2006) or more commonly to enriched CTC populations. Tumor cell–specific markers have included cytokeratins, prostate-specific antigen (PSA), mucin-1 (MUC-1), HER2, AFP (α-fetoprotein), and the CEA (carcinoembryonic antigen) gene family among others (Louha et al., 1997; de Cremoux et al., 2000; Hardingham et al., 2000; Mejean et al., 2000; Wu et al., 2006; Xi et al., 2007). However, significant challenges include the frequency of both false-positive and false-negative PCR results and the difficulty in quantitating relative levels of expression without prepurification of these extraordinarily rare cells (Paterlini-Brechot and Benali, 2007; Pantel et al., 2008).
Detection of CTCs based on their physical properties.
Although not absolute, several physical properties distinguish CTCs from most normal blood cells. These include the larger size of most epithelial cells and differences in density, charge, migratory properties, and some properties of specific cell types (e.g., melanocytic granules in melanoma cells). Differences in buoyant density have been used to separate mononucleated cells, including CTCs, from red blood cells through gradient centrifugation, although CTCs still comprise a minute fraction of mononucleated cells in the circulation (Gertler et al., 2003; Müller et al., 2005; Wülfing et al., 2006; Morgan et al., 2007).
Isolation of CTCs by virtue of their increased size, compared with leukocytes, has been applied using several different filtration-based approaches, such as isolation by size of epithelial tumor cells (Vona et al., 2000) and microelectromechanical systems (Zheng et al., 2007). Although most CTCs derived from epithelial cancers are in fact larger than leukocytes, there is a significant variation in their size range (Marrinucci et al., 2007). Several different pores and filter-based approaches have been developed to prevent clogging and facilitate the retrieval of CTCs (Mohamed et al., 2009; Tan et al., 2009). These approaches have shown significant promise in capturing cancer cell lines spiked into blood but will require further validation with clinical specimens, in which both heterogeneous CTC size and resistance to filtering shear stress are likely to be significant variables.
Capture of CTCs using antibodies against cell surface antigens.
The most widely used CTC isolation techniques rely on antibody-based capture of CTCs, which express epithelial cell surface markers that are absent from normal leukocytes. Among these, epithelial cell adhesion molecule (CAM [EpCAM]) is most commonly used because its expression is virtually universal (albeit at variable levels) in cells of epithelial origin, but it is absent in blood cells. Conjugation of antibodies against EpCAM to magnetic beads, followed by purification of captured cells through a magnetic field, has been used to enrich CTCs from the blood of patients with cancers of the breast, prostate, and colon. The CellSearch system (Veridex), a widely used FDA (Food and Drug Administration)–approved method, uses ferrofluids loaded with an EpCAM antibody to capture CTCs, which are subsequently visualized by staining with a cocktail of antibodies against the cytoplasmic epithelial cytokeratins (8, 18, and 19; Riethdorf et al., 2007). Staining for the leukocyte-specific marker CD45 is used as a control to exclude contaminating leukocytes. Surprisingly, a significant number of cells appears to stain both for cytokeratins and CD45; although the identity of these dual-positive cells is not well understood, they are excluded from enumeration, with CTCs defined as the subset of EpCAM-captured cells that are confirmed as both positive for cytokeratins and negative for CD45. Although this platform is the most standardized of any current technology and is now being tested for clinical applications (see CTCs as prognostic markers), it suffers from relatively low sensitivity: only a fraction of patients with metastatic cancer scores positive for any CTCs, with a median yield of approximately one CTC per milliliter and typically low purity (Allard et al., 2004; Attard et al., 2009).
Our own group has developed a microfluidic platform for single-step isolation of CTCs from unprocessed blood specimens (Nagrath et al., 2007; Maheswaran et al., 2008; Stott et al., 2010a). The CTC-chip is a silicon chamber etched with 78,000 microposts that are coated with an anti-EpCAM antibody. As 2–4 ml of whole blood flows through the chip, flow kinetics have been optimized for minimal shear stress on cells while enhancing contacts with the antibody-coated microposts. Captured CTCs attached to microposts are visualized by staining with antibodies against cytokeratin or tissue-specific markers (Fig. 1). For CTC enumeration, the entire device is imaged at multiple planes using a semiautomated imaging system while on-chip lysis allows for DNA and RNA extraction and molecular analyses (Fig. 2). The CTC-chip enables a high yield of capture (median, 50 CTCs per milliliter) and purity (ranging from 0.1 to 50%), most likely caused by the gentle one-step microfluidic processing, which may be critical when purifying rare delicate cell populations (Nagrath et al., 2007). Captured cells remain viable after capture, although the absence of cell fixation currently limits the time allowed between blood collection and microfluidic analysis to a few hours. Although the CTC-chip assay is technically difficult and not yet standardized for high throughput applications, the high number of cells captured provides a dynamic range that allows longitudinal monitoring of patients during therapy while improved (albeit still highly variable) purity of captured CTCs enables molecular characterization (see Molecular characterization of CTCs). Recently, we have improved the microfluidic CTC isolation approach using an enhanced platform, the herringbone (HB)-chip, which makes use of a microvortex mixing device (Stott et al., 2010b). Instead of using three-dimensional microposts to break up flow streamlines and enhance cell collisions with antibody-coated posts, the HB-chip uses calibrated microfluidic flow patterns to drive cells into contact with the antibody-coated walls of the device. In addition to increased target cell capture efficiency, the less complex design of the HB-chip is more amenable to high throughput manufacture and reliable coating of the inner surface with antibodies and allows for the chambers to be made out of transparent materials, which greatly enhance high resolution imaging, including the use of transmitted light microscopy.
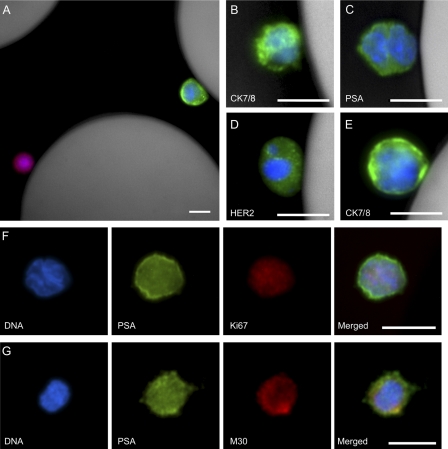
Figure 1.
Micrographs of CTCs captured from patients’ blood using an anti-EpCAM–coated CTC-chip. (A–C) Combined fluorescent and reflected light micrographs of a cytokeratin 7/8 (green)–stained CTC captured from breast cancer patient blood and a contaminating CD45-positive (red) white blood cell (A) and cytokeratin 7/8 (green, B)– and PSA (green, C)-stained CTCs captured from a prostate cancer patient. (D and E) HER2 (green, D)- and cytokeratin 7/8 (green, E)–stained CTCs captured from a breast cancer patient. (F and G) Individual and merged fluorescent micrographs of CTCs captured from prostate cancer patients’ blood stained positive for PSA (green) and Ki-67 (red, proliferative marker, F), and PSA (green) and M30 (red, apoptotic marker) demonstrated the heterogeneity in CTCs (G). In all panels, the nuclei are stained with DAPI (blue). CK7/8, cytokeratin 7/8. Bars, 10 µm.
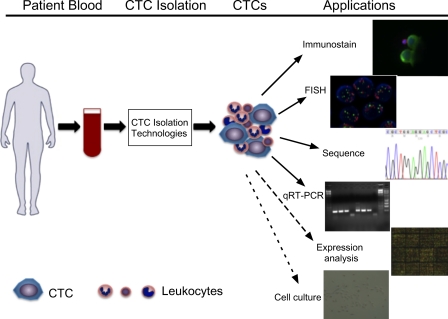
Figure 2.
Illustration of current and potential applications of CTC technologies. The peripheral blood of a cancer patient is collected and processed through various CTC isolation technologies. CTCs are captured along with contaminating leukocytes. Immunostaining for specific markers and FISH for genomic amplification and translocation can be applied to CTCs. DNA or RNA can be extracted from the CTCs and subjected to sequencing, quantitative RT-PCR (qRT-PCR), and potential expression profile analysis. Viable cells can be released and propagated in cell culture.
Additional cell surface marker–based CTC detection platforms are under development. Among these, standard flow cytometry (FACS) has been tested (Simpson et al., 1995; Cruz et al., 2005), although its sensitivity may be inadequate for detection of extraordinarily rare cells (Allan et al., 2005). A magnetic stir bar, the slowly rotating MagSweeper, coated with an antibody to EpCAM has also been tested to capture CTCs from blood samples (Talasaz et al., 2009). To avoid the bias of selecting cells by virtue of EpCAM expression, negative selection (i.e., removal of leukocytes, thereby leaving residual CTCs) has been advocated. Although this strategy has the potential to purify CTCs irrespective of presumed cell surface markers, the extremely low prevalence of CTCs limits the yield accomplished by negative selection (Tong et al., 2007; Tkaczuk et al., 2008; Balasubramanian et al., 2009; Yang et al., 2009).
Additional innovative CTC detection strategies.
Several additional innovative approaches have been developed with the goal of detecting these rare cells. Some of these make use of interesting physical or biological properties of epithelial cells. High throughput microscopic scanning approaches have been adapted to screen for CTCs. Among these, fiber-optic array–scanning technology involves deposition of nucleated cells on the surface of a large glass slide, with scanning of cells positive for epithelial or tumor-specific antigens (Krivacic et al., 2004; Marrinucci et al., 2007). Laser-scanning cytometry extends this approach by using a combination of fluorescent labeling and forward scatter to enhance identification of cells deposited on a glass slide (Pachmann et al., 2005b). Both fiber-optic array–scanning technology and laser-scanning cytometry enable cytological evaluation of CTCs without the preselection bias derived from reliance on their expression of specific cell surface markers, but the identification of unpurified viable CTCs is less conducive for further molecular and biological characterization. Multiphoton intravital flow cytometry detects CTCs tagged in vivo using injected fluorescent ligands as they flow through the vasculature (He et al., 2007). Dielectric properties of cells, a measure of their plasma membrane area affected by ruffles, folds, and microvilli, have also been used to isolate CTCs from mixed cell populations (Gascoyne et al., 2009). Photoacoustic flowmetry, making use of the broadband absorption spectrum of melanin, has been tested to detect melanoma cells (Weight et al., 2009) and has been combined with nanoparticles targeting cell surface antigens to broaden its applicability in CTC detection (Galanzha et al., 2009a, b). The proclivity of metastatic cells to ingest and invade through collagen has led to the development of a technique whereby mononucleated blood cells are seeded on fluorescent CAM-coated slides, and CTCs are visualized as they ingest CAM (Paris et al., 2009; Lu et al., 2010). Finally, staining of CTCs by virtue of detectable secreted products (epithelial immunospot) has been developed as a functional screen (Alix-Panabières et al., 2007, 2009). Of note, most new technologies are developed and tested using cancer cell lines spiked into control blood specimens. Extrapolation to the detection of real CTCs in blood specimens from cancer patients presents a significant challenge beyond cancer cell line experiments, given their heterogeneity, variable expression of cell surface markers, and reduced ability to survive intact through complex purification procedures. Hence, extensive validation of novel technologies using clinical specimens should be an integral part of assay development.
In summary, the existing CTC technologies rely on various properties of CTCs, with each having unique advantages and limitations. Additional technologies are under development, and future CTC capture platforms should aim for an efficient capture of the entire diverse spectrum of tumor cells that circulate in the blood, followed by their molecular and functional characterization. The inherent limitations of current CTC detection platforms should be considered when interpreting the literature about molecular properties of CTCs and their potential clinical applications.
CTCs as prognostic markers
Despite the limitation of the various CTC detection technologies, several studies with large cohorts of patients have been performed to evaluate the clinical utility of CTC enumeration. Given the need for high throughput standardization, most of these studies have used the immunomagnetic bead capture assay, which is commercially available. In general, these studies have concluded that the presence of detectable CTCs in the blood serves as an independent prognostic factor in patients with cancers of the breast, prostate, and colon. In patients with metastatic breast cancer, CTC counts above five CTCs per 7.5 ml of blood before the start of systemic therapy were associated with a shorter median progression-free survival and overall survival (Cristofanilli et al., 2004, 2008; Botteri et al., 2010). Additional studies extended these analyses to use molecular endpoints, such as HER2 staining, and to patients with invasive localized breast cancer receiving so-called neoadjuvant chemotherapy (Wülfing et al., 2006; Pierga et al., 2008). However, despite processing as much as 50 ml of blood, CTCs were detected in only half of the patients, with the number of HER2-positive cells ranging from one to eight CTCs per 50 ml. Thus, although promising, these approaches emphasize the critical need for increased sensitivity in CTC detection to enable clinical applications. As with breast cancer, a correlation between pretreatment CTC numbers and clinical prognosis has been reported for patients with colorectal (Cohen et al., 2006) and prostate cancers (Danila et al., 2007; de Bono et al., 2008; Okegawa et al., 2009). Although these major prognostic studies have focused on CTC enumeration using immunomagnetic bead capture, others have correlated patient outcomes with the RT-PCR analysis of unpurified mononucleated blood cell fractions to test for expression of epithelial markers, including various keratins and EpCAM (Weigelt et al., 2003; Masuda et al., 2005; Ignatiadis et al., 2007). Quantifying PCR-based analysis of epithelial marker expression without purification of rare CTCs from the massive number of normal leukocytes in the circulation presents a significant technical hurdle, although next-generation sequencing technologies may help address this challenge in the future.
Whichever technique for measuring CTCs is used, most have shown that baseline (i.e., pretreatment) CTC numbers among different cancer patients are not well correlated with standard measures of tumor mass, including tumor size determined radiographically by x ray or CT (computed tomography) scan or serum protein markers, such as PSA levels in prostate cancer (Budd et al., 2006; Scher et al., 2009; Stott et al., 2010a). Thus, the number of CTCs in the blood is not simply a measure of tumor volume, but instead, it may reflect additional biological features, possibly including tumor vascularity or invasiveness, both of which may have distinct prognostic contributions.
CTC analyses in monitoring responses to therapy
Whereas prognostic studies have focused on clinical outcomes for patients who have baseline CTC counts above or below an arbitrary threshold, correlating CTC numbers over time with response to therapy requires that the CTC detection method be sensitive enough to provide a dynamic range that can be followed over time. Using a microfluidic CTC capture device, we were able to show that the number of CTCs in patients with lung, prostate, and other cancers declined rapidly after the initiation of effective chemotherapy, hormonal therapy, or targeted kinase inhibition (Nagrath et al., 2007; Maheswaran et al., 2008; Stott et al., 2010a). Again, there was little correlation among different patients between baseline CTC counts and radiographical measures of tumor volume before therapy. However, treatment-induced changes in CTC numbers within individual patients followed longitudinally were well correlated with standard measures of tumor response, irrespective of baseline levels. Treatment-induced changes in CTC numbers have also been reported in some early breast cancer treated with neoadjuvant therapy using laser-scanning cytometry (Pachmann et al., 2005a) and CellSearch (Pierga et al., 2008), in adjuvant chemotherapy using OncoQuick (Müller et al., 2005), and in some metastatic breast and prostate cancers using immunomagnetic bead isolation (Hayes et al., 2006; Attard et al., 2009; Reid et al., 2010). Additional studies will be needed to determine whether monitoring the decline in CTC numbers after therapy is useful as an early marker of response or as a distinct predictor of overall therapeutic benefit. Eventually, CTC-based molecular analyses may have the added benefit of demonstrating direct inhibition of drug targets in viable tumor cells.
Although testing for chemotherapy-induced changes in CTC numbers may provide insight into treatment effectiveness for advanced cancers, monitoring the number of CTCs after curative surgical resection of localized cancers may identify cases that are at risk for disease recurrence. Measurements of CTCs in patients with localized cancer have been limited and require sensitive assays. In a pilot study of 19 patients with localized prostate cancer, we detected CTCs in eight cases (Stott et al., 2010a). In most of these, CTCs dropped to undetectable levels within 24 h of surgical resection, whereas in a few cases, a more delayed decline after surgery was evident. None of these patients have developed a recurrence at a 1-yr follow up. Although these studies are preliminary, they raise the possibility that localized cancers may invade the vasculature and shed CTCs before metastases are established, a concept that provides hope for early cancer detection applications. The prognostic significance of CTCs shed by localized cancers and the rate of decline of CTCs after surgical resection remain to be determined.
Molecular characterization of CTCs
Identification of molecular aberrations in CTCs.
Beyond the enumeration of CTCs, their molecular characterization provides a key to demonstrating their cellular origin from primary and metastatic tumor deposits, and it may also provide clues to their evolution during the course of cancer treatment.
FISH analysis performed on CTCs from breast, kidney, prostate, and colon cancer showed evidence of aneuploidy in some cells, which was consistent with their malignant origin (Fehm et al., 2002). However, these studies were somewhat limited by the low level of chromosome number variation combined with technical limitations in analysis of CTC and contaminating leukocytes. In more recent studies, demonstration of specific androgen receptor gene amplification, a known biomarker of castrate-resistant prostate cancer, has provided clear evidence that at least a subset of CTCs are tumor derived (Attard et al., 2009; Scher et al., 2009). Intragenic mutations in cancer genes have extended these findings, although given the presence of contaminating leukocytes in CTC populations, these analyses typically require allele-specific PCR assays. In nonsmokers’ lung cancer, EGF receptor (EGFR) mutations are particularly relevant because they are predictive of a dramatic clinical response to EGFR kinase inhibitors (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004). In our initial studies, EGFR mutations identified in lung CTCs were concordant with the mutations known to be present in the primary tumor in 12/13 cases (Maheswaran et al., 2008). Of note, during longitudinal follow up of these cases, continued therapy was associated with the emergence of a kinase inhibitor resistance-associated EGFR mutation, which was detectable at increasing allelic ratios in CTCs, coincident with the development of clinical drug resistance. In addition, CTC analyses after prolonged therapy showed the acquisition of additional EGFR mutations that were below detection in the primary tumor biopsy, which was suggestive of evolution in the dominant tumor clones as a result of treatment-induced selection (Maheswaran et al., 2008). Thus, serial analysis of tumor-derived samples may allow real-time monitoring of molecular evolution among tumor cells, which may be particularly relevant after the use of effective targeted cancer therapy that applies selective pressures toward the development of drug resistance mechanisms.
The use of CTCs to test for molecular evolution of tumors during the course of treatment has also been applied in breast cancer, in which some HER2-positive CTCs have been reported in breast cancer patients whose primary tumor was HER2 negative (Meng et al., 2004; Wülfing et al., 2006; Tewes et al., 2009) as well as some HER2-negative CTCs emerging in HER2-positive breast cancers subjected to anti-HER2 therapy (Hayes et al., 2002). The true frequency of such genotype conversion events remains to be established in larger studies. In prostate cancer, the TMPRSS2-ERG translocation has been detected in CTCs either by FISH or by translocation-specific RT-PCR analysis (Attard et al., 2009; Stott et al., 2010a). Improving CTC isolation technologies has allowed selective RNA-based expression analyses (Ghossein et al., 1995; Helzer et al., 2009), but CTC impurity remains a major hurdle for all detailed molecular analyses of CTC genomic and transcriptional profiles. For instance, FISH analyses of CTCs can be challenging when scoring four individual CTCs surrounded by 1,000 leukocytes with a 4% false-positive rate (Attard et al., 2009). The use of fine needle microselection of individual CTCs has provided the level of purity required for loss of heterozygosity studies in some cases of prostate cancer (Schmidt et al., 2006). However, collectively, comprehensive molecular analyses of CTCs will require enhanced isolation methods that result in a higher degree of captured cell purity as well as the use of technologies optimized for minute amounts of nucleic acids, including the single-cell level.
Evaluation of signaling, proliferation, and apoptosis in CTCs.
Dynamic studies of CTCs after therapeutic interventions may eventually allow real-time monitoring of treatment responses, a major goal of drug development. The signaling markers studied include phospho-PI3K and phospho-Akt in breast cancer CTCs (Kallergi et al., 2008) and the DNA damage marker phosphorylated γ-H2AX in patients treated with chemotherapeutic agents (Wang et al., 2010). Evidence of apoptosis in CTCs has also been tested using morphological criteria as well as flow cytometric analysis of M30-stained cells (Larson et al., 2004; Hou et al., 2009; Swennenhuis et al., 2009). Up-regulation of the antiapoptotic BCL2-α transcript in CTCs captured from mice harboring PC3 xenografts has also been reported (Helzer et al., 2009). Collectively, monitoring intracellular signaling pathways is likely to significantly enhance the utility of CTC analyses, although early studies have been limited both by the challenge of low amounts of RNA or protein recovery from these rare cells and by the need for dynamic analysis over time using a sufficient number of CTCs within each clinical specimen.
Although the use of apoptotic markers may be complicated by the effect of ex vivo processing, analysis of cellular proliferation markers appears to be particularly promising using the microfluidic chip. In early studies, costaining of prostate cancer CTCs for the tumor-specific marker PSA together with the proliferation marker Ki-67 revealed significant variation in the proliferative index of CTCs (Stott et al., 2010a). In a pilot study of metastatic prostate cancer, CTCs from patients who were responding to hormone withdrawal therapy exhibited a low proliferative index (1–2%), whereas a much higher Ki-67–positive fraction was scored in CTCs from treatment-resistant patients (69–73%; Stott et al., 2010a). These results need to be verified using a larger cohort of patients but suggest that a significant heterogeneity in the proliferative index of CTCs may reflect the intrinsic biological properties of tumors, with potential applications in assessing therapeutic responses.
Molecular characterization of CTCs may also provide insight into the biology of these cells and their link to the process of metastasis. Mouse models and in vitro experiments have suggested that EMT, together with stemlike properties in cancer cells, is associated with increased cellular migration and resistance to therapy (Mani et al., 2008; Polyak and Weinberg, 2009). Although definitive studies of EMT and stem cell markers in CTCs are lacking, some studies of tumor cells isolated from bone marrow biopsies (so-called DTCs) have suggested the expression of characteristic EMT markers, either in the cells themselves or in derivative cell lines (Putz et al., 1999; Schardt et al., 2005; Willipinski-Stapelfeldt et al., 2005). PCR assays have suggested a possible expression of some EMT inducers in CTC-enriched blood specimens (Aktas et al., 2009). However, an intrinsic challenge in studying EMT in CTCs is the standard use of an antibody to EpCAM to capture these cells, thus preselecting for cells that have retained significant expression of epithelial markers. A recent study using the CellSearch system has shown that a basal-like breast cancer cell line with features of EMT expresses EpCAM levels that are too low to allow capture using such antibodies (Sieuwerts et al., 2009). Development of alternative CTC isolation strategies, as well as refining the imaging parameters for EMT and other cell fate markers, will be essential. Finally, the successful isolation of viable CTCs capable of extended proliferation in vitro will enable detailed molecular and functional studies, providing important insight into the process of metastasis in human cancers.
CTC clusters.
A recent and surprising insight derived from CTC analyses has been the finding of clusters of CTCs within the circulation of patients with known cancer (Stott et al., 2010b). Predominant models of cancer metastasis have focused on the presumed role of individual cells with increased mobility through EMT, migrating into the vasculature and exiting at distant sites to initiate a metastatic tumor deposit. The finding of aggregated tumor cells in the vasculature raises the possibility that such tumor emboli may also contribute to the metastatic process. Work in animal models has shown that intravenously injected tumor cell clumps have a greater tendency to form metastases than comparable numbers of single tumor cells (Watanabe, 1954; Fidler, 1973). Increased metastatic potential has also been correlated with tumor cell clumps in mouse models of fibrosarcoma, melanoma, and lung carcinoma (Liotta et al., 1974, 1976; Glaves, 1983). Intravascular aggregation of cancer cells with apparent attachment to the endothelium has been described after injection of breast and prostate cancer cells as well as fibrosarcoma cells (Al-Mehdi et al., 2000; Glinsky et al., 2003).
The finding of CTC clusters in human blood samples might be limited by the technologies used for their isolation, most of which use harsh batch purification approaches likely to disrupt such cellular aggregates. Early studies in advanced prostate, breast, and colorectal cancers relied on gradient centrifugation in combination with magnetic bead selection (Brandt et al., 1996, 1998; Wang et al., 2000; Molnar et al., 2001). Our own recent study identified CTC clusters, ranging from 3 to 14 cells, in several prostate and lung cancer patients (Fig. 3; Stott et al., 2010b). These were readily identified using the microfluidic HB-chip, which appears to preserve multicellular aggregates, whereas they were never found with our micropost CTC-chip, in which tight distances between the microposts presumably prevent the passage of large cellular clusters. Interestingly, when stained with hematoxylin and eosin, most CTC clusters have an increased staining intensity relative to individual CTCs, suggesting less autolysis in the CTC clusters. This raises the intriguing possibility that cells within microclusters might be relatively protected either from anoikis associated with loss of basement membrane attachment (Zhang et al., 2004, 2008) or perhaps from the harsh environment and shear stresses of the vascular circulation. Collectively, the clinical relevance of CTC clusters versus single CTCs within the circulation and their potential implications for metastatic spread warrant further investigation.
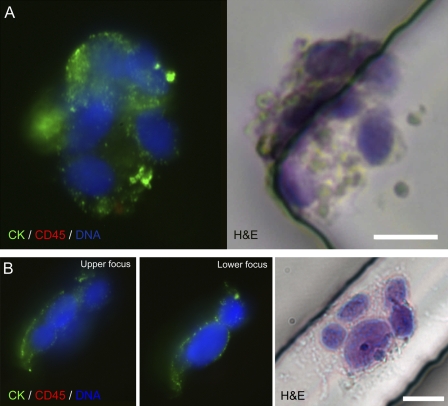
Figure 3.
CTC clusters captured from lung cancer patients’ blood using an anti-EpCAM–coated HB-chip. (A and B) Fluorescent micrographs of cytokeratin 7/8 (CK; green)–, CD45 (red)-, and DAPI (blue)-stained CTC clusters taken at different focal planes and corresponding hematoxylin and eosin (H&E) stains are shown. Bars, 10 µm.
Future perspectives
The molecular characterization of CTCs has the potential to revolutionize our understanding of cancer metastasis. However, technological platforms need significant optimization to enable highly sensitive and reliable CTC isolation before the full array of currently available molecular tools may be applied. Such technologies are likely to emerge in the near future as the broad field of microfluidic bioengineering and other new approaches are applied to the challenge of rare tumor cell detection in blood specimens. Although increasingly sophisticated analytic approaches will be required to tackle the characteristics of CTCs, we can already discern a broad range of potential applications, from the noninvasive monitoring of tumor genotypes to the identification and analysis of metastatic precursor cell subpopulations. As the technology evolves further, applications in the early detection of invasive but localized cancer may even be contemplated. Together, these advances are likely to have important implications for our understanding of cancer metastasis and our treatment of patients with cancer.
Footnotes
Abbreviations used in this paper:
- CAM
- cell adhesion molecule
- CTC
- circulating tumor cell
- DTC
- disseminated tumor cell, EGFR, EGF receptor
- EMT
- epithelial–mesenchymal transition
- EpCAM
- epithelial CAM
- HB
- herringbone
- PSA
- prostate-specific antigen
References
- Aktas B., Tewes M., Fehm T., Hauch S., Kimmig R., Kasimir-Bauer S. 2009. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 11:R46 10.1186/bcr2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabières C., Vendrell J.P., Pellé O., Rebillard X., Riethdorf S., Müller V., Fabbro M., Pantel K. 2007. Detection and characterization of putative metastatic precursor cells in cancer patients. Clin. Chem. 53:537–539 10.1373/clinchem.2006.079509 [DOI] [PubMed] [Google Scholar]
- Alix-Panabières C., Vendrell J.P., Slijper M., Pellé O., Barbotte E., Mercier G., Jacot W., Fabbro M., Pantel K. 2009. Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. 11:R39 10.1186/bcr2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan A.L., Vantyghem S.A., Tuck A.B., Chambers A.F., Chin-Yee I.H., Keeney M. 2005. Detection and quantification of circulating tumor cells in mouse models of human breast cancer using immunomagnetic enrichment and multiparameter flow cytometry. Cytometry A. 65:4–14 [DOI] [PubMed] [Google Scholar]
- Allard W.J., Matera J., Miller M.C., Repollet M., Connelly M.C., Rao C., Tibbe A.G., Uhr J.W., Terstappen L.W. 2004. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10:6897–6904 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- Al-Mehdi A.B., Tozawa K., Fisher A.B., Shientag L., Lee A., Muschel R.J. 2000. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: a new model for metastasis. Nat. Med. 6:100–102 10.1038/71429 [DOI] [PubMed] [Google Scholar]
- Ashworth T.R. 1869. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust. Med. J. 14:146–149 [Google Scholar]
- Attard G., Swennenhuis J.F., Olmos D., Reid A.H., Vickers E., A’Hern R., Levink R., Coumans F., Moreira J., Riisnaes R., et al. 2009. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69:2912–2918 10.1158/0008-5472.CAN-08-3667 [DOI] [PubMed] [Google Scholar]
- Balasubramanian P., Yang L., Lang J.C., Jatana K.R., Schuller D., Agrawal A., Zborowski M., Chalmers J.J. 2009. Confocal images of circulating tumor cells obtained using a methodology and technology that removes normal cells. Mol. Pharm. 6:1402–1408 10.1021/mp9000519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovskaya O., Schimmer A.D., Glinskii A.B., Pinilla C., Hoffman R.M., Reed J.C., Glinsky G.V. 2005. Increased expression of apoptosis inhibitor protein XIAP contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 65:2378–2386 10.1158/0008-5472.CAN-04-2649 [DOI] [PubMed] [Google Scholar]
- Botteri E., Sandri M.T., Bagnardi V., Munzone E., Zorzino L., Rotmensz N., Casadio C., Cassatella M.C., Esposito A., Curigliano G., et al. 2010. Modeling the relationship between circulating tumour cells number and prognosis of metastatic breast cancer. Breast Cancer Res. Treat. 122:211–217 10.1007/s10549-009-0668-7 [DOI] [PubMed] [Google Scholar]
- Brandt B., Junker R., Griwatz C., Heidl S., Brinkmann O., Semjonow A., Assmann G., Zänker K.S. 1996. Isolation of prostate-derived single cells and cell clusters from human peripheral blood. Cancer Res. 56:4556–4561 [PubMed] [Google Scholar]
- Brandt B., Roetger A., Heidl S., Jackisch C., Lelle R.J., Assmann G., Zänker K.S. 1998. Isolation of blood-borne epithelium-derived c-erbB-2 oncoprotein-positive clustered cells from the peripheral blood of breast cancer patients. Int. J. Cancer. 76:824–828 [DOI] [PubMed] [Google Scholar]
- Budd G.T., Cristofanilli M., Ellis M.J., Stopeck A., Borden E., Miller M.C., Matera J., Repollet M., Doyle G.V., Terstappen L.W., Hayes D.F. 2006. Circulating tumor cells versus imaging—predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 12:6403–6409 10.1158/1078-0432.CCR-05-1769 [DOI] [PubMed] [Google Scholar]
- Cohen S.J., Alpaugh R.K., Gross S., O’Hara S.M., Smirnov D.A., Terstappen L.W., Allard W.J., Bilbee M., Cheng J.D., Hoffman J.P., et al. 2006. Isolation and characterization of circulating tumor cells in patients with metastatic colorectal cancer. Clin. Colorectal Cancer. 6:125–132 10.3816/CCC.2006.n.029 [DOI] [PubMed] [Google Scholar]
- Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., Hayes D.F. 2004. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351:781–791 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- Cristofanilli M., Reuben J., Uhr J. 2008. Circulating tumor cells in breast cancer: fiction or reality? J. Clin. Oncol. 26:3656–3657 10.1200/JCO.2008.18.0356 [DOI] [PubMed] [Google Scholar]
- Cruz I., Ciudad J., Cruz J.J., Ramos M., Gómez-Alonso A., Adansa J.C., Rodríguez C., Orfao A. 2005. Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples. Am. J. Clin. Pathol. 123:66–74 10.1309/WP3QWKVJFYDHHXQD [DOI] [PubMed] [Google Scholar]
- Danila D.C., Heller G., Gignac G.A., Gonzalez-Espinoza R., Anand A., Tanaka E., Lilja H., Schwartz L., Larson S., Fleisher M., Scher H.I. 2007. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin. Cancer Res. 13:7053–7058 10.1158/1078-0432.CCR-07-1506 [DOI] [PubMed] [Google Scholar]
- de Bono J.S., Scher H.I., Montgomery R.B., Parker C., Miller M.C., Tissing H., Doyle G.V., Terstappen L.W., Pienta K.J., Raghavan D. 2008. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14:6302–6309 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- de Cremoux P., Extra J.M., Denis M.G., Pierga J.Y., Bourstyn E., Nos C., Clough K.B., Boudou E., Martin E.C., Müller A., et al. 2000. Detection of MUC1-expressing mammary carcinoma cells in the peripheral blood of breast cancer patients by real-time polymerase chain reaction. Clin. Cancer Res. 6:3117–3122 [PubMed] [Google Scholar]
- Fehm T., Sagalowsky A., Clifford E., Beitsch P., Saboorian H., Euhus D., Meng S., Morrison L., Tucker T., Lane N., et al. 2002. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin. Cancer Res. 8:2073–2084 [PubMed] [Google Scholar]
- Fidler I.J. 1973. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur. J. Cancer. 9:223–227 [DOI] [PubMed] [Google Scholar]
- Galanzha E.I., Shashkov E.V., Kelly T., Kim J.W., Yang L., Zharov V.P. 2009a. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells. Nat. Nanotechnol. 4:855–860 10.1038/nnano.2009.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanzha E.I., Shashkov E.V., Spring P.M., Suen J.Y., Zharov V.P. 2009b. In vivo, noninvasive, label-free detection and eradication of circulating metastatic melanoma cells using two-color photoacoustic flow cytometry with a diode laser. Cancer Res. 69:7926–7934 10.1158/0008-5472.CAN-08-4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne P.R., Noshari J., Anderson T.J., Becker F.F. 2009. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 30:1388–1398 10.1002/elps.200800373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler R., Rosenberg R., Fuehrer K., Dahm M., Nekarda H., Siewert J.R. 2003. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 162:149–155 [DOI] [PubMed] [Google Scholar]
- Ghossein R.A., Scher H.I., Gerald W.L., Kelly W.K., Curley T., Amsterdam A., Zhang Z.F., Rosai J. 1995. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J. Clin. Oncol. 13:1195–1200 [DOI] [PubMed] [Google Scholar]
- Glaves D. 1983. Correlation between circulating cancer cells and incidence of metastases. Br. J. Cancer. 48:665–673 10.1038/bjc.1983.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky V.V., Glinsky G.V., Glinskii O.V., Huxley V.H., Turk J.R., Mossine V.V., Deutscher S.L., Pienta K.J., Quinn T.P. 2003. Intravascular metastatic cancer cell homotypic aggregation at the sites of primary attachment to the endothelium. Cancer Res. 63:3805–3811 [PubMed] [Google Scholar]
- Gormally E., Caboux E., Vineis P., Hainaut P. 2007. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. 635:105–117 10.1016/j.mrrev.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Hardingham J.E., Hewett P.J., Sage R.E., Finch J.L., Nuttall J.D., Kotasek D., Dobrovic A. 2000. Molecular detection of blood-borne epithelial cells in colorectal cancer patients and in patients with benign bowel disease. Int. J. Cancer. 89:8–13 [DOI] [PubMed] [Google Scholar]
- Hayes D.F., Walker T.M., Singh B., Vitetta E.S., Uhr J.W., Gross S., Rao C., Doyle G.V., Terstappen L.W. 2002. Monitoring expression of HER-2 on circulating epithelial cells in patients with advanced breast cancer. Int. J. Oncol. 21:1111–1117 [DOI] [PubMed] [Google Scholar]
- Hayes D.F., Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Miller M.C., Matera J., Allard W.J., Doyle G.V., Terstappen L.W. 2006. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 12:4218–4224 10.1158/1078-0432.CCR-05-2821 [DOI] [PubMed] [Google Scholar]
- He W., Wang H., Hartmann L.C., Cheng J.X., Low P.S. 2007. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc. Natl. Acad. Sci. USA. 104:11760–11765 10.1073/pnas.0703875104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helzer K.T., Barnes H.E., Day L., Harvey J., Billings P.R., Forsyth A. 2009. Circulating tumor cells are transcriptionally similar to the primary tumor in a murine prostate model. Cancer Res. 69:7860–7866 10.1158/0008-5472.CAN-09-0801 [DOI] [PubMed] [Google Scholar]
- Hou J.M., Greystoke A., Lancashire L., Cummings J., Ward T., Board R., Amir E., Hughes S., Krebs M., Hughes A., et al. 2009. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am. J. Pathol. 175:808–816 10.2353/ajpath.2009.090078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E.W., Leung S.C., Yuen H.F., Chua C.W., Lee D.T., Chan K.W., Wang X., Wong Y.C. 2008. Decreased adhesiveness, resistance to anoikis and suppression of GRP94 are integral to the survival of circulating tumor cells in prostate cancer. Clin. Exp. Metastasis. 25:497–508 10.1007/s10585-008-9157-3 [DOI] [PubMed] [Google Scholar]
- Hüsemann Y., Geigl J.B., Schubert F., Musiani P., Meyer M., Burghart E., Forni G., Eils R., Fehm T., Riethmüller G., Klein C.A. 2008. Systemic spread is an early step in breast cancer. Cancer Cell. 13:58–68 10.1016/j.ccr.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Igetei R., Otegbayo J.A., Ndububa D.A., Lesi O.A., Anumudu C.I., Hainaut P., Gormally E. 2008. Detection of p53 codon 249 mutation in Nigerian patients with hepatocellular carcinoma using a novel evaluation of cell-free DNA. Ann. Hepatol. 7:339–344 [PubMed] [Google Scholar]
- Ignatiadis M., Xenidis N., Perraki M., Apostolaki S., Politaki E., Kafousi M., Stathopoulos E.N., Stathopoulou A., Lianidou E., Chlouverakis G., et al. 2007. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J. Clin. Oncol. 25:5194–5202 10.1200/JCO.2007.11.7762 [DOI] [PubMed] [Google Scholar]
- Kallergi G., Agelaki S., Kalykaki A., Stournaras C., Mavroudis D., Georgoulias V. 2008. Phosphorylated EGFR and PI3K/Akt signaling kinases are expressed in circulating tumor cells of breast cancer patients. Breast Cancer Res. 10:R80 10.1186/bcr2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.N., Riba R.D., Zacharoulis S., Bramley A.H., Vincent L., Costa C., MacDonald D.D., Jin D.K., Shido K., Kerns S.A., et al. 2005. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 438:820–827 10.1038/nature04186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Oskarsson T., Acharyya S., Nguyen D.X., Zhang X.H., Norton L., Massagué J. 2009. Tumor self-seeding by circulating cancer cells. Cell. 139:1315–1326 10.1016/j.cell.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivacic R.T., Ladanyi A., Curry D.N., Hsieh H.B., Kuhn P., Bergsrud D.E., Kepros J.F., Barbera T., Ho M.Y., Chen L.B., et al. 2004. A rare-cell detector for cancer. Proc. Natl. Acad. Sci. USA. 101:10501–10504 10.1073/pnas.0404036101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C.J., Moreno J.G., Pienta K.J., Gross S., Repollet M., O’Hara S.M., Russell T., Terstappen L.W. 2004. Apoptosis of circulating tumor cells in prostate cancer patients. Cytometry A. 62A:46–53 10.1002/cyto.a.20073 [DOI] [PubMed] [Google Scholar]
- Leary R.J., Kinde I., Diehl F., Schmidt K., Clouser C., Duncan C., Antipova A., Lee C., McKernan K., De La Vega F.M., et al. 2010. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2:ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta L.A., Kleinerman J., Saidel G.M. 1974. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res. 34:997–1004 [PubMed] [Google Scholar]
- Liotta L.A., Saidel M.G., Kleinerman J. 1976. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 36:889–894 [PubMed] [Google Scholar]
- Louha M., Poussin K., Ganne N., Zylberberg H., Nalpas B., Nicolet J., Capron F., Soubrane O., Vons C., Pol S., et al. 1997. Spontaneous and iatrogenic spreading of liver-derived cells into peripheral blood of patients with primary liver cancer. Hepatology. 26:998–1005 10.1002/hep.510260430 [DOI] [PubMed] [Google Scholar]
- Lu J., Fan T., Zhao Q., Zeng W., Zaslavsky E., Chen J.J., Frohman M.A., Golightly M.G., Madajewicz S., Chen W.T. 2010. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int. J. Cancer. 126:669–683 10.1002/ijc.24814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350:2129–2139 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- Maheswaran S., Sequist L.V., Nagrath S., Ulkus L., Brannigan B., Collura C.V., Inserra E., Diederichs S., Iafrate A.J., Bell D.W., et al. 2008. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 359:366–377 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrinucci D., Bethel K., Bruce R.H., Curry D.N., Hsieh B., Humphrey M., Krivacic R.T., Kroener J., Kroener L., Ladanyi A., et al. 2007. Case study of the morphologic variation of circulating tumor cells. Hum. Pathol. 38:514–519 10.1016/j.humpath.2006.08.027 [DOI] [PubMed] [Google Scholar]
- Masuda T.A., Kataoka A., Ohno S., Murakami S., Mimori K., Utsunomiya T., Inoue H., Tsutsui S., Kinoshita J., Masuda N., et al. 2005. Detection of occult cancer cells in peripheral blood and bone marrow by quantitative RT-PCR assay for cytokeratin-7 in breast cancer patients. Int. J. Oncol. 26:721–730 [PubMed] [Google Scholar]
- Mejean A., Vona G., Nalpas B., Damotte D., Brousse N., Chretien Y., Dufour B., Lacour B., Bréchot C., Paterlini-Bréchot P. 2000. Detection of circulating prostate derived cells in patients with prostate adenocarcinoma is an independent risk factor for tumor recurrence. J. Urol. 163:2022–2029 10.1016/S0022-5347(05)67621-5 [DOI] [PubMed] [Google Scholar]
- Meng S., Tripathy D., Shete S., Ashfaq R., Haley B., Perkins S., Beitsch P., Khan A., Euhus D., Osborne C., et al. 2004. HER-2 gene amplification can be acquired as breast cancer progresses. Proc. Natl. Acad. Sci. USA. 101:9393–9398 10.1073/pnas.0402993101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed H., Murray M., Turner J.N., Caggana M. 2009. Isolation of tumor cells using size and deformation. J. Chromatogr. A. 1216:8289–8295 10.1016/j.chroma.2009.05.036 [DOI] [PubMed] [Google Scholar]
- Molnar B., Ladanyi A., Tanko L., Sréter L., Tulassay Z. 2001. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin. Cancer Res. 7:4080–4085 [PubMed] [Google Scholar]
- Morgan T.M., Lange P.H., Vessella R.L. 2007. Detection and characterization of circulating and disseminated prostate cancer cells. Front. Biosci. 12:3000–3009 10.2741/2290 [DOI] [PubMed] [Google Scholar]
- Müller V., Stahmann N., Riethdorf S., Rau T., Zabel T., Goetz A., Jänicke F., Pantel K. 2005. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin. Cancer Res. 11:3678–3685 10.1158/1078-0432.CCR-04-2469 [DOI] [PubMed] [Google Scholar]
- Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., et al. 2007. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 450:1235–1239 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa T., Nutahara K., Higashihara E. 2009. Prognostic significance of circulating tumor cells in patients with hormone refractory prostate cancer. J. Urol. 181:1091–1097 10.1016/j.juro.2008.11.015 [DOI] [PubMed] [Google Scholar]
- Pachmann K., Camara O., Kavallaris A., Schneider U., Schünemann S., Höffken K. 2005a. Quantification of the response of circulating epithelial cells to neodadjuvant treatment for breast cancer: a new tool for therapy monitoring. Breast Cancer Res. 7:R975–R979 10.1186/bcr1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachmann K., Clement J.H., Schneider C.P., Willen B., Camara O., Pachmann U., Höffken K. 2005b. Standardized quantification of circulating peripheral tumor cells from lung and breast cancer. Clin. Chem. Lab. Med. 43:617–627 10.1515/CCLM.2005.107 [DOI] [PubMed] [Google Scholar]
- Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 304:1497–1500 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- Pantel K., Brakenhoff R.H., Brandt B. 2008. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer. 8:329–340 10.1038/nrc2375 [DOI] [PubMed] [Google Scholar]
- Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. 2004. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA. 101:13306–13311 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou E., Davilas E., Sotiriou V., Georgakopoulos E., Georgakopoulou S., Koliopanos A., Aggelakis F., Dardoufas K., Agnanti N.J., Karydas I., Nasioulas G. 2006. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Ann. NY Acad. Sci. 1075:235–243 10.1196/annals.1368.032 [DOI] [PubMed] [Google Scholar]
- Paris P.L., Kobayashi Y., Zhao Q., Zeng W., Sridharan S., Fan T., Adler H.L., Yera E.R., Zarrabi M.H., Zucker S., et al. 2009. Functional phenotyping and genotyping of circulating tumor cells from patients with castration resistant prostate cancer. Cancer Lett. 277:164–173 10.1016/j.canlet.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Paterlini-Brechot P., Benali N.L. 2007. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 253:180–204 10.1016/j.canlet.2006.12.014 [DOI] [PubMed] [Google Scholar]
- Pierga J.Y., Bidard F.C., Mathiot C., Brain E., Delaloge S., Giachetti S., de Cremoux P., Salmon R., Vincent-Salomon A., Marty M. 2008. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin. Cancer Res. 14:7004–7010 10.1158/1078-0432.CCR-08-0030 [DOI] [PubMed] [Google Scholar]
- Polyak K., Weinberg R.A. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Putz E., Witter K., Offner S., Stosiek P., Zippelius A., Johnson J., Zahn R., Riethmüller G., Pantel K. 1999. Phenotypic characteristics of cell lines derived from disseminated cancer cells in bone marrow of patients with solid epithelial tumors: establishment of working models for human micrometastases. Cancer Res. 59:241–248 [PubMed] [Google Scholar]
- Reid A.H., Attard G., Danila D.C., Oommen N.B., Olmos D., Fong P.C., Molife L.R., Hunt J., Messiou C., Parker C., et al. 2010. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J. Clin. Oncol. 28:1489–1495 10.1200/JCO.2009.24.6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S., Fritsche H., Müller V., Rau T., Schindlbeck C., Rack B., Janni W., Coith C., Beck K., Jänicke F., et al. 2007. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13:920–928 10.1158/1078-0432.CCR-06-1695 [DOI] [PubMed] [Google Scholar]
- Schardt J.A., Meyer M., Hartmann C.H., Schubert F., Schmidt-Kittler O., Fuhrmann C., Polzer B., Petronio M., Eils R., Klein C.A. 2005. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell. 8:227–239 10.1016/j.ccr.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Scher H.I., Jia X., de Bono J.S., Fleisher M., Pienta K.J., Raghavan D., Heller G. 2009. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 10:233–239 10.1016/S1470-2045(08)70340-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., DeAngelis G., Eltze E., Gockel I., Semjonow A., Brandt B. 2006. Asynchronous growth of prostate cancer is reflected by circulating tumor cells delivered from distinct, even small foci, harboring loss of heterozygosity of the PTEN gene. Cancer Res. 66:8959–8965 10.1158/0008-5472.CAN-06-1722 [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H., Chun F.K., Lange I., Carpenter S., Gottberg M., Erbersdobler A., Friedrich M.G., Huland H., Pantel K. 2007. Detection of tumor-specific DNA in blood and bone marrow plasma from patients with prostate cancer. Int. J. Cancer. 120:1465–1471 10.1002/ijc.22470 [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H., Alix-Panabières C., Müller I., Letang N., Vendrell J.P., Rebillard X., Pantel K. 2009. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin. Cancer Res. 15:1032–1038 10.1158/1078-0432.CCR-08-1910 [DOI] [PubMed] [Google Scholar]
- Sieuwerts A.M., Kraan J., Bolt J., van der Spoel P., Elstrodt F., Schutte M., Martens J.W., Gratama J.W., Sleijfer S., Foekens J.A. 2009. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 101:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson S.J., Vachula M., Kennedy M.J., Kaizer H., Coon J.S., Ghalie R., Williams S., Van Epps D. 1995. Detection of tumor cells in the bone marrow, peripheral blood, and apheresis products of breast cancer patients using flow cytometry. Exp. Hematol. 23:1062–1068 [PubMed] [Google Scholar]
- Stott S.L., Lee R.J., Nagrath S., Yu M., Miyamoto D.T., Ulkus L., Inserra E.J., Ulman M., Springer S., Nakamura Z., et al. 2010a. Isolation and characterization of circulating tumor cells from localized and metastatic prostate cancer patients. Sci. Transl. Med. 2:25ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott S.L., Hsu C.H., Tsukrov D.I., Yu M., Miyamoto D.T., Waltman B.A., Rothenberg S.M., Shah A.M., Smas M.E., Korir G.K., et al. 2010b. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA. 107:18392–18397 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swennenhuis J.F., Tibbe A.G., Levink R., Sipkema R.C., Terstappen L.W. 2009. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry A. 75:520–527 [DOI] [PubMed] [Google Scholar]
- Talasaz A.H., Powell A.A., Huber D.E., Berbee J.G., Roh K.H., Yu W., Xiao W., Davis M.M., Pease R.F., Mindrinos M.N., et al. 2009. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA. 106:3970–3975 10.1073/pnas.0813188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.J., Yobas L., Lee G.Y., Ong C.N., Lim C.T. 2009. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices. 11:883–892 10.1007/s10544-009-9305-9 [DOI] [PubMed] [Google Scholar]
- Tewes M., Aktas B., Welt A., Mueller S., Hauch S., Kimmig R., Kasimir-Bauer S. 2009. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res. Treat. 115:581–590 10.1007/s10549-008-0143-x [DOI] [PubMed] [Google Scholar]
- Tkaczuk K.H., Goloubeva O., Tait N.S., Feldman F., Tan M., Lum Z.P., Lesko S.A., Van Echo D.A., Ts’o P.O. 2008. The significance of circulating epithelial cells in Breast Cancer patients by a novel negative selection method. Breast Cancer Res. Treat. 111:355–364 10.1007/s10549-007-9771-9 [DOI] [PubMed] [Google Scholar]
- Tong X., Yang L., Lang J.C., Zborowski M., Chalmers J.J. 2007. Application of immunomagnetic cell enrichment in combination with RT-PCR for the detection of rare circulating head and neck tumor cells in human peripheral blood. Cytometry B Clin. Cytom. 72:310–323 [DOI] [PubMed] [Google Scholar]
- Tsuji T., Ibaragi S., Shima K., Hu M.G., Katsurano M., Sasaki A., Hu G.F. 2008. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res. 68:10377–10386 10.1158/0008-5472.CAN-08-1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T., Ibaragi S., Hu G.F. 2009. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 69:7135–7139 10.1158/0008-5472.CAN-09-1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona G., Sabile A., Louha M., Sitruk V., Romana S., Schütze K., Capron F., Franco D., Pazzagli M., Vekemans M., et al. 2000. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am. J. Pathol. 156:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H., Pfister T.D., Parchment R.E., Kummar S., Rubinstein L., Evrard Y.A., Gutierrez M.E., Murgo A.J., Tomaszewski J.E., Doroshow J.H., Kinders R.J. 2010. Monitoring drug-induced gammaH2AX as a pharmacodynamic biomarker in individual circulating tumor cells. Clin. Cancer Res. 16:1073–1084 10.1158/1078-0432.CCR-09-2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.P., Eisenberger M.A., Carducci M.A., Partin A.W., Scher H.I., Ts’o P.O. 2000. Identification and characterization of circulating prostate carcinoma cells. Cancer. 88:2787–2795 [DOI] [PubMed] [Google Scholar]
- Watanabe S. 1954. The metastasizability of tumor cells. Cancer. 7:215–223 [DOI] [PubMed] [Google Scholar]
- Weigelt B., Bosma A.J., Hart A.A., Rodenhuis S., van’t Veer L.J. 2003. Marker genes for circulating tumour cells predict survival in metastasized breast cancer patients. Br. J. Cancer. 88:1091–1094 10.1038/sj.bjc.6600868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weight R.M., Dale P.S., Viator J.A. 2009. Detection of circulating melanoma cells in human blood using photoacoustic flowmetry. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009:106–109 [DOI] [PubMed] [Google Scholar]
- Willipinski-Stapelfeldt B., Riethdorf S., Assmann V., Woelfle U., Rau T., Sauter G., Heukeshoven J., Pantel K. 2005. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin. Cancer Res. 11:8006–8014 10.1158/1078-0432.CCR-05-0632 [DOI] [PubMed] [Google Scholar]
- Wu C.H., Lin S.R., Yu F.J., Wu D.C., Pan Y.S., Hsieh J.S., Huang S.Y., Wang J.Y. 2006. Development of a high-throughput membrane-array method for molecular diagnosis of circulating tumor cells in patients with gastric cancers. Int. J. Cancer. 119:373–379 10.1002/ijc.21856 [DOI] [PubMed] [Google Scholar]
- Wülfing P., Borchard J., Buerger H., Heidl S., Zänker K.S., Kiesel L., Brandt B. 2006. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin. Cancer Res. 12:1715–1720 10.1158/1078-0432.CCR-05-2087 [DOI] [PubMed] [Google Scholar]
- Xi L., Nicastri D.G., El-Hefnawy T., Hughes S.J., Luketich J.D., Godfrey T.E. 2007. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin. Chem. 53:1206–1215 10.1373/clinchem.2006.081828 [DOI] [PubMed] [Google Scholar]
- Yang L., Lang J.C., Balasubramanian P., Jatana K.R., Schuller D., Agrawal A., Zborowski M., Chalmers J.J. 2009. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol. Bioeng. 102:521–534 10.1002/bit.22066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.A., Park S., Lee S.H., Kim J.H., Lee J.S. 2009. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J. Mol. Diagn. 11:182–185 10.2353/jmoldx.2009.080098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti-Dällenbach R., Wight E., Fan A.X., Lapaire O., Hahn S., Holzgreve W., Zhong X.Y. 2008. Positive correlation of cell-free DNA in plasma/serum in patients with malignant and benign breast disease. Anticancer Res. 28:921–925 [PubMed] [Google Scholar]
- Zhang Y., Lu H., Dazin P., Kapila Y. 2004. Squamous cell carcinoma cell aggregates escape suspension-induced, p53-mediated anoikis: fibronectin and integrin alphav mediate survival signals through focal adhesion kinase. J. Biol. Chem. 279:48342–48349 10.1074/jbc.M407953200 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Han L., Cao L., Liang X., Liu Y., Liu H., Du J., Qu Z., Zhu C., Liu S., et al. 2008. Aggregation formation mediated anoikis resistance of BEL7402 hepatoma cells. Folia Histochem. Cytobiol. 46:331–336 10.2478/v10042-008-0042-3 [DOI] [PubMed] [Google Scholar]
- Zheng S., Lin H., Liu J.Q., Balic M., Datar R., Cote R.J., Tai Y.C. 2007. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J. Chromatogr. A. 1162:154–161 10.1016/j.chroma.2007.05.064 [DOI] [PubMed] [Google Scholar]