FGF signaling in neurons is regulated by Survival Motor Neuron, a component of a complex that regulates snRNP biogenesis and FGF receptor expression.
Abstract
Spinal muscular atrophy (SMA), a devastating neurodegenerative disorder characterized by motor neuron loss and muscle atrophy, has been linked to mutations in the Survival Motor Neuron (SMN) gene. Based on an SMA model we developed in Drosophila, which displays features that are analogous to the human pathology and vertebrate SMA models, we functionally linked the fibroblast growth factor (FGF) signaling pathway to the Drosophila homologue of SMN, Smn. Here, we characterize this relationship and demonstrate that Smn activity regulates the expression of FGF signaling components and thus FGF signaling. Furthermore, we show that alterations in FGF signaling activity are able to modify the neuromuscular junction defects caused by loss of Smn function and that muscle-specific activation of FGF is sufficient to rescue Smn-associated abnormalities.
Introduction
Spinal muscular atrophy (SMA) is an inherited neurodegenerative disease causing progressive deterioration of motor functions and loss of motor neurons (Azzouz et al., 2004). After cystic fibrosis, SMA is the most common autosomal recessive disorder in humans with an incidence of 1 in 6,000 and defines the most common genetic cause of infant mortality. SMA is caused by the loss of Survival Motor Neuron (SMN1), a ubiquitously expressed gene that encodes a key component of the SMN complex, which is essential for snRNP biogenesis. Biochemical studies established that SMN mediates the accuracy of interactions between RNA binding proteins and their target snRNAs in the cytoplasm (Massenet et al., 2002; Meister et al., 2002; Paushkin et al., 2002; Wan et al., 2005; Battle et al., 2006; Eggert et al., 2006; Zhang et al., 2008).
The human genome harbors two homologous, nearly identical genes encoding SMN, SMN1, and SMN2. However, under normal conditions, SMN1 accounts for 90% of cellular SMN expression due to a splicing mutation in SMN2 that results in the production of only a small fraction (∼10%) of full-length functional SMN (Lefebvre et al., 1997; Wolstencroft et al., 2005). Thus, though SMA is caused by mutations that impair SMN1 function, the severity of the disease is modulated by SMN2 copy number, which varies in the human population (McAndrew et al., 1997). As SMN2 copy number increases, the amount of full-length SMN protein also increases, rendering loss of SMN1 less pathogenic. Therefore, cellular processes as well as single genes capable of augmenting SMN protein activity may be therapeutically relevant. To identify such processes/targets and gain insights into fundamental aspects of SMA, several different organisms, including Drosophila, are currently being used to model this disease (Schrank et al., 1997; Miguel-Aliaga et al., 1999, 2000; Frugier et al., 2000; Hannus et al., 2000; Hsieh-Li et al., 2000; Monani et al., 2000; Owen et al., 2000; Paushkin et al., 2000; Chan et al., 2003; McWhorter et al., 2003; Rajendra et al., 2007; Chang et al., 2008; Briese et al., 2009; Kong et al., 2009).
The Drosophila genome encodes a single orthologue of SMN, the Survival motor neuron (Smn) protein, which is ubiquitously expressed and localizes to nuclear gems (Chan et al., 2003; Liu et al., 2006; Chang et al., 2008), similar to the distribution observed in vertebrates (Monani, 2005). In Drosophila, Smn loss-of-function mutations result in reduced viability and decreased motility as well as muscular atrophy in the adult thorax, phenotypes analogous to the human pathology (Chan et al., 2003; Rajendra et al., 2007; Chang et al., 2008). Moreover, neuromuscular junction (NMJ) defects are associated with both vertebrate and invertebrate models (Chan et al., 2003; Chang et al., 2008; Kariya et al., 2008). In addition to its canonical subcellular distribution, Smn is also clearly concentrated in the postsynaptic region of the larval NMJ (Chang et al., 2008) and has been reported to localize to sarcomeres of adult myofibrils (Rajendra et al., 2007). Despite this, tissue-specific reduction of Smn demonstrates that normal NMJ morphology requires Smn activity in both muscles and neurons (Chang et al., 2008). Finally, an observation of critical importance to the Drosophila model is that the morphology and the physiology of the NMJ are sensitive to levels of Smn (Chang et al., 2008; unpublished data), mirroring the SMN2 dosage dependence observed in SMA patients.
Taking advantage of the dosage sensitivity of Smn loss-of-function phenotypes, we performed systematic genetic screens to identify modifiers of Smn activity (Chang et al., 2008). Among the genes identified in this manner was the breathless locus, which encodes one of the two Drosophila FGF receptors (Glazer and Shilo, 1991). In general, the FGF pathway has been demonstrated to be involved in a diverse range of cellular and developmental processes, including proliferation, migration, differentiation, and apoptosis (Itoh and Ornitz, 2004; Huang and Stern, 2005). In Drosophila, this pathway has been demonstrated to control the development of the tracheal system (Ghabrial et al., 2003) and the musculature (Shishido et al., 1993, 1997; Beiman et al., 1996; Gisselbrecht et al., 1996; Michelson et al., 1998; Vincent et al., 1998; Schulz and Gajewski, 1999; Stathopoulos et al., 2004). In contrast, the role of FGF in the Drosophila nervous system remains poorly characterized (García-Alonso et al., 2000; Forni et al., 2004).
In this study, we investigate the relationship between Smn and several components of the FGF pathway, demonstrating a clear link between Smn and FGF. Epistasis analysis reveals that Smn regulates FGF signaling output, and molecular studies indicate that Smn activity influences FGF receptor transcript levels. Furthermore, we show that activation of FGF signaling can restore Smn-associated NMJ defects, thus raising the possibility that FGF can act as a protective modifier of SMA.
Results
The FGF signaling pathway and Smn
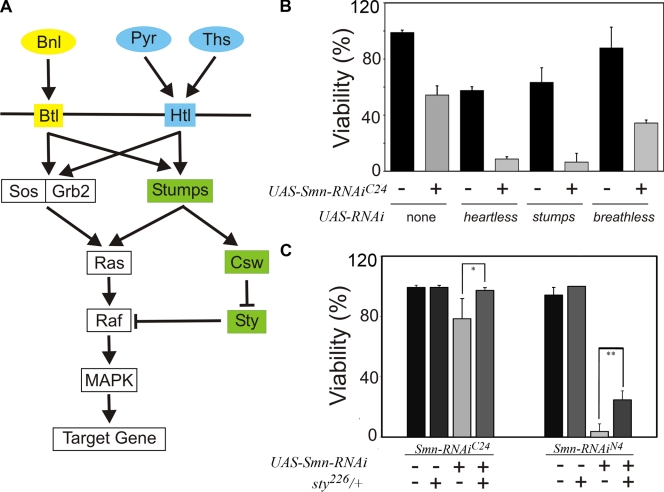
breathless (btl), which encodes one of the two known Drosophila FGF receptors, was identified in a genetic screen as a modifier of Smn-dependent lethality (Chang et al., 2008), suggesting a connection between the FGF pathway (Fig. 1 A) and Smn. We extended this finding by determining the effect of different btl mutations on Smn-dependent viability using an inducible RNAi allele of Smn, UAS-Smn-RNAiFL26B (FL26B), which displays reduced viability when ubiquitously expressed by the tubulinGAL4 (tubGAL4) driver (Chang et al., 2008). This phenotype was modified by multiple btl alleles (btlf02864, btldev1, and UAS-λbtl) as judged by our survival assay (Fig. S1, A and B). These genetic results confirm btl as a bona fide modifier of Smn loss-of-function mutations, thereby validating our initial observations.
Figure 1.
Multiple FGF pathway components modify Smn-dependent viability. (A) Schematic diagram depicting the FGF signaling pathway in Drosophila. In Drosophila, pathway activation is mediated by the two known FGF receptor orthologues, breathless (btl) and heartless (htl) (Glazer and Shilo, 1991). btl, which functions in the tracheal system, is activated by its ligand branchless (bnl) (Sutherland et al., 1996), whereas htl, which functions in the mesoderm and muscles, is activated either by the thisbe (ths) (Kerr et al., 2003) or pyramus (pyr) (Stathopoulos et al., 2004) ligands. Both receptors act through Sos-Grb2 to activate Ras/Raf/MAP kinase signaling. Additional regulation of Ras/Raf/MAP kinase signaling occurs through stumps (Vincent et al., 1998), which regulates the phosphatase corkscrew (csw) (Petit et al., 2004). In turn, Csw negatively regulates sprouty (sty), itself a negative regulator of Raf, thereby leading to MAPK activation (Jarvis et al., 2006). (B) The lethal phenotype associated with mesoderm-specific how24BGAL4-directed expression of UAS-Smn-RNAiC24 is enhanced by the reduction of the FGF signaling pathway components htl, stumps, and breathless. (C) sprouty alleles suppress the how24BGAL4 UAS-Smn-RNAi lethal phenotype. Significant differences are indicated (*, P < 0.05; **, P < 0.01).
If, as the above analysis of btl implies, FGF signaling can modulate Smn activity, we expect other genetic elements of the FGF pathway to behave as Smn modifiers as well. We chose to examine this relationship in the mesoderm, as the activity of the FGF signaling pathway has been shown to be important for the development and the maintenance of muscles (Shishido et al., 1993; Beiman et al., 1996; Gisselbrecht et al., 1996; Michelson et al., 1998; Vincent et al., 1998; Schulz and Gajewski, 1999; Stathopoulos et al., 2004). The mesoderm-specific how24BGAL4 driver was used to control expression of inducible RNAi transgenes that specifically target either of the two Drosophila FGF receptors, btl or heartless (htl), and a specific FGF signaling effector, stumps. We monitored the effects of these mutations on two additional Smn RNAi strains, UAS-Smn-RNAiC24 (C24) and UAS-Smn-RNAiN4 (N4), which, based on phenotypic analyses and Smn expression levels, are of increasing allelic strength with respect to the FL26B allele (Chang et al., 2008).
In control experiments, we observe an Smn-independent effect on viability in backgrounds in which Btl, Htl, or Stumps expression were reduced, whereas no effect on viability was observed upon removal of one copy of sprouty (sty), an inhibitor of the pathway (Fig. 1, B and C). However, when Smn activity is reduced in each of these backgrounds, a further decrease in viability is detected (Fig. 1, B and C). Moreover, loss of function for the FGF pathway antagonist sty suppresses Smn-induced lethality (Fig. 1 C). Based on these observations, we conclude that a genetic link between Smn activity and the FGF signaling cascade exists in the mesoderm.
Reduction of FGF signaling in muscles causes NMJ defects
Previous analyses established that the vast majority of mutations in genes that altered the viability of Smn loss-of-function mutations (Chang et al., 2008) were also accompanied by structural defects in the larval NMJ (Chang et al., 2008). As multiple alleles of several FGF signaling elements modify Smn-dependent lethality, we assessed the significance of this interaction at the third larval instar NMJ in several different genetic backgrounds.
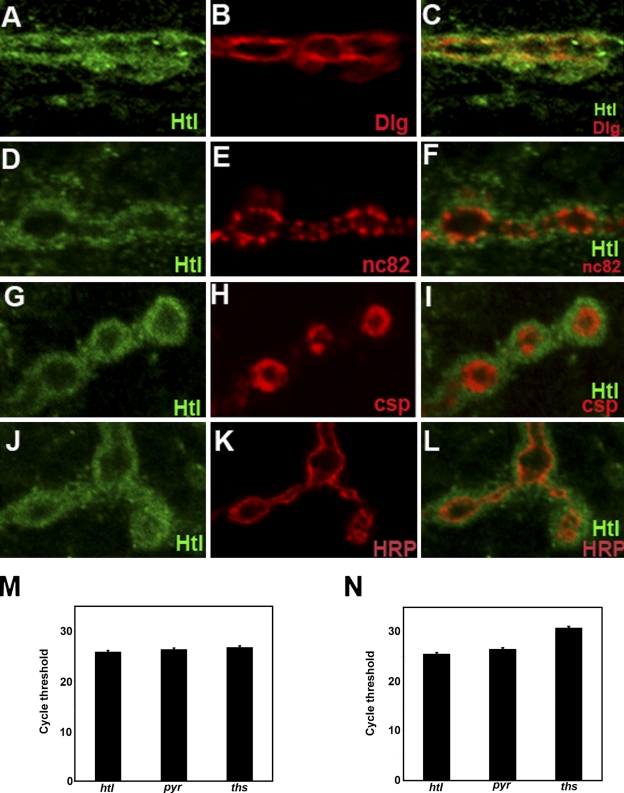
An initial examination of the distribution of Htl revealed that it is specifically expressed at the NMJ during the third instar (Fig. 2, A–L; and Fig. S2). This expression appears to be primarily in the muscle, as it coincides with the postsynaptic marker Discs Large 1 (Dlg; Fig. 2 B) and does not obviously overlap with the presynaptic marker nc82 (Bruchpilot; Fig. 2 E). This is corroborated by the localization of additional presynaptic markers, Cysteine string protein (Csp; Fig. 2 H) and anti-horseradish peroxidase (anti-HRP; Fig. 2 K), which label synaptic vesicles and neuronal membranes, respectively, and appear to be distinct from Htl expression. Together, these data suggest that Htl is predominantly postsynaptic, as indeed is Smn.
Figure 2.
Heartless localizes to the postsynaptic region of the Drosophila larval NMJ. All panels show wild-type NMJs derived from larval muscle 4. (A, D, G, and J) Htl (green) expression in the NMJ boutons. (B) Dlg (red) marks the postsynaptic region of the NMJ. (C) Htl (green) and Dlg (red) expression coincide at the larval NMJ. (F) Htl (green) and the presynaptic nc82 (red) expression do not overlap at the NMJ boutons. (I) Mutually exclusive expression of Htl (green) and presynaptic marker Csp (red) at the larval NMJ. (L) Htl (green) expression does not colocalize with presynaptic HRP staining (red). (M and N) qPCR from mRNA derived from tissues extracted from third instar larvae reveals the expression of htl and its ligands, ths and pyr, in the brain (M) and muscle (N). Bar, 5 µm.
The presence of the htl ligands, pyramus (pyr) and thisbe (ths) at the NMJ would suggest that Htl is active, but the localization of the ligands has not been previously described. In embryos, it is known that the ligands are expressed in the epithelia adjacent to the mesoderm, which expresses htl (Stathopoulos, et al., 2004). However, because antibodies that recognize the ligands are not available, we used qPCR to determine that each is expressed to varying degrees in the third instar larva (Fig. 2, M and N).
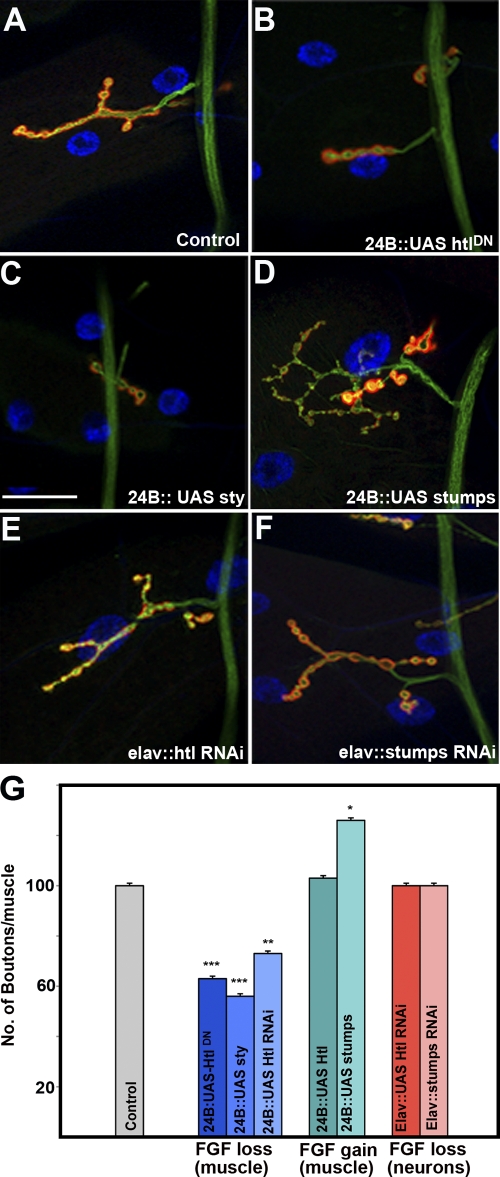
Although the above analysis is compatible with the notion that the postsynaptic expression of Htl is functionally relevant, it does not have the resolution necessary to provide reliable evidence for Htl activity. To explore this possibility directly, we examined the NMJs of larvae in which htl activity is reduced through the muscle-directed expression of an htl dominant-negative transgenic construct (how24BGAL4/UAS-htlDN; Michelson et al., 1998). This manipulation caused significant alterations in the NMJ synaptic terminals (Fig. 3 B) as compared with sibling controls (Fig. 3 A). The defects were quantified by counting the number of boutons per muscle and normalized to muscle surface area, revealing a significant (∼40%) reduction in synaptic size (Fig. 3 G and Fig. S3 A).
Figure 3.
heartless signaling regulates NMJ morphology. (A) Wild-type NMJ derived from larval muscle 4. (B) Drastic reduction in NMJ size in how24BGAL4 animals driving expression of a transgenic construct carrying a dominant-negative heartless (UAS-htlDN). The how24BGAL4 driver expresses GAL4 predominantly in the muscles. (C) Reduction in NMJ size by overexpression of Sprouty. (D) Expansion of the NMJ in animals overexpressing Stumps by how24BGAL4. (E) Neuronal overexpression of htl RNAi by elavGAL4 has no effect on NMJ size or morphology. (F) Neuronal overexpression of stumps RNAi has no effect on NMJ size and/or morphology. (G) Quantitation of bouton number/muscle in animals of the indicated genotypes, normalized to muscle surface area (MSA) as a percentage of wild type (WT). how24BGAL4/+ animals are used as controls. The ANOVA multiple comparison test was used for statistical analysis of the bouton number/muscle. P ≤ 0.05. Bar, 50 µm. n = 40.
To corroborate these observations and ensure that the structural defects we observe at the larval NMJ are associated with the FGF pathway, we modulated the activities of htl and its downstream effectors, stumps (dof) and sprouty (sty) using GAL4-inducible transgenic strains to determine whether further modifications in FGF signaling result in structural defects at the larval NMJ. Muscle-specific expression of sty, a dominant inhibitor of the pathway (how24BGAL4:UAS-sty; Fig. 3, C and G; and Fig. S3 A), or a transgenic UAS-htl RNAi construct caused a similar NMJ phenotype (how24BGAL4:UAS-htl-RNAi; Fig. 3 G and Fig. S3 A).
Though overexpression of just htl in muscles had no discernable impact on NMJ growth (how24BGAL4:UAS-htl; Fig. 3 G and Fig. S3 A), expression of stumps, which acts upstream of Ras, caused a pronounced synaptic overgrowth and an overelaboration of synaptic terminals (how24BGAL4:UAS-stumps; Fig. 3, D and G; and Fig. S3 A). Quantification of the number of synaptic boutons, which reflects synaptic size, revealed that muscle-specific stumps overexpression resulted in a 15–20% increase in the number of synaptic boutons at muscle 4 (Fig. 3 G and Fig. S3 A) and a 35% increase at the muscle 6/7 A3 synapse (Fig. S4) relative to controls. Though there was a significant effect on NMJ size, there were no gross morphological defects as the localization of pre- (anti-HRP) and postsynaptic markers (anti-Dlg) were not detectably altered, and there was no major variation in muscle size or morphology. Thus, the effects of FGF loss-of-function (Fig. 3, B, C, and G; and Fig. S3 A) are opposite to those observed for FGF gain-of-function (Fig. 3, D and G; and Fig. S3 A) in the regulation of synaptic elaboration on muscle 4, a strong genetic argument in favor of a functional role for FGF signaling at the NMJ.
In contrast, presynaptic expression of RNAi transgenes for htl and stumps using the elavGAL4 driver did not result in any measurable changes in synaptic size (elavGAL4:UAS-htl or elavGAL4:UAS-stumps; Fig. 3, E–G; and Fig. S3 A), suggesting that htl and stumps are not active presynaptically. It is important to point out, however, that we cannot exclude a presynaptic role for these elements of the FGF pathway given the possibility that RNAi in neurons may not have been effective. Despite this caveat, these experiments demonstrate that activation of the FGF pathway in the muscle is required to regulate the size of the NMJ.
FGF signaling in muscles affects responsiveness to presynaptic transmitter release
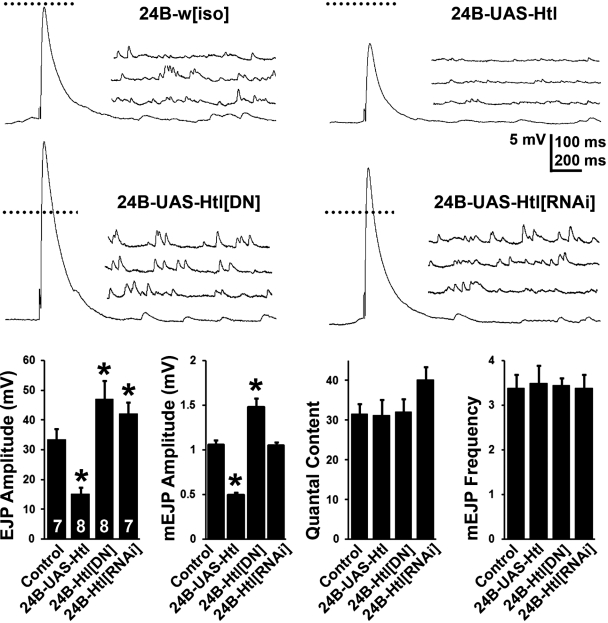
The preceding experiments show that altering FGF components in the muscle influences presynaptic morphology, which is likely accompanied by changes in either transmitter release or postsynaptic receptivity. To directly test this possibility, we performed electrophysiological measurements of evoked excitatory junction potentials (EJPs) under conditions in which we either increased or decreased FGF signaling selectively in the muscle using the how24BGAL4 driver. Perturbation of FGF signaling in muscles primarily leads to altered mEJP (miniature EJP) amplitude or quantal size (Fig. 4), a phenotype most often associated with the postsynaptic compartment (Petersen et al., 1997). Thus, increasing the expression of wild-type Htl decreases the average mEJP amplitude by more than 50%, whereas inhibition of FGF signaling through expression of the dominant-negative htl transgene increases mEJP amplitude by 50%. Interestingly, such reciprocal regulation of quantal size by htl is also mirrored in our EJP measurements (Fig. 4, A and B). As a result, quantal content (defined as the number of synaptic vesicles released per action potential and estimated by dividing the mean EJP response by the mean mEJP amplitude) remains essentially unchanged across genotypes. In addition, there are no observed changes in the frequencies of spontaneous release. It is noteworthy that changes in EJP and mEJP values are contrary to those observed for presynaptic bouton number, an observation most parsimoniously explained through feedback mechanisms that are known to operate at the larval NMJ to control its structural and functional properties (see Discussion). Therefore, the morphological changes associated with FGF signal modulation at the NMJ are accompanied by functional abnormalities.
Figure 4.
Postsynaptic FGF signaling regulates the quantal size of transmitter release. (top) Representative recordings of EJP and mEJP at 0.5 mM extracellular Ca2+ are shown for control (how24BGAL4-w[iso]), muscle expression of wild-type Htl (how24BGAL4-UAS-htl), muscle expression of a Htl dominant-negative Htl (how24BGAL4-UAS-htl[DN]), and muscle expression of an RNAi targeted against Htl (how24BGAL4-UAS-htl[RNAi]). Whereas Htl expression reduces both EJP and mEJP amplitude, Htl inhibition leads to significantly larger EJP and mEJP amplitudes. Horizontal scale bar is 100 ms for EJPs and 200 ms for mEJPs. Dotted line represents magnitude of control EJP. (bottom) Quantification of EJP amplitude, mEJP amplitude, quantal content, and mEJP frequency in the four genotypes. Both EJP and mEJP amplitude are altered after experimental perturbation in Htl signaling in the muscle. Quantal content of transmitter release, however, remains unchanged. Similarly, the frequency of spontaneous release is comparable across genotypes. Asterisks denote P < 0.01 (ANOVA). The number of animals recorded for each genotype is shown within the first graph.
Synergy of Smn and FGF signaling in muscles regulates NMJ growth
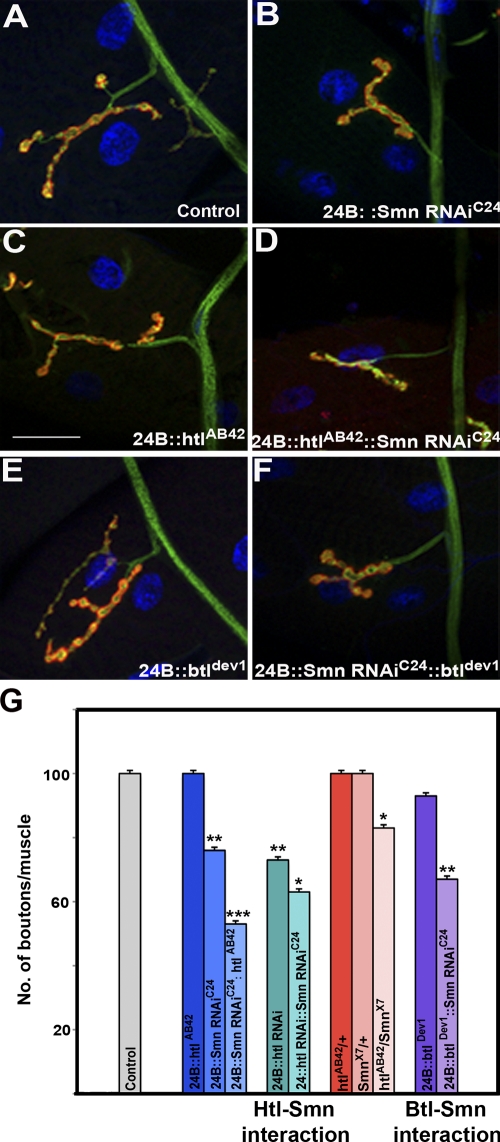
Given that the effects of loss of FGF signaling on NMJ morphology are independent of Smn, it is possible that the observed interactions between this pathway and Smn arise from the cumulative, rather than synergistic, effects of these mutations. If these interactions were due to additive effects, then the introduction of a single recessive allele of either btl or htl would not be expected to significantly alter the NMJ phenotype of Smn mutants. Therefore, we assayed whether either a strong hypomorphic allele of btl (btldev1) or a null allele of htl (htlAB42) could modify as heterozygotes the reduction of the NMJ size caused by the muscle-specific expression of the UAS-Smn-RNAiC24 allele.
Examination of larvae heterozygous for either of the two FGF receptors (how24BGAL4/btldev1 or how24BGAL4/htlAB42) revealed the introduction of these mutations had no significant effect on the NMJ size of animals from the how24BGAL4 driver genetic background (how24BGAL4/+) as assayed by the number of synaptic boutons per unit of muscle surface area (MSA; Fig. 5, A, C, E, and G; and Fig. S3 B). In contrast, when Smn function is reduced by the introduction of the UAS-Smn-RNAiC24 allele into these genetic backgrounds (how24BGAL4 UAS-Smn-RNAiC24/btldev1 and how24BGAL4 UAS-Smn-RNAiC24/htlAB42), these heterozygous FGF receptor mutations elicited a drastic decrease in synaptic size when compared with loss of Smn alone (how24BGAL4 UAS-Smn-RNAiC24/+; Fig. 5, B, D, and G; and Fig. S3 B).
Figure 5.
Synergy between Smn and FGF signaling regulates NMJ morphology. (A) Wild-type NMJ derived from larval muscle 4. how24BGAL4 expresses GAL4 predominantly in the mesoderm (muscles). (B) Reduction in NMJ size resulting from muscle-specific (how24BGAL4) reduction of Smn (UAS-Smn-RNAiC24). (C) Heterozygotes carrying the null allele htlAB42 show no effect on the number of synaptic boutons. (D) Reduction in NMJ size in how24BGAL4 UAS-Smn-RNAiC24/htlAB42. (E) how24BGAL4; btldev1/+ animals do not show any significant effects on the NMJ. (F) how24BGAL4 UAS-Smn-RNAiC24/btldev1 transheterozygous animals have reduced synapses. (G) Quantitation of bouton number/muscle in animals of indicated genotypes, normalized per muscle surface area (MSA) as a percentage of wild type (WT). how24BGAL4/+ is used as control. The ANOVA multiple comparison test was used for statistical analysis of the bouton number/muscle. P ≤ 0.05. Bar, 50 µm. n = 40. All preparations were stained with anti-HRP (red) and anti-Dlg (green). The muscle nucleus was labeled using DAPI.
Because these results indicate the link between FGF signaling and Smn activity is synergistic rather than additive, we applied a further, more stringent assay to determine the sensitivity of the NMJ to this interaction. Examination of transheterozygous larvae carrying a combination of null alleles of Smn and htl (SmnX7/htlAB42) revealed that simultaneously reducing the dosage of each locus by half leads to a statistically significant decrease in the number of synaptic boutons per unit MSA when compared with each heterozygote alone (SmnX7/+ and +/htlAB42; Fig. 5 G and Fig. S3 B). Thus, this experiment provides formal evidence that the relationship we have uncovered is synergistic in nature and is not merely due to the additive effects of two mutations that independently affect the NMJ.
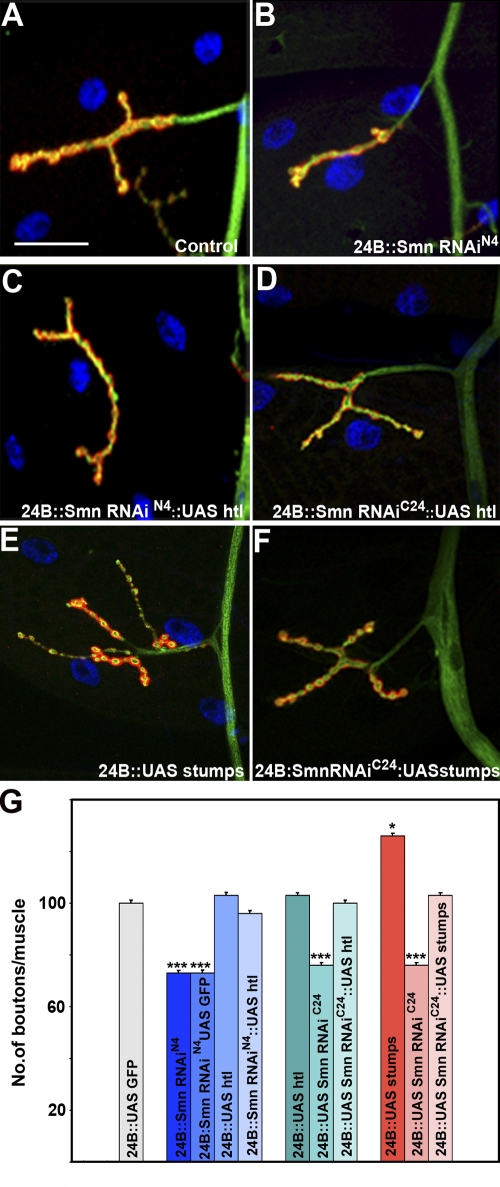
Activation of FGF signaling in muscles rescues synaptic defects caused by Smn RNAi
Because reduction of FGF signaling clearly exacerbates the NMJ defects caused by Smn loss, we examined whether activation of this pathway could reverse these effects. As shown in Fig. 6, muscle-specific overexpression of wild-type Htl completely rescues the NMJ phenotypes associated with both the UAS-Smn-RNAiC24 allele and the stronger UAS-Smn-RNAiN4 allele (Fig. 6, B–D and G; Fig. S3 C). In control crosses, expression of Stumps alone resulted in an expansion of synaptic branching (Fig. 6 E); however, as observed for Htl, expression of Stumps completely suppressed the NMJ defects observed in the how24BGAL4; UAS-Smn-RNAiC24 background (Fig. 6 F and G; and Fig. S3 C). These results suggest that Smn is situated upstream of the FGF pathway. Based on this genetic behavior, however, we could not distinguish whether Smn directly regulates htl or any other pathway component.
Figure 6.
Overexpression of Htl rescues NMJ defects caused by reduced Smn. how24BGAL4 expresses GAL4 predominantly in the mesoderm (muscles). (A) Wild type NMJ derived from larval muscle 4. (B) Reduction in NMJ size in how24BGAL4 UAS-Smn-RNAiN4/+ animals. (C) The how24BGAL4 UAS-Smn-RNAiN4/+ and (D) how24BGAL4 UAS-Smn-RNAiC24/+ NMJ size defects are rescued by overexpression of Htl (UAS-htlwt). (E) how24BGAL4/UAS-stumps individuals show an expansion in NMJ size. (F) The how24BGAL4 UAS-Smn-RNAiC24/+ NMJ size defects are rescued by overexpression of Stumps (UAS-stumps). (G) Quantitation of bouton number/muscle in animals of different genotypes, normalized per muscle surface area (MSA) as a percentage of wild type (WT). how24BGAL4; UASGFP is used as control. The ANOVA multiple comparison test was used for statistical analysis of the bouton number/muscle. P ≤ 0.05. Bar, 50 µm. n = 40. All preparations were stained with anti-HRP (red) and anti-Dlg (green). The muscle nucleus was labeled using DAPI.
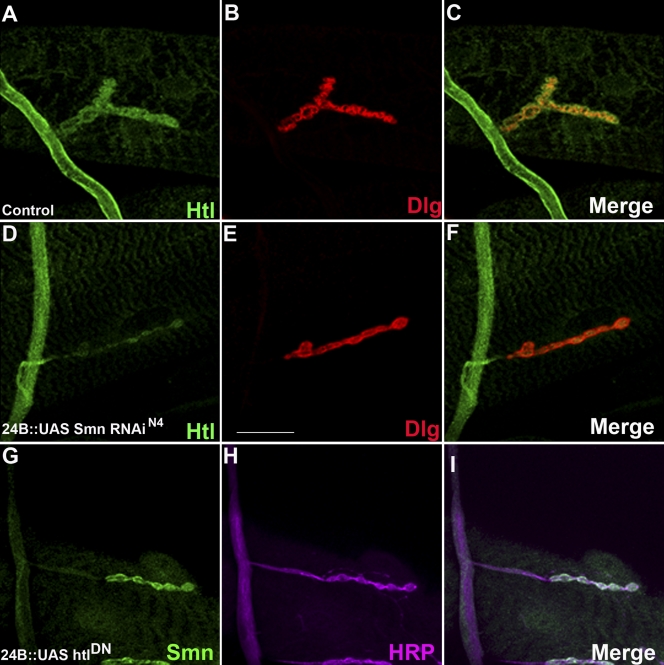
Smn regulates expression of the FGF receptor, Htl, and its downstream effector Stumps
To further explore the relationship between Smn and FGF signaling pathway components, we first tested whether reducing Smn expression impacts expression of Htl and Stumps. Using the how24B driver to direct expression of the UAS-Smn-RNAiN4 transgenic construct in the musculature resulted in a drastic postsynaptic reduction in Htl levels at the NMJ (Fig. 7, D–F) as well as a more general loss of Stumps staining throughout the muscle (Fig. S5). Conversely, we did not detect any obvious changes in the distribution or levels of the Smn protein when htl activity was reduced postsynaptically (how24BGAL4/UAS-htlDN; Fig. 7, G–I), indicating that FGF activity does not influence Smn expression. These observations corroborate the relationship observed above and indicate that Smn regulates elements of the FGF pathway.
Figure 7.
Effects of Smn on the expression of FGF signaling pathway component at the NMJ. All panels show NMJs derived from larval muscle 4 from wild-type (A–C) and how24BGAL4 UAS-Smn-RNAiN4/+ (D–F) individuals. (A) Htl (green) expression is detected in the neuron as well as in the NMJ boutons. (B) Dlg (red) marks the postsynaptic region of the NMJ. (C) An overlap of A and B showing that Htl (green) expression overlaps with Dlg (red) in the postsynaptic region of the NMJ. (D) GAL4-directed muscle-specific reduction of Smn results in a loss of Htl (green) expression in postsynaptic boutons. (E) Dlg (red) marks the postsynaptic region of the NMJ. (F) An overlap of D and E showing that Htl expression is lost from the postsynaptic portion of the NMJ in a how24BGAL4 UAS-Smn-RNAiN4/+ background. (G) Smn (green) expression in NMJ in animals expressing a dominant-negative heartless construct in the musculature (how24BGAL4; UAS-htlDN). (H) The same NMJ co-stained with HRP. (I) Overlap between G and H. Bar, 50 µm. n = 40.
Ideally, the aforementioned observations, which address the issue of autonomy in the Smn–FGF pathway interaction, would have included data derived from classical alleles of Smn. Unfortunately, however, it is practically impossible to generate the motor neuron or muscle mitotic clones necessary to examine this question. For this reason, we extended our observations to include the Drosophila wing imaginal disc, a tissue that is well suited to this type of experiment and where the distribution and expression levels of elements of the FGF pathway have been well characterized (Sato and Kornberg, 2002). An additional benefit of analyzing this relationship in the wing disc is that it allows us to determine whether the ability of Smn to regulate Htl is not specific for the NMJ but is conserved across tissues.
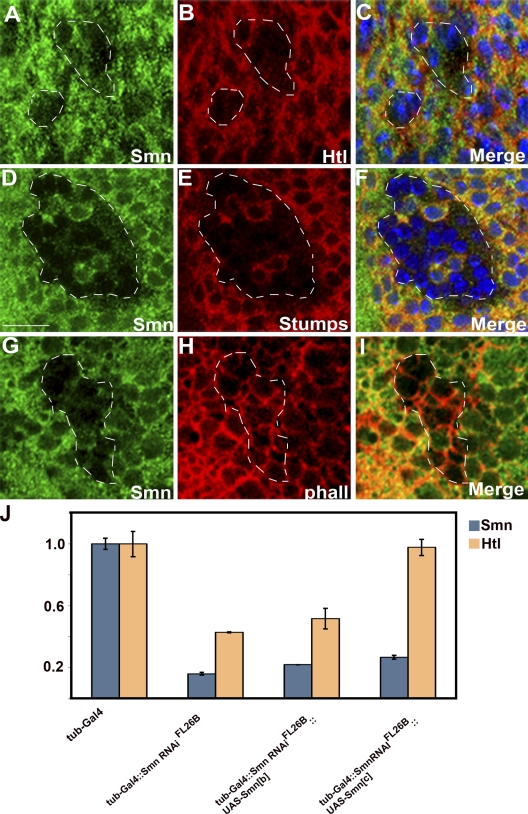
Therefore, we monitored Htl and Stumps expression in Smn loss-of-function mitotic clones that were generated using several different classical alleles of Smn. In these Smn−/− cells, both Htl and Stumps expression were lost (Fig. 8, A–F). htl expression is unaffected in stumps mutant embryos, and conversely, stumps expression is unaffected in htl mutant embryos (Vincent et al., 1998), and we thus attribute these observations in the wing imaginal disc to the loss of Smn activity. Together, these results both corroborate and extend our investigation of the epistatic relationship established between Smn and FGF signaling.
Figure 8.
Effects of Smn on the expression of FGF signaling pathway components in the wing disc. (A–I) Smn73Ao/Smn73Ao mitotic recombination clones in third instar wing imaginal discs stained for Smn (A, D, and G), Htl (B), Stumps (E), and F-actin (H). Smn, Htl, and Stumps expression were monitored using anti-Smn (green), anti-Htl, and anti-Stumps antibodies (red). In C and F, DAPI (blue) was used to identify nuclei. Notice that Htl (B) and Stumps (E) expression is not detected in Smn73Ao/Smn73Ao clones, whereas removal of Smn activity has no effect on F-actin distribution (H). (C, F, and I) Merged images of A and B, D and E, and G and H, respectively, in which nuclei are also detected (blue). Note the presence of two Smn+ cells that also express Stumps are located in the center of the clone depicted in D, E, and F. (J) Quantitative RT-PCR from third instar larval brains demonstrates that altering the level of Smn dosage results in changes in htl transcript levels. The following genotypes are depicted: w1118; tubulinGAL4 (tubGAL4)/+ (columns 1 and 2), w1118; UAS-Smn-RNAiFL26B/+; tubGAL4/+ (columns 3 and 4), w1118; UAS-Smn-RNAiFL26B/UAS-Smn-FLAGB; tubGAL4/+ (columns 5 and 6) and w1118; UAS-Smn-RNAiFL26B/+; tubGAL4/UAS-Smn-FLAGC (columns 7 and 8). Levels of Rp49 were used for normalization. Bar, 10 µm.
As Smn is required for basic RNA metabolism, removal of its activity might be expected to affect expression of multiple proteins. Therefore, to test the specificity of the molecular relationship between Smn and FGF, we monitored the effects of loss of Smn function on the expression of F-actin and the transcription factor, Cut, a gene whose expression overlaps that of Htl in the developing wing. Our results indicate that removal of Smn activity had no effect on the distribution or levels of either cytoskeletal actin (Fig. 8, G–I) or Cut (not depicted) in the wing imaginal disc. Consistent with this notion, reduction of Smn at the NMJ had no discernable effect on Dlg expression (Fig. 7, D–F).
RNAi-induced knockdown of Smn affects htl transcript levels
Having established that reduction of Smn at the NMJ or removal of Smn activity in the wing imaginal disc reduces or eliminates Htl expression, respectively (Fig. 7 D, Fig. 8 B), indicating that the SMN–FGF relationship is not specific for the NMJ, we were interested in exploring the underlying mechanism. Given the role of Smn in snRNP biogenesis, it is possible that reduction of Smn could alter levels of htl transcript, so we tested this using mRNA isolated from third larval instar brains. As shown in Fig. 8 J, knockdown of Smn (tubulinGAL4::UAS-Smn-RNAiFL26B) results in Smn transcript levels being reduced to 16% of wild type (tubulinGAL4). Concomitantly, htl transcript is reduced to 45% of the wild-type value. Interestingly, coexpression in the aforementioned genetic background of two independent insertions of Smn transgenes (UAS-Smn-FLAGB and UAS-Smn-FLAGC) that partially rescue Smn lethality (Chang et al., 2008) results in slight increases in Smn transcripts. It is noteworthy that, depending on the construct used, we observed significantly different changes in htl levels. Remarkably, expression of UAS-Smn-FLAGC, which led to only a slightly higher increase in Smn levels (27% of wild type) when compared with the expression of UAS-Smn-FLAGB (21% of wild type), fully restored htl transcript levels to the wild-type value, as opposed to the small rescue effect we see with UAS-Smn-FLAGB. These results demonstrate that reduction of Smn activity acts to modulate the levels of htl mRNA. Furthermore, it appears that a small increase in Smn may lead to a large change in the levels of htl, which in turn influence the survival of the animal.
Discussion
Given the variability of the SMA phenotype and the proven relationship between the severity of the disease and small changes in wild-type SMN activity, there is a significant possibility that any modifiers of SMN activity, either direct or indirect, will have therapeutic value. To systematically explore the genome for genes that are capable of modulating SMN function in vivo, we took advantage of the existence of an SMA model offered by Drosophila to search for Smn genetic interactors (Chang et al., 2008). The model we developed is based on the lethality and an associated neuromuscular junction phenotype linked to loss of Smn function, a phenotype remarkably similar to the NMJ phenotype reported for human patients (Kariya et al., 2008). Though the role of SMN in biogenesis of snRNPs has been well documented, its regulators and downstream effectors have not been systematically delineated, nor has the link between mutations in SMN and the specific loss of motor neurons seen in SMA patients been uncovered. It may be the case that the specificity of this phenotype is reflective of either specialized SMN functions at the NMJ or a particular sensitivity of motor neurons to the loss of SMN activity (McWhorter et al., 2003; Carrel et al., 2006; Kariya et al., 2008; Murray et al., 2008, 2010; Kong et al., 2009). Among the genes our genetic strategy revealed as Smn loss of function modifiers was breathless, encoding an FGF receptor, thus establishing a link between Smn and the FGF pathway (Chang et al., 2008).
Importantly, in addition to this link, we also found that FGF signaling is independently involved in NMJ morphogenesis, a function demonstrated in vertebrates (Fox et al., 2007) but not previously attributed to this pathway in Drosophila despite extensive characterization of its essential role in branching morphogenesis of the tracheal system, migration of multiple cell types, as well as the proper patterning of the mesoderm (Shishido et al., 1993, 1997; Beiman et al., 1996; Gisselbrecht et al., 1996; Michelson et al., 1998; Vincent et al., 1998; Schulz and Gajewski, 1999; Ghabrial et al., 2003; Stathopoulos et al., 2004). The morphological effects we observe, caused by the modulation of several pathway elements, plainly reveal an involvement of FGF signaling at the NMJ, a role confirmed by the electrophysiological analyses. The down-regulation of FGF signals in muscle results in a reduction of bouton numbers and is associated with increased mEJP amplitudes. The opposite effect is observed when FGF signaling is increased in muscles, suggesting that FGF signaling inversely regulates quantal size. Thus, FGF perturbation in muscle alters both presynaptic growth and specific aspects of synaptic transmission. These observations imply the existence of functional trans-synaptic homeostatic mechanisms, which have been previously shown to compensate for similar changes by increasing presynaptic bouton numbers and transmitter release (Davis et al., 1998; Sigrist et al., 2000, 2002; Menon et al., 2004). However, in this specific instance, only synaptic growth (bouton number) but not transmitter release (quantal content) is affected, the precise mechanisms for which remain unclear. Moreover, the fact that mEJP amplitudes are affected suggests that postsynaptic receptivity to glutamate release from the presynapse is altered. Similar quantal size phenotypes have been observed in several instances previously. For instance, postsynaptic PKA and NF-κB are known to regulate quantal size (Davis et al., 1998; Heckscher et al., 2007) through changes in DGluRs. Directly altering the expression of various GluR subunits also predictably influences quantal size (Petersen et al., 1997; Marrus et al., 2004; Featherstone et al., 2005). The genetic interaction we have demonstrated between FGF and Smn can be described as an epistatic relationship in which the FGF pathway functions downstream of Smn and is consistent with the observation that neuromuscular defects associated with loss of Smn function in muscle can be rescued by muscle-specific activation of FGF signaling. Intriguingly, the relationship we describe here between Smn and FGF is valid beyond the NMJ, as loss of Smn function genetic mosaics in the wing disc clearly result in the down-regulation of FGF signaling. Although the precise molecular mechanism underlying this relationship is still elusive, Smn activity affects transcript and protein levels of the FGF receptor, as well as the expression of additional elements of the FGF pathway. Whether this defines a cascade of interrelated events or whether each of these changes reflects an independent Smn-related regulatory event remains to be determined. Given the fact that Smn mutants in Drosophila display altered postsynaptic currents and severely compromised postsynaptic receptor clustering in muscles (Chan et al., 2003), it is conceivable that FGF signaling represents a link between Smn activity and postsynaptic glutamate receptor levels.
Here it should be noted that a link between SMN and the FGF pathway has been suggested by a series of studies in vertebrates where a molecular interaction between an FGF-2 isoform and the SMN protein has been described (Claus et al., 2003, 2004; Bruns et al., 2009).These studies raise the possibility that FGF-2 may negatively interfere with SMN complex function through SMN itself. Such observations would, on first appearance, suggest that the epistatic relationship between SMN and FGF signaling in vertebrate cells may be the reverse of what we observe in Drosophila. In point of fact however, the differences in the experimental parameters and approaches between these studies do not allow meaningful comparisons.
An important question raised by the above phenotypic analyses is whether the abnormalities associated with FGF and/or Smn perturbations reflect developmental or maintenance issues. It may be the case that the larval system in Drosophila is not ideally suited to differentiate between these alternatives as larval tissue is destined to undergo programmed cell death (histolysis) during metamorphosis. One advantage that flies do offer, however, is the ability to dissociate the development of the adult neuromuscular system from its maintenance as the entirety of its development occurs during the pupal stage, before emergence of the adult (Fernandes and Keshishian, 1998; Consoulas et al., 2002; Hebbar and Fernandes, 2004). Thus, the Drosophila pupa/adult may provide a platform to address these issues, as Drosophila displays Smn-dependent adult phenotypes (unpublished data; Rajendra et al., 2007). In light of the relationship we established between Smn and FGF signaling and the known involvement of FGF signaling in the development of both the larval and adult musculature (Emori and Saigo, 1993; Shishido et al., 1993, 1997; Vincent et al., 1998; Imam et al., 1999; Schulz and Gajewski, 1999; Stathopoulos et al., 2004; Dutta et al., 2005; Wilson et al., 2005; Kadam et al., 2009), it will be particularly interesting to examine the effects of modulating FGF activity on the aforementioned processes. Such studies may be of particular relevance to SMA where it is quite difficult to discern the developmental consequences of SMN loss in humans, as neurodegenerative symptoms displayed by patients may obscure basic problems resulting from altered developmental programs such as neuronal pathfinding, initial NMJ formation, etc (Simic et al., 2008; Liu et al., 2010).
In vertebrates, synaptic development and maintenance use at least three distinct signaling mechanisms: the TGF-β, wingless, and FGF pathways. In Drosophila, it is noteworthy that the first two have been demonstrated to function in a similar fashion at the NMJ (Packard et al., 2002; McCabe et al., 2003). Remarkably, our genetic screens involving Smn have identified elements of all three of these pathways as modifiers of Smn-related phenotypes (Chang, et al., 2008; unpublished data). We consider these connections particularly significant as they raise the possibility that Smn may serve as a node, integrating signaling events crucial for NMJ function, potentially leaving this structure particularly vulnerable to the loss of Smn. Though further correspondence between the Drosophila model and the human condition remains to be determined, the Smn–FGF relationship we observe in Drosophila raises the possibility that pharmacological manipulation of FGF signals might mitigate SMN motor neuron–related abnormalities.
Materials and methods
Drosophila stocks and culture
All fly stocks were maintained on standard fly medium at 25°C. The btldev1, htlAB42, P{w+mC=UAS-htl.DN.M}33-B40; P{w+mC=UAS-htl.DN.M}33-B61 (UAS-htlDN), P{w+mC=UAS-htl.M}YYDFR-F16 (UAS-htlwt), styΔ5, and sty226 alleles were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). The btlf02864 allele was from the Exelixis collection at Harvard Medical School (Boston, MA), and the UAS-RNAi-btl27106, UAS-RNAi-htl6692, UAS-RNAi-stumps21317, and UAS-RNAi-ths24536 alleles were from the Vienna Drosophila RNAi Center (Vienna, Austria). The UAS-λbtl, UAS-sty, and UAS-stumps (UAS-dof) transgenic lines were gifts from Denise Montell (Johns Hopkins University School of Medicine, Baltimore, MD; Cousins et al., 1996; Lee et al., 1996), Mark Krasnow (Stanford University School of Medicine, Stanford, CA; Sutherland et al., 1996), and Maria Leptin (European Molecular Biology Laboratory, Heidelberg, Germany; Vincent et al., 1998), respectively. UAS-FLAG-Smn, UAS-Smn-RNAiC24, UAS-Smn-RNAiFL26B, and UAS-Smn-RNAiN4 transgenic strains were used to modulate the Smn expression level (Chang et al., 2008). The actGAL4 (Ito et al., 1997), elavGAL4 (Luo et al., 1994), how24B-GAL4, ptcGAL4 (Brand and Perrimon, 1993), and tubGAL4 (Lee and Luo, 1999) strains were used to drive the genes placed under the UAS promoter.
Viability assay
Three driver GAL4 males were crossed with four UAS transgenic females and cultured on standard fly media overnight. Flies were transferred to fresh media and allowed to lay eggs for 2 d. Adults were subsequently discarded and the progeny were cultured at 25°C. For all crosses, the percentage of viability was calculated as (1) the total number of adults divided by the total number of pupae (defined as Adults); or (2) the total number of dead pupae 4–5 d after puparium formation plus the number of individuals that reached adult stages divided by the total number of pupae (defined as Late Pupal Viability). Viability was calculated based on four independent crosses for each genotype.
Antibody staining
NMJ preparation and analysis: third instar larvae were dissected in cold 1x phosphate-buffered saline (PBS) and fixed at room temperature (RT) for 20 min in 4% paraformaldehyde (PFA). The samples were washed in 0.1% Triton X-100 in PBS (PTX) and incubated overnight at 4°C with primary antibody. The primary antibody was washed off with PTX at RT. The samples were incubated at RT with secondary antibody for 90 min. This was followed by PTX wash, and the tissues were mounted in Vectashield mounting media with DAPI (Vector Laboratories). Bouton numbers were counted using a microscope (TE2000; Nikon), based on the Discs large and anti-HRP staining in the A3 segment muscle 4 as indicated. The muscle area for every animal was measured, and no significant difference was observed among different genotypes. At least 20–25 animals of each genotype were dissected for the bouton analysis. The ANOVA multiple comparison test was used for statistical analysis of the bouton number/muscle. The images were pseudo-colored using Adobe Photoshop CS2 (v9.0.2). Wing imaginal disc preparation and analysis: third instar larvae were dissected and fixed as described previously (Kankel et al., 2004). Discs were stained at RT with the following primary antibodies in PBS Triton X-100 (PBSTx): mouse anti-Smn (Chang et al., 2008) at 1:500; rabbit anti-Htl (Shishido et al., 1997) at 1:3,000; rabbit anti-Stumps (Vincent et al., 1998) at 1:2,000; and mouse anti-Cut (Developmental Studies Hybridoma Bank) at 1:10; and were visualized with Alexa Fluor 488 goat anti–mouse (green) and Alexa Fluor 594 goat anti–rabbit (red), both at 1:1,000 (Invitrogen). Alexa Fluor 594 conjugated to phalloidin was used at 1:100 (Invitrogen). Discs were mounted in Vectashield with DAPI.
Microscopy
All images were collected with a spectral point scanning confocal (model C1si; Nikon) connected to an inverted microscope (TE2000; Nikon) equipped with DIC, phase, and epi-fluorescence optics; 40x Plan Fluor NA 1.4 objective lens; the Perfect Focus System for continuous maintenance of focus; 100-mW mercury arc lamp illumination for viewing fluorescence by eye; and confocal scanning using solid-state diode lasers (Melles Griot): 405 nm, 488 nm (10 mW), and 561 nm (10 mW). The image acquisition software used was Nikon EZ-C1. All samples were mounted and imaged in Vectashield mounting medium with DAPI (Vector Laboratories) at room temperature. Adobe Photoshop CS5 was used to pseudocolor images.
Electrophysiology
Electrophysiological recordings were made from muscle 6 in segment A3 from wandering third instar larvae in modified HL3 saline (0.5 mM Ca2+) as described previously (Stewart et al., 1994; Sanyal et al., 2002; Kim et al., 2009). In brief, electrodes with tip resistances between 25–30 MΩ were used to record evoked excitatory junction potentials (EJPs) after a stimulus train delivered at 0.5 Hz to the segmental nerve such that both units were consistently recruited. Only recordings where the resting membrane potential was more polarized than −60 mV were selected for analysis. Because EJPs were larger than 10 mV in amplitude, Martin’s correction for nonlinear summation was applied to all recordings (McLachlan and Martin, 1981; Kim et al., 2009). Spontaneous events (mEJPs) were recorded for a total duration of 2 min and were analyzed using MiniAnalysis software (Synaptosoft, Inc.). EJPs were low-pass filtered and analyzed in Clampfit and representative images were generated in Microsoft Excel and Adobe Photoshop. Quantal content is determined by dividing the average corrected EJP amplitude by the average mEJP amplitude in each instance. Statistical significance was determined using ANOVA.
Strain construction
Smn FRT 79D-F chromosomes were constructed by recombining the Smn73Ao (Chan et al., 2003), Smnf05960 (Thibault et al., 2004), and SmnX7 (Chang et al., 2008) alleles onto the ru1 h1 th1 st1 P{FRT(whs)}2A chromosome obtained from the Bloomington Stock Center. Multiple recombinant strains for each allele were balanced using the TM6C, Sb Tb chromosome. Independent recombinant lines were then selected from candidate strains on the basis of a failure to complement SmnX7, accompanied by loss of ru1 h1 th1 st1 markers while maintaining the w+ expression indicative of the presence of the FRT.
Somatic mitotic clones
Clones of Smn tissue were induced by mitotic recombination using the FLP-FRT technique (Golic, 1991; Xu and Rubin, 1993). Mutant Smn clones in wing imaginal discs were identified by loss of GFP driven by the ubiquitin reporter (Ubi-GFP). The crosses were as follows: y w hsFLP122; Ubi-GFP FRT 79D-F females were crossed to SmnX7 FRT 79D-F/TM6C, Sb Tb, Smn73Ao FRT 79D-F/TM6C, Sb Tb and Smnf05960 FRT 79D-F/TM6C, Sb Tb males. Approximately 20 y w hsFLP122; Ubi-GFP FRT 79D-F females were crossed to Smn FRT 79D-F/TM6C, Sb Tb males and allowed to lay eggs for 3 d at 25°C; vials were then heat-shocked at 37°C for 2 h on two consecutive days to induce somatic clones. The somatic clones produced using the SmnX7 allele produced very small Smn somatic clones and were not used for assessing Htl or Stumps expression. Smn expression was not detected in somatic clones generated using the Smn73Ao allele, suggesting that it behaves as a molecular null. In Smnf05960 somatic clones, Smn levels were strongly reduced, but not eliminated, suggesting that it is hypomorphic in nature. Consistent with this notion, Smn and Stumps expression were reduced, but not eliminated in Smnf05960 somatic clones.
RNA extraction, quality control, and reverse transcription
RNA was extracted following standard procedure. In brief, third instar larvae were dissected in ice-cold PBS, and brains and wing imaginal discs (∼100 brains/sample) were collected into TRIzol (Life Sciences). Samples were homogenized, and RNA was isolated after purification on an RNeasy mini column (QIAGEN). Quality control of RNA was assessed with a Bioanalyzer 2100 (Agilent Technologies) and only high quality samples were chosen for further analysis of gene expression. 200 ng of total RNA were reverse transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems) following the manufacturer’s instructions.
RT-PCR
The following oligonucleotides were designed for RT-PCR assays: htl sense: 5′-CGGAAGGGATCAGGATAGGG-3′ and antisense: 5′-CCTCGCCAGTCCAAAATCAG-3′; Smn sense: 5′-TGGGATGACTCCTTGCTGGT-3′ and antisense: 5′-GAGCAACACCTCCTGCTCGT-3′; Rp49/RpL32 sense: 5′-CGACGCTTCAAGGGACAGTATC-3′ and antisense: 5′-TCCGACCAGGTTACAAGAACTCTC-3′.
Quantitative PCR
TaqMan gene expression assays (Applied Biosystems) were used to assess the expression of Smn (Dm01822923_s1) and heartless (Dm02373745_s1). The ribosomal protein gene Rp49/RpL32 (Dm02151827_g1) was used as a calibrator to normalize cDNA input. 6 ng of equivalent RNA was used for each reaction and samples were processed in triplicate. To determine relative gene expression levels, data were processed using the 2-DDCt method (Livak and Schmittgen, 2001) normalized to the tub-GAL4 control sample. All calculations and plots were generated using Excel (Microsoft).
Online supplemental material
Fig. S1 shows that Smn lethality is modified by mutations in the Drosophila FGF receptor breathless, and Smn knockdown alters levels of downstream targets of FGF pathway. Fig. S2 shows that postsynaptic expression of htl RNAi results in a loss of Htl protein localization. Fig. S3 shows bouton counts (non-normalized) of larval NMJs from genotypes depicted in Figs. 3 G, 5 G, and 6 G. Fig. S4 shows expansion of the NMJ in animals overexpressing Stumps by how24BGAL4. Fig. S5 shows reduction of Stumps staining in how24BGAL4/UAS Smn RNAi. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004016/DC1.
Acknowledgments
We thank Mark Krasnow, Maria Leptin, and Denise Montell for fly lines. The anti-Htl antibody was a gift from Testuya Kojima, and anti-Stumps antibody was a gift from Maria Leptin. This work was supported by a grant from the SMA Foundation to S. Artavanis-Tsakonas, Anne Hart, and Davie van Vactor, whom we thank for extensive input throughout the course of this work.
A. Sen was supported by a postdoctoral fellowship from the Families of Spinal Muscular Atrophy. S. Sanyal acknowledges support from grant 1R03DA027979 from the NIDA, National Institutes of Health. The authors thank the Nikon Imaging Center at Harvard Medical School for help with light microscopy.
Footnotes
Abbreviations used in this paper:
- btl
- breathless
- htl
- heartless
- EJP
- excitatory junction potential
- MSA
- muscle surface area
- NMJ
- neuromuscular junction
- SMA
- spinal muscular atrophy
- SMN
- Survival Motor Neuron
- sty
- sprouty
References
- Azzouz M., Le T., Ralph G.S., Walmsley L., Monani U.R., Lee D.C., Wilkes F., Mitrophanous K.A., Kingsman S.M., Burghes A.H., Mazarakis N.D. 2004. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J. Clin. Invest. 114:1726–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle D.J., Kasim M., Yong J., Lotti F., Lau C.K., Mouaikel J., Zhang Z., Han K., Wan L., Dreyfuss G. 2006. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 71:313–320 10.1101/sqb.2006.71.001 [DOI] [PubMed] [Google Scholar]
- Beiman M., Shilo B.Z., Volk T. 1996. Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 10:2993–3002 10.1101/gad.10.23.2993 [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Briese M., Esmaeili B., Fraboulet S., Burt E.C., Christodoulou S., Towers P.R., Davies K.E., Sattelle D.B. 2009. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet. 18:97–104 10.1093/hmg/ddn320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns A.F., van Bergeijk J., Lorbeer C., Nölle A., Jungnickel J., Grothe C., Claus P. 2009. Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc. Natl. Acad. Sci. USA. 106:12747–12752 10.1073/pnas.0900122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. 2006. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 26:11014–11022 10.1523/JNEUROSCI.1637-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trülzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. 2003. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 12:1367–1376 10.1093/hmg/ddg157 [DOI] [PubMed] [Google Scholar]
- Chang H.C., Dimlich D.N., Yokokura T., Mukherjee A., Kankel M.W., Sen A., Sridhar V., Fulga T.A., Hart A.C., Van Vactor D., Artavanis-Tsakonas S. 2008. Modeling spinal muscular atrophy in Drosophila. PLoS One. 3:e3209 10.1371/journal.pone.0003209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus P., Doring F., Gringel S., Muller-Ostermeyer F., Fuhlrott J., Kraft T., Grothe C. 2003. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J. Biol. Chem. 278:479–485 10.1074/jbc.M206056200 [DOI] [PubMed] [Google Scholar]
- Claus P., Werner S., Timmer M., Grothe C. 2004. Expression of the fibroblast growth factor-2 isoforms and the FGF receptor 1-4 transcripts in the rat model system of Parkinson’s disease. Neurosci. Lett. 360:117–120 10.1016/j.neulet.2004.01.046 [DOI] [PubMed] [Google Scholar]
- Consoulas C., Restifo L.L., Levine R.B. 2002. Dendritic remodeling and growth of motoneurons during metamorphosis of Drosophila melanogaster. J. Neurosci. 22:4906–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins D.J., Staynov D.Z., Lee T.H. 1996. Regulation of cytokine genes implicated in asthma and atopy. Monogr. Allergy. 33:138–152 [PubMed] [Google Scholar]
- Davis G.W., DiAntonio A., Petersen S.A., Goodman C.S. 1998. Postsynaptic PKA controls quantal size and reveals a retrograde signal that regulates presynaptic transmitter release in Drosophila. Neuron. 20:305–315 10.1016/S0896-6273(00)80458-4 [DOI] [PubMed] [Google Scholar]
- Dutta D., Shaw S., Maqbool T., Pandya H., Vijayraghavan K. 2005. Drosophila Heartless acts with Heartbroken/Dof in muscle founder differentiation. PLoS Biol. 3:e337 10.1371/journal.pbio.0030337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert C., Chari A., Laggerbauer B., Fischer U. 2006. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 12:113–121 10.1016/j.molmed.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Emori Y., Saigo K. 1993. Distinct expression of two Drosophila homologs of fibroblast growth factor receptors in imaginal discs. FEBS Lett. 332:111–114 10.1016/0014-5793(93)80494-F [DOI] [PubMed] [Google Scholar]
- Featherstone D.E., Rushton E., Rohrbough J., Liebl F., Karr J., Sheng Q., Rodesch C.K., Broadie K. 2005. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J. Neurosci. 25:3199–3208 10.1523/JNEUROSCI.4201-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.J., Keshishian H. 1998. Nerve-muscle interactions during flight muscle development in Drosophila. Development. 125:1769–1779 [DOI] [PubMed] [Google Scholar]
- Forni J.J., Romani S., Doherty P., Tear G. 2004. Neuroglian and FasciclinII can promote neurite outgrowth via the FGF receptor Heartless. Mol. Cell. Neurosci. 26:282–291 10.1016/j.mcn.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Fox M.A., Sanes J.R., Borza D.B., Eswarakumar V.P., Fässler R., Hudson B.G., John S.W., Ninomiya Y., Pedchenko V., Pfaff S.L., et al. 2007. Distinct target-derived signals organize formation, maturation, and maintenance of motor nerve terminals. Cell. 129:179–193 10.1016/j.cell.2007.02.035 [DOI] [PubMed] [Google Scholar]
- Frugier T., Tiziano F.D., Cifuentes-Diaz C., Miniou P., Roblot N., Dierich A., Le Meur M., Melki J. 2000. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 9:849–858 10.1093/hmg/9.5.849 [DOI] [PubMed] [Google Scholar]
- García-Alonso L., Romani S., Jiménez F. 2000. The EGF and FGF receptors mediate neuroglian function to control growth cone decisions during sensory axon guidance in Drosophila. Neuron. 28:741–752 10.1016/S0896-6273(00)00150-1 [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Luschnig S., Metzstein M.M., Krasnow M.A. 2003. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19:623–647 10.1146/annurev.cellbio.19.031403.160043 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S., Skeath J.B., Doe C.Q., Michelson A.M. 1996. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 10:3003–3017 10.1101/gad.10.23.3003 [DOI] [PubMed] [Google Scholar]
- Glazer L., Shilo B.Z. 1991. The Drosophila FGF-R homolog is expressed in the embryonic tracheal system and appears to be required for directed tracheal cell extension. Genes Dev. 5:697–705 10.1101/gad.5.4.697 [DOI] [PubMed] [Google Scholar]
- Golic K.G. 1991. Site-specific recombination between homologous chromosomes in Drosophila. Science. 252:958–961 10.1126/science.2035025 [DOI] [PubMed] [Google Scholar]
- Hannus S., Bühler D., Romano M., Seraphin B., Fischer U. 2000. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 9:663–674 10.1093/hmg/9.5.663 [DOI] [PubMed] [Google Scholar]
- Hebbar S., Fernandes J.J. 2004. Pruning of motor neuron branches establishes the DLM innervation pattern in Drosophila. J. Neurobiol. 60:499–516 10.1002/neu.20031 [DOI] [PubMed] [Google Scholar]
- Heckscher E.S., Fetter R.D., Marek K.W., Albin S.D., Davis G.W. 2007. NF-kappaB, IkappaB, and IRAK control glutamate receptor density at the Drosophila NMJ. Neuron. 55:859–873 10.1016/j.neuron.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. 2000. A mouse model for spinal muscular atrophy. Nat. Genet. 24:66–70 10.1038/71709 [DOI] [PubMed] [Google Scholar]
- Huang P., Stern M.J. 2005. FGF signaling in flies and worms: more and more relevant to vertebrate biology. Cytokine Growth Factor Rev. 16:151–158 10.1016/j.cytogfr.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Imam F., Sutherland D., Huang W., Krasnow M.A. 1999. stumps, a Drosophila gene required for fibroblast growth factor (FGF)-directed migrations of tracheal and mesodermal cells. Genetics. 152:307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D. 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 124:761–771 [DOI] [PubMed] [Google Scholar]
- Itoh N., Ornitz D.M. 2004. Evolution of the Fgf and Fgfr gene families. Trends Genet. 20:563–569 10.1016/j.tig.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Jarvis L.A., Toering S.J., Simon M.A., Krasnow M.A., Smith-Bolton R.K. 2006. Sprouty proteins are in vivo targets of Corkscrew/SHP-2 tyrosine phosphatases. Development. 133:1133–1142 10.1242/dev.02255 [DOI] [PubMed] [Google Scholar]
- Kadam S., McMahon A., Tzou P., Stathopoulos A. 2009. FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development. 136:739–747 10.1242/dev.027904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Duncan D.M., Duncan I. 2004. A screen for genes that interact with the Drosophila pair-rule segmentation gene fushi tarazu. Genetics. 168:161–180 10.1534/genetics.104.027250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. 2008. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 17:2552–2569 10.1093/hmg/ddn156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr D.A., Lladó J., Shamblott M.J., Maragakis N.J., Irani D.N., Crawford T.O., Krishnan C., Dike S., Gearhart J.D., Rothstein J.D. 2003. Human embryonic germ cell derivatives facilitate motor recovery of rats with diffuse motor neuron injury. J. Neurosci. 23:5131–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.M., Kumar V., Lin Y.Q., Karunanithi S., Ramaswami M. 2009. Fos and Jun potentiate individual release sites and mobilize the reserve synaptic vesicle pool at the Drosophila larval motor synapse. Proc. Natl. Acad. Sci. USA. 106:4000–4005 10.1073/pnas.0806064106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marcé M., Griffin J.W., Rich M.M., Sumner C.J. 2009. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 29:842–851 10.1523/JNEUROSCI.4434-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lee T., Hacohen N., Krasnow M., Montell D.J. 1996. Regulated Breathless receptor tyrosine kinase activity required to pattern cell migration and branching in the Drosophila tracheal system. Genes Dev. 10:2912–2921 10.1101/gad.10.22.2912 [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16:265–269 10.1038/ng0797-265 [DOI] [PubMed] [Google Scholar]
- Liu H., Shafey D., Moores J.N., Kothary R. 2010. Neurodevelopmental consequences of Smn depletion in a mouse model of spinal muscular atrophy. J. Neurosci. Res. 88:111–122 10.1002/jnr.22189 [DOI] [PubMed] [Google Scholar]
- Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G. 2006. The Drosophila melanogaster Cajal body. J. Cell Biol. 172:875–884 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y.J., Jan L.Y., Jan Y.N. 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8:1787–1802 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Marrus S.B., Portman S.L., Allen M.J., Moffat K.G., DiAntonio A. 2004. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J. Neurosci. 24:1406–1415 10.1523/JNEUROSCI.1575-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S., Pellizzoni L., Paushkin S., Mattaj I.W., Dreyfuss G. 2002. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 22:6533–6541 10.1128/MCB.22.18.6533-6541.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. 1997. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 60:1411–1422 10.1086/515465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe B.D., Marqués G., Haghighi A.P., Fetter R.D., Crotty M.L., Haerry T.E., Goodman C.S., O’Connor M.B. 2003. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 39:241–254 10.1016/S0896-6273(03)00426-4 [DOI] [PubMed] [Google Scholar]
- McLachlan E.M., Martin A.R. 1981. Non-linear summation of end-plate potentials in the frog and mouse. J. Physiol. 311:307–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. 2003. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 162:919–931 10.1083/jcb.200303168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Eggert C., Fischer U. 2002. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12:472–478 10.1016/S0962-8924(02)02371-1 [DOI] [PubMed] [Google Scholar]
- Menon K.P., Sanyal S., Habara Y., Sanchez R., Wharton R.P., Ramaswami M., Zinn K. 2004. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 44:663–676 10.1016/j.neuron.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Michelson A.M., Gisselbrecht S., Zhou Y., Baek K.H., Buff E.M. 1998. Dual functions of the heartless fibroblast growth factor receptor in development of the Drosophila embryonic mesoderm. Dev. Genet. 22:212–229 [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Culetto E., Walker D.S., Baylis H.A., Sattelle D.B., Davies K.E. 1999. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum. Mol. Genet. 8:2133–2143 10.1093/hmg/8.12.2133 [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Chan Y.B., Davies K.E., van den Heuvel M. 2000. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett. 486:99–102 10.1016/S0014-5793(00)02243-2 [DOI] [PubMed] [Google Scholar]
- Monani U.R. 2005. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 48:885–896 10.1016/j.neuron.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. 2000. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 9:333–339 10.1093/hmg/9.3.333 [DOI] [PubMed] [Google Scholar]
- Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. 2008. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 17:949–962 10.1093/hmg/ddm367 [DOI] [PubMed] [Google Scholar]
- Murray L.M., Lee S., Bäumer D., Parson S.H., Talbot K., Gillingwater T.H. 2010. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum. Mol. Genet. 19:420–433 10.1093/hmg/ddp506 [DOI] [PubMed] [Google Scholar]
- Owen N., Doe C.L., Mellor J., Davies K.E. 2000. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum. Mol. Genet. 9:675–684 10.1093/hmg/9.5.675 [DOI] [PubMed] [Google Scholar]
- Packard M., Koo E.S., Gorczyca M., Sharpe J., Cumberledge S., Budnik V. 2002. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell. 111:319–330 10.1016/S0092-8674(02)01047-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Charroux B., Abel L., Perkinson R.A., Pellizzoni L., Dreyfuss G. 2000. The survival motor neuron protein of Schizosaccharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 275:23841–23846 10.1074/jbc.M001441200 [DOI] [PubMed] [Google Scholar]
- Paushkin S., Gubitz A.K., Massenet S., Dreyfuss G. 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14:305–312 10.1016/S0955-0674(02)00332-0 [DOI] [PubMed] [Google Scholar]
- Petersen S.A., Fetter R.D., Noordermeer J.N., Goodman C.S., DiAntonio A. 1997. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 19:1237–1248 10.1016/S0896-6273(00)80415-8 [DOI] [PubMed] [Google Scholar]
- Petit V., Nussbaumer U., Dossenbach C., Affolter M. 2004. Downstream-of-FGFR is a fibroblast growth factor-specific scaffolding protein and recruits Corkscrew upon receptor activation. Mol. Cell. Biol. 24:3769–3781 10.1128/MCB.24.9.3769-3781.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra T.K., Gonsalvez G.B., Walker M.P., Shpargel K.B., Salz H.K., Matera A.G. 2007. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J. Cell Biol. 176:831–841 10.1083/jcb.200610053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S., Sandstrom D.J., Hoeffer C.A., Ramaswami M. 2002. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 416:870–874 10.1038/416870a [DOI] [PubMed] [Google Scholar]
- Sato M., Kornberg T.B. 2002. FGF is an essential mitogen and chemoattractant for the air sacs of the Drosophila tracheal system. Dev. Cell. 3:195–207 10.1016/S1534-5807(02)00202-2 [DOI] [PubMed] [Google Scholar]
- Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 94:9920–9925 10.1073/pnas.94.18.9920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R.A., Gajewski K. 1999. Ventral neuroblasts and the heartless FGF receptor are required for muscle founder cell specification in Drosophila. Oncogene. 18:6818–6823 10.1038/sj.onc.1203081 [DOI] [PubMed] [Google Scholar]
- Shishido E., Higashijima S., Emori Y., Saigo K. 1993. Two FGF-receptor homologues of Drosophila: one is expressed in mesodermal primordium in early embryos. Development. 117:751–761 [DOI] [PubMed] [Google Scholar]
- Shishido E., Ono N., Kojima T., Saigo K. 1997. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 124:2119–2128 [DOI] [PubMed] [Google Scholar]
- Sigrist S.J., Thiel P.R., Reiff D.F., Lachance P.E., Lasko P., Schuster C.M. 2000. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 405:1062–1065 10.1038/35016598 [DOI] [PubMed] [Google Scholar]
- Sigrist S.J., Thiel P.R., Reiff D.F., Schuster C.M. 2002. The postsynaptic glutamate receptor subunit DGluR-IIA mediates long-term plasticity in Drosophila. J. Neurosci. 22:7362–7372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic G., Mladinov M., Seso Simic D., Jovanov Milosevic N., Islam A., Pajtak A., Barisic N., Sertic J., Lucassen P.J., Hof P.R., Kruslin B. 2008. Abnormal motoneuron migration, differentiation, and axon outgrowth in spinal muscular atrophy. Acta Neuropathol. 115:313–326 10.1007/s00401-007-0327-1 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A., Tam B., Ronshaugen M., Frasch M., Levine M. 2004. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 18:687–699 10.1101/gad.1166404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wang J., Wu C.F. 1994. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. [A]. 175:179–191 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
- Sutherland D., Samakovlis C., Krasnow M.A. 1996. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 87:1091–1101 10.1016/S0092-8674(00)81803-6 [DOI] [PubMed] [Google Scholar]
- Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Vincent S., Wilson R., Coelho C., Affolter M., Leptin M. 1998. The Drosophila protein Dof is specifically required for FGF signaling. Mol. Cell. 2:515–525 10.1016/S1097-2765(00)80151-3 [DOI] [PubMed] [Google Scholar]
- Wan L., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G. 2005. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25:5543–5551 10.1128/MCB.25.13.5543-5551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Vogelsang E., Leptin M. 2005. FGF signalling and the mechanism of mesoderm spreading in Drosophila embryos. Development. 132:491–501 10.1242/dev.01603 [DOI] [PubMed] [Google Scholar]
- Wolstencroft E.C., Mattis V., Bajer A.A., Young P.J., Lorson C.L. 2005. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 14:1199–1210 10.1093/hmg/ddi131 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G.M. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 133:585–600 10.1016/j.cell.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]