Abstract
Background and Aims
Genotype by environment (G × E) interactions are important for the long-term persistence of plant species in heterogeneous environments. It has often been suggested that disease is a key factor for the maintenance of genotypic diversity in plant populations. However, empirical evidence for this contention is scarce. Here virus infection is proposed as a possible candidate for maintaining genotypic diversity in their host plants.
Methods
The effects of White clover mosaic virus (WClMV) on the performance and development of different Trifolium repens genotypes were analysed and the G × E interactions were examined with respect to genotype-specific plant responses to WClMV infection. Thus, the environment is defined as the presence or absence of the virus.
Key Results
WClMV had a negative effect on plant performance as shown by a decrease in biomass and number of ramets. These effects of virus infection differ greatly among host genotypes, representing a strong G × E interaction. Moreover, the relative fitness and associated ranking of genotypes changed significantly between control and virus treatments. This shift in relative fitness among genotypes suggests the potential for WClMV to provoke differential selection on T. repens genotypes, which may lead to negative frequency-dependent selection in host populations.
Conclusions
The apparent G × E interaction and evident repercussions for relative fitness reported in this study stress the importance of viruses for ecological and evolutionary processes and suggest an important role for viruses in shaping population dynamics and micro-evolutionary processes.
Keywords: Disease, genotypic diversity, G × E interactions, Trifolium repens, White clover mosaic virus
INTRODUCTION
Genotypic diversity is essential for the long-term maintenance of species in natural environments (Hedrick et al., 1976; Gillespie and Turelli, 1989; Vellend, 2006). It is the primary substrate on which natural selection acts (Fisher, 1958; Endler, 1986), and genotypic diversity can profoundly impact ecological processes at the population, community and ecosystem level (Hughes et al., 2008). Genotypes often differ in their responses to environmental conditions. Such genotype by environment (G × E) interactions represent qualitative or quantitative variation in phenotypic plasticity of individual genotypes (Conover and Schultz, 1995; Via et al., 1995; Zhivotovsky et al., 1996; Pigliucci, 2005; Fordyce et al., 2006) and are commonly visualized by non-parallel reaction norms (Conover and Schultz, 1995; Sultan, 2007). Genotype-specific responses to environmental variation are of primary importance for the coexistence of genotypes (Silander, 1985; Gillespie and Turelli, 1989), as they fuel micro-evolutionary processes in natural environments with spatio-temporally complex selection regimes (Sultan, 2000; Fordyce, 2006).
Disease has repeatedly been proposed as a key factor for the maintenance of genotypic diversity in plant populations (Haldane, 1949; Burdon, 1987; Kirchner and Roy, 2001; Summers et al., 2003; Bradley et al., 2008). Pathogens are believed to exert strong selection pressure on plants (Jarosz and Davelos, 1995) and they can profoundly affect the structure, diversity and functioning of plant populations (Dobson and Crawley, 1994; Godfree et al., 2007; Bradley et al., 2008). Viruses may play a crucial role (Malmstrom et al., 2005) in shaping micro-evolutionary processes and genotypic diversity in plants (Gilbert, 2002; Burdon et al., 2006). Plant viruses are virtually ubiquitous in the field and they can strongly decrease host performance and fitness (Hull and Davies, 1992; Bosque-Pérez et al., 1998; Strange and Scott, 2005) and affect the host's competitive ability (Pagan et al., 2009). However, viruses are not of necessity exclusively pathogens; they may also confer upon their hosts ecological benefits such as improved drought tolerance (Xu et al., 2008) and protection from herbivores (Gibbs, 1980). As a consequence of spatial and temporal variation in virus presence within plant populations, some genotypes will be exposed to virus infections whereas others will not. Here, we consider the presence or absence of the virus as two environmental conditions in which the host plant can grow. Variable selection, caused by genotype-specific responses to viral infections (thus G × E interactions) is likely to counteract selection forces which tend to depress host plant diversity.
Three conditions should be met if viruses are to preserve genotypic diversity in their host plants via G × E interactions (Mitchell-Olds, 1992). First, there should be genotypic variation in components determining plant fitness. Secondly, the ranking of genotypes in terms of performance and fitness should change between different patches of the environment, preventing a single genotype from dominating multiple environments. Thirdly, plant populations should experience environmental heterogeneity in the sense of variation in virus incidence. In this study we investigate whether plant viruses have the potential to preserve host genotypic diversity via G × E interactions and may hence be a common, yet underappreciated, player influencing patterns and dynamics of genotypic diversity in wild plants.
Most studies on plant–virus interactions have been performed on annual crops plants, providing valuable knowledge about the negative consequences of virus infections for plant fitness and the mechanisms underlying these interactions. However, the effect of virus infections on natural plant species are far less understood (Gilbert, 2002; Cooper and Jones, 2006, and references therein). Therefore, in this study, we used natural genotypes of the stoloniferous herb Trifolium repens, which is a common species of the temperate regions of the world, occurring in many different habitats at a range of altitudes (Daday, 1958). Clonally propagating, perennial plant species from wild populations may interact differently with their viral pathogens compared with seed-producing annual plants (Stuefer et al., 2004; van Mölken and Stuefer, 2008), and this study complements current knowledge on plant–virus interactions in annual and crop plants. Owing to the clonal mode of reproduction, genotypes of T. repens can be replicated under various experimental conditions, providing us with an excellent tool to test whether the first and second conditions (Mitchell-Olds, 1992) described above can be met.
This study on the potential of plant viruses to promote genotypic diversity in T. repens focuses mainly on the first and second conditions, since other studies clearly demonstrated that the third condition is valid for our system. Sherwood (1997) has shown that the incidence of White clover mosaic virus (WClMV) fluctuates considerably in populations of the stoloniferous herb T. repens. The same study reports that 30 % of all plants were infected by WClMV and infection levels varied from 1 to 96 % between different sites. In another study, 1 % of the plants were found to be infected with WClMV at one site, while infection rates ranged from 9 to 46 % at another site (Coutts and Jones, 2002).
The first and second conditions proposed by Mitchell-Olds (1992) were experimentally investigated by testing the following specific hypotheses: (a) genotypes of T. repens vary significantly with respect to fitness-related traits; (b) virus infection has negative effects on fitness-related traits and plant performance; and (c) the ranking of genotypes changes in response to WClMV infection. In order to test these hypotheses, we examined the growth and performance of genetically distinct individuals of T. repens in control and virus treatments, and we evaluated G × E interactions in terms of genotype-specific plant responses to WClMV infection. We report substantial G × E interactions, which resulted in significant shifts in the relative fitness of host genotypes grown in control and virus-infected conditions, respectively.
MATERIALS AND METHODS
Study organisms
The stoloniferous herb Trifolium repens L. was used for this study. Trifolium repens can propagate vegetatively through the production of genetically identical offspring (ramets) which develop at the nodes of horizontally growing stems (stolons), or by sexual reproduction. Each individual ramet consists of a single leaf, an internode, and meristems which can develop into roots, branches and flowers. In 2001, T. repens plants were randomly collected in riverine grasslands along the river Waal near Ewijk (The Netherlands, 51°52′54′′N, 5°45′00′′E), and the genetic identity of the genotypes was established by amplified fragment length polymorphism (AFLP; for details, see Weijschedé et al., 2006). The plants were maintained under common garden and greenhouse conditions for 2 years before this experiment was conducted. Eleven genotypes were randomly selected from this collection and used for this experiment. Whereas measuring production of viable seed is a clear and easily accomplished method to estimate plant fitness in annual plants, fitness of clonally propagated plants is more difficult to study. The most important reason for this is that many T. repens genotypes do not produce seed, and vegetative reproduction should be taken into account when assessing lifetime plant fitness (Pan and Price, 2001). In general, fitness can be defined as ‘the rate of change in number of units carrying a certain allele or allele complex’ (Wikberg, 1995). In sexually reproducing plants these ‘units’ are provided by seed, and seed production increases the number of units that carry the parental genetic material. In analogy, the number of units carrying the parental genetic material of clonal plants increases with the production of new ramets. Just like seeds, each ramet can produce roots and leaves and therefore has the potential for autonomous growth. Therefore, clonal growth represented by the total number of clonal offspring (ramets) is the closest measure of fitness available for clonal plants that show no (or only partial) sexual reproduction (Sackville-Hamilton et al., 1987; Winkler and Fisher, 1999; Pan and Price, 2001). Therefore, the number of ramets will be used as an indicator of plant fitness throughout this paper.
White clover mosaic virus (necrosis strain, originally isolated from T. repens in Denmark) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ; Braunschweig, Germany). This virus is a member of the genus Potexvirus and is transmitted mechanically between hosts. WClMV is not transmitted by insect vectors such as aphids (Tapio, 1970).
Experimental design
In April 2005 the experiment was started with rooted, apical cuttings consisting of six ramets each. Fourteen cuttings per genotype were individually planted in plastic trays (15 × 23 × 5 cm) filled with SERAMIS clay granules (Masterfoods GMbH, Verden, Germany). These cuttings (subsequently referred to as ‘plants’) were grown in a greenhouse with a 16 h light and 8 h dark period at 19/18 °C. High pressure sodium lamps (Hortilux-Schréder 600 W, Monster, The Netherlands) were switched on automatically whenever the irradiance dropped below 250 µmol m−2 s−1. Stolons that grew out of the trays were bent back to facilitate root formation. At 19 and 32 d after transplanting the cuttings, each tray received 50 mL of half-strength Hoagland nutrient solution. All plants were nodulated with rhizobium bacteria.
Seven replicates of each genotype were randomly assigned to the control treatment (no virus infection) and to the infection treatment (experimental virus infection), respectively. Ten days after planting, all plants in the virus treatment were inoculated with WClMV on the third and fourth youngest ramets. Inoculation was performed mechanically with cell sap prepared by grinding calcium chloride-dried WClMV-infected plant material in inoculation buffer (50 mm Na2HPO4 buffer, 1 mm EDTA, set to pH 7·0 with HCl). Leaves on the third and fourth ramets were dusted with carborundum (500 mesh), and 10 µL of virus suspension was rubbed on each leaf by hand. Control plants were mock-inoculated with inoculation buffer only. This standard inoculation procedure results in systemic WClMV transport throughout the clonal plant network. All plants were re-inoculated on the third and fourth youngest ramets (newly formed) after 17 d with fresh WClMV-infected material (Phaseolus vulgaris leaves infected with WClMV obtained from DSMZ), using the same procedure as described above.
All plant material was harvested 50 d after the first inoculation, and the fourth youngest leaf was sampled for enzyme-linked immunosorbent assay (ELISA) testing. The length of the primary stolon was measured and the number of ramets on the primary stolon, number of ramets on the branches, number of branches and the number of flowers were counted. All plant material was dried at 70 °C for 72 h, and dry weights of stolons, leaves, flowers and roots were measured separately. The total number of ramets and the total biomass of plants were used to calculate relative fitness values for genotypes within the control and virus treatment. The relative fitness was calculated as the mean genotypic trait value divided by the overall mean (i.e. mean of all genotypes) for the same trait within the control or the virus treatment, respectively.
ELISA testing
Qualitative analysis of the presence of WClMV was tested by double antibody sandwich (DAS)-ELISA (based on Clark and Adams, 1977) on the ninth oldest ramet on the primary stolon. White clover mosaic virus proved to be present in all tested leaf samples, showing that virus application was successful (data not shown).
Data analysis
A two-way analysis of variance (ANOVA) was performed to determine the effects of WClMV on development, growth and flowering of T. repens and to analyse genotype × virus interactions for relative fitness, using genotype and virus infection as main factors. Genotype was regarded as a random factor. The effect of WClMV on flowering was analysed only for those four genotypes that produced flowers (i.e. A15, B51, D129 and D134). To meet assumptions for normality and homoscedasticity, log transformations were applied whenever necessary. All tests were carried out with SAS, version 9·1 (SAS Institute Inc., Cary, NC, USA).
RESULTS
Genotypic variation
Genotypic variation was strong and significant with respect to the components determining fitness in clonal plants, i.e. total number of ramets and total biomass. All other traits showed strong genotypic variation as well (Table 1A, B, genotype effects).
Table 1.
Statistical analysis of the effects of WClMV, genotype and their interaction (ANOVA) on (A) different developmental and architectural traits, (B) absolute biomass of different plant parts and biomass allocation (% biomass) to various plant parts and (C) different flowering traits
| Error |
Genotype |
Virus |
Genotype × virus |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | d.f.* | MS† | d.f.* | MS† | F‡ | P | d.f.* | MS† | F‡ | P | d.f.* | MS† | F‡ | P |
| (A) | ||||||||||||||
| Total no. of ramets | 130 | 466·39 | 10 | 5662·92 | 12·14 | <0·0001 | 1 | 8496·31 | 6·83 | 0·0259 | 10 | 1244·55 | 2·67 | 0·0053 |
| No. of ramets on pr. stolon | 130 | 5·47 | 10 | 84·07 | 15·36 | <0·0001 | 1 | 110·61 | 2·76 | 0·1276 | 10 | 40·11 | 7·33 | <0·0001 |
| No. of ramets on branches | 130 | 412·15 | 10 | 4806·41 | 11·66 | <0·0001 | 1 | 6668·07 | 6·74 | 0·0267 | 10 | 990·16 | 2·40 | 0·0119 |
| % Branches on pr. stolon | 130 | 159·80 | 10 | 5117·61 | 32·02 | <0·0001 | 1 | 2215·88 | 5·89 | 0·0356 | 10 | 376·21 | 2·35 | 0·0137 |
| Root–shoot ratio | 130 | 0·00 | 10 | 0·09 | 25·68 | <0·0001 | 1 | 0·00 | 0·44 | 0·5225 | 10 | 0·01 | 2·20 | 0·0214 |
| Length of pr. stolon | 130 | 0·07 | 10 | 1·70 | 23·16 | <0·0001 | 1 | 2·42 | 4·86 | 0·0521 | 10 | 0·50 | 6·78 | <0·0001 |
| Length of branches | 130 | 76·91 | 10 | 655·57 | 8·52 | <0·0001 | 1 | 1489·68 | 2·98 | 0·1151 | 10 | 500·54 | 6·51 | <0·0001 |
| (B) | ||||||||||||||
| Total biomass | 130 | 0·08 | 10 | 0·99 | 12·59 | <0·0001 | 1 | 1·87 | 5·58 | 0·0398 | 10 | 0·34 | 4·25 | <0·0001 |
| Biomass of roots | 130 | 0·01 | 10 | 0·09 | 17·25 | <0·0001 | 1 | 0·13 | 5·16 | 0·0465 | 10 | 0·03 | 5·05 | <0·0001 |
| Biomass of stolons | 130 | 0·01 | 10 | 0·08 | 16·26 | <0·0001 | 1 | 0·14 | 3·30 | 0·0994 | 10 | 0·04 | 8·32 | <0·0001 |
| Biomass of leaves | 130 | 0·02 | 10 | 0·22 | 10·81 | <0·0001 | 1 | 0·35 | 7·82 | 0·0189 | 10 | 0·05 | 2·22 | 0·0203 |
| Biomass per ramet | 130 | 0·00 | 10 | 0·00 | 37·97 | <0·0002 | 1 | 0·06 | 1·24 | 0·2914 | 10 | 0·00 | 3·66 | 0·0002 |
| % Biomass of roots | 130 | 8·56 | 10 | 237·91 | 27·81 | <0·0001 | 1 | 6·02 | 0·31 | 0·5908 | 10 | 19·54 | 2·28 | 0·0168 |
| % Biomass of stolons | 130 | 9·06 | 10 | 283·15 | 31·26 | <0·0001 | 1 | 15·77 | 0·59 | 0·4586 | 10 | 26·55 | 2·93 | 0·0024 |
| % Biomass of leaves | 130 | 29·77 | 10 | 367·11 | 12·33 | <0·0001 | 1 | 40·80 | 0·94 | 0·3552 | 10 | 43·44 | 1·46 | 0·1619 |
| (C) | ||||||||||||||
| Total no. of flowers | 47 | 0·94 | 3 | 21·85 | 23·35 | <0·0001 | 1 | 1·07 | 0·56 | 0·5084 | 3 | 1·92 | 2·05 | 0·1199 |
| % Flowers on pr. stolon | 47 | 21·03 | 3 | 488·45 | 23·23 | <0·0001 | 1 | 21·70 | 0·40 | 0·5698 | 3 | 53·62 | 2·55 | 0·0669 |
| Biomass of flowers | 47 | 0·00 | 3 | 0·08 | 36·18 | <0·0001 | 1 | 0·00 | 1·36 | 0·3274 | 3 | 0·00 | 0·85 | 0·471 |
| % Biomass of flowers | 47 | 58·84 | 3 | 1578·89 | 26·83 | <0·0001 | 1 | 0·17 | 0·00 | 0·963 | 3 | 66·25 | 1·13 | 0·3482 |
Pr. stolon: the primary stolon.
* Degrees of freedom.
† Mean square.
‡ F-statistics.
Virus effects on plant performance
White clover mosaic virus infection had a clear negative effect on plant growth and development (Table 1A, virus effects). The total number of ramets was reduced by 25 %, and WClMV caused a 17 % decrease in the branching probability of primary stolons. White clover mosaic virus infection caused a reduction in the biomass of roots (28 %) and leaves (32 %), as well as in the total plant biomass (30 %), but had no significant effect on stolon biomass or proportional biomass allocation to plant organs (Table 1B). WClMV infection did not change any of the recorded flowering traits (Table 1C).
Genotype × environment interactions
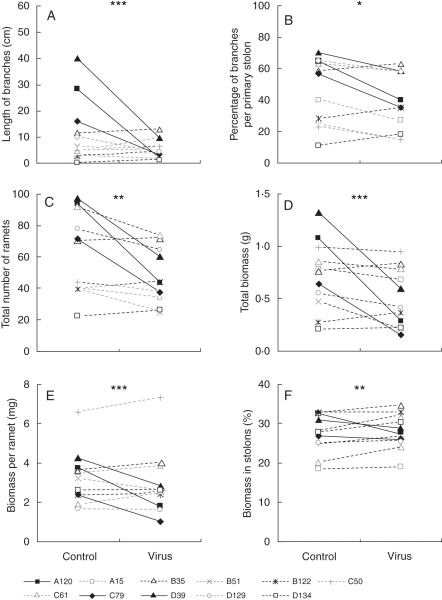
Genotypes differed greatly in their response to WClMV infection (Table 1A, genotype × virus interaction). In several genotypes (i.e. A120, C79 and D39) WClMV caused a dramatic decrease in the length of branches, while other genotypes (i.e. B35, B122 and D134) showed no response to the virus treatment (Fig. 1A). Similar patterns were recorded for the percentage of branches on the primary stolon (Fig. 1B) and for the total number of vegetative offspring produced during the experiment (Fig. 1C).
Fig. 1.
Genotypic variation in the effect of WClMV infection for (A) length of the branches, (B) percentage of branches on the primary stolon, (C) total number of ramets, (D) total biomass, (E) average biomass per ramet and (F) biomass allocation to the stolons, i.e. % biomass stolons. All traits show a significant genotype × virus interaction (two-way ANOVA: *P < 0·05, **P < 0·01 and ***P < 0·001), i.e. the genotypes are affected differently by the virus infection.
With the exception of biomass allocation to leaves, all biomass production and allocation traits showed genotypic variation in the effect of WClMV infection (Table 1B). Total biomass values (Fig. 1D) decreased strongly for some genotypes (i.e. A120, C79 and D39), while they remained equal for others. Average biomass per ramet (Fig. 1E) and percentage biomass allocation to the stolons (Fig. 1F) decreased in some infected genotypes, and increased in others.
The flowering probability of primary stolons showed a non-significant trend to differ among genotypes after infection with WClMV (Table 1C). There was no significant interaction effect between virus infection and genotype for any of the flowering traits.
Mean values ± s.e. of all traits mentioned above are given for control and virus-infected treatments per genotype in Supplementary Data Table S1 (available online).
Relative fitness
The relative fitness in terms of the total number of ramets and total biomass shows a strong genotype × virus interaction (Table 2) leading to significant shifts in genotype ranking between the two experimental conditions (Fig. 2A, B). For example, some of the highest ranking genotypes in the control conditions, such as genotypes A120 and D39, clearly occupied lower ranks in the virus treatment (Fig. 2A, B).
Table 2.
Statistical analysis (ANOVA) of the relative fitness of the different genotypes in both treatments
| Error |
Genotype |
Virus |
Genotype × virus |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relative fitness | d.f.* | MS† | d.f.* | MS† | F‡ | P | d.f.* | MS† | F‡ | P | d.f.* | MS† | F‡ | P |
| Total no. of ramets | 130 | 0·1477 | 10 | 1·8000 | 12·19 | <0·0001 | 1 | 0·0013 | 0·00 | 0·9523 | 10 | 0·3342 | 2·26 | 0·0179 |
| Total biomass | 130 | 0·2220 | 10 | 2·7417 | 12·35 | <0·0001 | 1 | 0·0000 | 0·00 | 0·9947 | 10 | 0·9259 | 4·17 | <0·0001 |
* Degrees of freedom.
† Mean square.
‡ F-statistics.
Fig. 2.
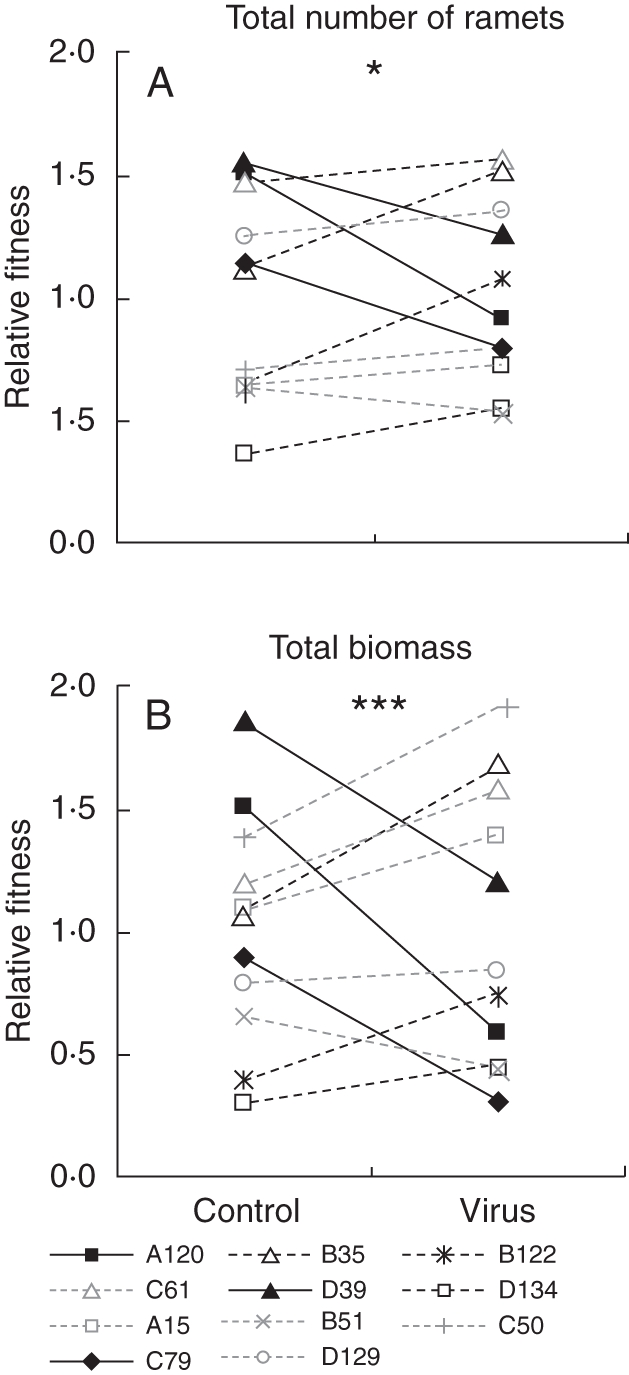
G × E interactions for relative fitness of the different genotypes, represented as reaction norm plots. The relative fitness of each genotype is calculated relative to the mean fitness within an environment (i.e. control or virus), where fitness is expressed by both the total number of ramets (A) and total biomass (B). The relative fitness of genotypes differs significantly between the virus-free and virus-prone environments (two-way ANOVA: *P < 0·05, **P < 0·01 and ***P < 0·001).
DISCUSSION
Genotype × environment interactions can sustain genotypic diversity in natural environments, thereby promoting long-term coexistence (Gillespie and Turelli, 1989) and enhancing system stability (Thompson, 1991). Here we demonstrate that virus infections can substantially shift the ranking of plant genotypes with respect to relative fitness (in terms of total number of ramets and total biomass) between control and virus treatments. Based on these findings, and on general predictions from evolutionary theory, we suggest that viruses may play an important yet unrecognized role in the long-term maintenance of genotypic diversity in their host populations through variable selection and G × E interactions.
For pathogen-caused G × E interactions to occur, infections should significantly affect plant performance and fitness. In our study, WClMV compromised biomass accumulation, retarded vegetative propagation and curtailed the spatial expansion capabilities of infected as compared with non-infected plants. These findings are in accordance with other studies reporting negative effects of virus infection on plant performance (Jones, 1992; Funayama et al., 1997; Dudas et al., 1998; Godfree et al., 2007; Pagan et al., 2007).
The effects of virus infection on host plants showed conspicuous levels of genotypic variation for most development- and growth-related traits recorded in this experiment. Consequently, the genotypes which performed best in the control treatment did not occupy high ranks in the virus treatment, and vice versa. This suggests that virus infections can cause significant alterations in genotype frequencies within host populations that depend mainly on vegetative reproduction for growth. The observed G × E interactions indicate genotypic dissimilarities in host plant sensitivity to viral infection, which may be caused by variation in virulence levels. Virulence can be understood as pathogen-caused reduction of host fitness (Brown et al., 2006) and is mainly a function of the activity of the host tissue (Hull, 2004) and the degree of host resistance. Pathogen virulence can vary considerably among host genotypes (Godfree et al., 2007). Fast-growing and hence larger genotypes are likely to experience higher virulence levels than slow-growing, smaller genotypes (Morrison, 1996) owing to their superior metabolic activity which promotes virus replication.
Mitchell-Olds (1992) postulated three conditions for the maintenance of genotypic variation through G × E interactions. The first condition demands genotypic variation in fitness: here we demonstrated strong genotypic variation in closely fitness-related traits such as clonal offspring production and total plant biomass. These results are consistent with other studies showing genotypic variation for many fitness-associated traits in T. repens (Turkington, 1989; Weijschedé et al., 2006). The second condition requires genotype fitness to vary between environments: the performance of genotypes differed greatly between virus-free and virus-prone environments in our study, resulting in a marked shift in the ranking of genotypes between these environments. These results are in agreement with Pagan et al. (2008) who show that different accessions of Arabidopsis thaliana vary in their response on growth investment to infection with Cucumber mosaic virus. The third condition requires environmental heterogeneity in virus prevalence which has been clearly shown by others (Sherwood, 1997; Norton and Johnstone, 1998; Coutts and Jones, 2002) for the plant–virus system used in this study. They demonstrate that virus incidence shows considerable fluctuations both within and between populations of T. repens. Marked heterogeneity in disease incidence has also been described for other plant viruses (Bosque-Pérez et al., 1998; Godfree et al., 2007). We hence conclude that viruses are excellent candidates for maintaining genotypic variation in their hosts, and that their virtual omnipresence in nature may render them prime biotic agents counteracting declines of genotypic diversity in natural plant populations.
The shift in relative fitness among genotypes indicates the existence of trade-offs between plant performance in control and virus treatments. As a result, genotypes successful in the control condition perform relatively much worse in the virus treatment. This suggests the potential for WClMV to provoke differential selection on T. repens genotypes, which may lead to negative frequency-dependent selection in host populations. Such negative frequency-dependent selection occurs when common genotypes as compared with less common genotypes suffer from a fitness disadvantage in virus-prone environments (Haldane, 1949; Brunet and Mundt, 2000; Rueffler et al., 2006).
The maintenance of genotypic diversity by viruses may depend on the ecological conditions. For example, virus infections may play a more prominent role in species with strong genotypic variation in response to virus infection, as compared with species with low genotypic variation. The mechanism behind this genotypic variation is not clear, but may depend on the effectiveness of defence mechanisms, the degree of tolerance or the metabolic rate of the plant, since virus replication depends on the activity of the host tissue (Hull, 2004). The latter may partly explain why some of the fast-growing genotypes in our experiment were most affected by the virus infection. Other factors such as plant competition, abiotic factors, tripartite interactions with herbivores or geographical distance are expected to play a role as well. Ahmad et al. (2007), for example, showed that the geographic distance between some sugarcane cultivars can explain variation in effects of Sugarcane yellow leaf virus plant growth.
Although there is ample evidence of significant negative effects of virus infection on plant vigour, there is surprisingly little information about their potential role as selective agents. The hypothesis that pathogens can maintain genotypic variation in their hosts has often been proposed, but has hardly ever been studied empirically. Our data suggest that virus infections may be excellent candidates for promoting genotypic diversity in their host plants and call for empirical studies that analyse virus-induced frequency-dependent selection. The apparent negative effects on plant performance, significant G × E interaction and evident repercussions for relative fitness reported in this study clearly stress the significance of virus infections for ecological and evolutionary processes and identify viruses as possible key factors for driving population dynamics and selection in the wild.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Arjen Biere, Hans de Kroon and Eric Visser for useful comments on a previous version of the manuscript. The experiments described in this paper comply with current Dutch laws.
LITERATURE CITED
- Ahmad YA, Girard J-C, Fernandez E, et al. Variation in virus populations and growth characteristics of two sugarcane cultivars naturally infected by Sugarcane yellow leaf virus in different geographical locations. Plant Pathology. 2007;56:743–754. [Google Scholar]
- Bosque-Perez NA, Olojede SO, Buddenhagen IW. Effect of maize streak virus disease on the growth and yield of maize as influenced by varietal resistance levels and plant stage at time of challenge. Euphytica. 1998;101:307–317. [Google Scholar]
- Bradley DJ, Gilbert GS, Martiny JBH. Pathogens promote plant diversity through a compensatory response. Ecology Letters. 2008;11:461–469. doi: 10.1111/j.1461-0248.2008.01162.x. [DOI] [PubMed] [Google Scholar]
- Brown NF, Wickham ME, Coombes BK, Finlay BB. Crossing the line: selection and evolution of virulence traits. PLoS Pathogens. 2006;2:e42. doi: 10.1371/journal.ppat.0020042. doi:10.1371/journal.ppat.0020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet J, Mundt CC. Disease, frequency-dependent selection, and genetic polymorphism: experiments with stripe rust and wheat. Evolution. 2000;54:406–415. doi: 10.1111/j.0014-3820.2000.tb00043.x. [DOI] [PubMed] [Google Scholar]
- Burdon JJ. Disease and plant population biology. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- Burdon JJ, Thrall PH, Ericson L. The current and future dynamics of disease in plant communities. Annual Review of Phytopathology. 2006;44:19–39. doi: 10.1146/annurev.phyto.43.040204.140238. [DOI] [PubMed] [Google Scholar]
- Clark MF, Adams AN. Characteristics of the microplate method of enzyme-linked immunosorbent assay for detection of plant viruses. Journal of General Virology. 1977;34:475–483. doi: 10.1099/0022-1317-34-3-475. [DOI] [PubMed] [Google Scholar]
- Conover DO, Schultz ET. Phenotypic similarity and the evolutionary significance of countergradient variation. Trends in Ecology and Evolution. 1995;10:248–252. doi: 10.1016/S0169-5347(00)89081-3. [DOI] [PubMed] [Google Scholar]
- Cooper I, Jones RAC. Wild plants and viruses: under-investigated ecosystems. Advances in Virus Research. 2006;67:1–47. doi: 10.1016/S0065-3527(06)67001-2. [DOI] [PubMed] [Google Scholar]
- Coutts BA, Jones RAC. Temporal dynamics of spread of four viruses within mixed species perennial pastures. Annals of Applied Biology. 2002;140:37–52. [Google Scholar]
- Daday H. Gene frequencies in wild populations of Trifolium repens L. III. World distribution. Heredity. 1958;12:169–184. [Google Scholar]
- Dobson A, Crawley W. Pathogens and the structure of plant-communities. Trends in Ecology and Evolution. 1994;9:393–398. doi: 10.1016/0169-5347(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Dudas B, Woodfield DR, Tong PM, et al. Estimating the agronomic impact of white clover mosaic virus on white clover performance in the North Island of New Zealand. New Zealand Journal of Agricultural Research. 1998;41:171–178. [Google Scholar]
- Endler JA. Natural selection in the wild. Princeton, NJ: Princeton University Press; 1986. [Google Scholar]
- Fisher RA. The genetical theory of natural selection. 2nd edn. New York: Dover Publications Inc; 1958. [Google Scholar]
- Fordyce JA. The evolutionary consequences of ecological interactions mediated through phenotypic plasticity. Journal of Experimental Biology. 2006;209:2377–2383. doi: 10.1242/jeb.02271. [DOI] [PubMed] [Google Scholar]
- Funayama S, Hikosaka K, Yahara T. Effects of virus infection and growth irradiance on fitness components and photosynthetic properties of Eupatorium makinoi (Compositae) American Journal of Botany. 1997;84:823–829. [PubMed] [Google Scholar]
- Gibbs A. A plant virus that partially protects its wild legume host against herbivores. Intervirology. 1980;13:42–47. doi: 10.1159/000149105. [DOI] [PubMed] [Google Scholar]
- Gilbert GS. Evolutionary ecology of plant diseases in natural ecosystems. Annual Review of Phytopathology. 2002;40:13–43. doi: 10.1146/annurev.phyto.40.021202.110417. [DOI] [PubMed] [Google Scholar]
- Gillespie JH, Turelli M. Genotype–environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfree RC, Thrall PH, Young AG. Enemy release after introduction of disease-resistant genotypes into plant–pathogen systems. Proceedings of the National Academy of Sciences, USA. 2007;104:2756–2760. doi: 10.1073/pnas.0608356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Disease and evolution. Supplement to La Ricerca Scientifica. 1949;19:68–76. [Google Scholar]
- Hedrick PW, Ginevan ME, Ewing EP. Genetic polymorphism in heterogeneous environments. Annual Review of Ecology and Systematics. 1976;7:1–32. [Google Scholar]
- Hughes AR, Inouye BD, Johnson MTJ, Underwood M, Vellend M. Ecological consequences of genetic diversity. Ecology Letters. 2008;11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- Hull R, Davies JW. Approaches to nonconventional control of plant-virus diseases. Critical Reviews in Plant Science. 1992;11:17–33. [Google Scholar]
- Hull R. Matthews' plant virology. 4th edn. Oxford: Elsevier Academic Press; 2004. [Google Scholar]
- Jarosz AM, Davelos AL. Effects of disease in wild plant-populations and the evolution of pathogen aggressiveness. New Phytologist. 1995;129:371–387. [Google Scholar]
- Jones RAC. Further studies on losses in productivity caused by infection of annual pasture legumes with three viruses. Australian Journal of Agriculural Research. 1992;43:1229–1241. [Google Scholar]
- Kirchner JW, Roy BA. Evolutionary implications of host–pathogen specificity: the fitness consequences of host life history traits. Evolutionary Ecology. 2001;14:665–692. [Google Scholar]
- Malmstrom CM, Hughes CC, Newton LA, Stoner CJ. Virus infection in remnant native bunchgrasses from invaded California grasslands. New Phytologist. 2005;168:217–230. doi: 10.1111/j.1469-8137.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. Does environmental variation maintain genetic-variation – a question of scale. Trends in Ecology and Evolution. 1992;7:397–398. doi: 10.1016/0169-5347(92)90017-6. [DOI] [PubMed] [Google Scholar]
- Morrison JA. Infection of Juncus dichotomus by the smut fungus Cintractia junci: an experimental field test of the effects of neighbouring plants, environment, and host plant genotype. Journal of Ecology. 1996;84:691–702. [Google Scholar]
- Norton MR, Johnstone GR. Occurrence of alfalfa mosaic, clover yellow vein, subterranean clover red leaf, and white clover mosaic viruses in white clover throughout Australia. Australian Journal of Agriculural Research. 1998;49:723–728. [Google Scholar]
- Pagan I, Alonso-Blanco C, García-Arenal F. The relationship of within-host multiplication and virulence in a plant–virus system. PLoS One. 2007;2:e786. doi: 10.1371/journal.pone.0000786. doi:10.1371/journal.pone.0000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan I, Alonso-Blanco C, García-Arenal F. Host responses in life-history traits and tolerance to virus infection in Arabidopsis thaliana. PLoS Pathogens. 2008;4:e1000124. doi: 10.1371/journal.ppat.1000124. doi:10.1371/journal.ppat.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan I, Alonso-Blanco C, García-Arenal F. Differential tolerance to direct, indirect density-dependent costs of viral infection in Arabidopsis thaliana. PLoS Pathologens. 2009;5:e1000531. doi: 10.1371/journal.ppat.1000531. doi:10.1371/journal.ppat.1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JJ, Price JS. Fitness and evolution in clonal plants: the impact of clonal growth. Evolutionary Ecology. 2001;15:583–600. [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends in Ecology and Evolution. 2005;20:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Rueffler C, Van Dooren TJM, Leimar O, Abrams PA. Disruptive selection and then what? Trends in Ecology and Evolution. 2006;21:238–245. doi: 10.1016/j.tree.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Sackville-Hamilton NRS, Schmid B, Harper JL. Life-history concepts and the population biology of clonal organisms. Proceedings of the Royal Society B: Biological Sciences. 1987;232:35–57. [Google Scholar]
- Sherwood RT. Viruses of white clover in pastures of Pennsylvania, New York, and Vermont. Plant Disease. 1997;817:817–820. doi: 10.1094/PDIS.1997.81.7.817. [DOI] [PubMed] [Google Scholar]
- Silander JA. Microevolution in clonal plants. In: Jackson JBC, Buss LW, Cook RE, editors. Population biology and evolution of clonal organisms. New Haven, CT: Yale University Press; 1985. pp. 107–152. [Google Scholar]
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annual Review of Phytopathology. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- Stuefer JF, Gómez S, Van Mölken T. Clonal integration beyond resource sharing: implications for defence signalling and disease transmission in clonal plant networks. Evolutionary Ecology. 2004;18:647–667. [Google Scholar]
- Sultan SE. Phenotypic plasticity for plant development, function and life history. Trends in Plant Sciences. 2000;5:537–542. doi: 10.1016/s1360-1385(00)01797-0. [DOI] [PubMed] [Google Scholar]
- Sultan SE. Development in context: the timely emergence of eco-devo. Trends in Ecology and Evolution. 2007;22:575–582. doi: 10.1016/j.tree.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Summers K, McKeon S, Sellars J, et al. Parasitic exploitation as an engine of diversity. Biological Reviews of the Cambridge Philosophical Society. 2003;78:639–675. doi: 10.1017/s146479310300616x. [DOI] [PubMed] [Google Scholar]
- Tapio E. Virus diseases of legumes in Finland and in the Scandinavian countries. Annales Agriculturae Fenniae. 1970;9:1–97. [Google Scholar]
- Thompson JD. Phenotypic plasticity as a component of evolutionary change. Trends in Ecology and Evolution. 1991;6:246–249. doi: 10.1016/0169-5347(91)90070-E. [DOI] [PubMed] [Google Scholar]
- Turkington R. The growth, distribution and neighbour relationships of Trifolium repens in a permanent pasture. VI. Conditioning effects by neighbours. Journal of Ecology. 1989;77:734–746. [Google Scholar]
- Van Mölken T, Stuefer JF. Virulence in clonal plants: conflicting selection pressures at work? Evolutionary Ecology. 2008;22:467–470. [Google Scholar]
- Vellend M. The consequences of genetic diversity in competitive communities. Ecology. 2006;87:304–311. doi: 10.1890/05-0173. [DOI] [PubMed] [Google Scholar]
- Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Schlichting CD, Van Tienderen PH. Adaptive phenotypic plasticity – consensus and controversy. Trends in Ecology and Evolution. 1995;10:212–217. doi: 10.1016/s0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Weijschede J, Martinkova J, De Kroon H, Huber H. Shade avoidance in Trifolium repens: costs and benefits of plasticity in petiole length and leaf size. New Phytologist. 2006;172:655–666. doi: 10.1111/j.1469-8137.2006.01885.x. [DOI] [PubMed] [Google Scholar]
- Wikberg S. Fitness in clonal plants. Oikos. 1995;72:293–297. [Google Scholar]
- Winkler E, Fischer M. Two fitness measures for clonal plants and the importance of spatial aspects. Plant Ecology. 1999;141:191–199. [Google Scholar]
- Xu P, Cheng F, Mannas JP, Feldman T, Sumner LW, Roossinck MJ. Virus infection improves drought tolerance. New Phytologist. 2008;180:911–921. doi: 10.1111/j.1469-8137.2008.02627.x. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Feldman MW, Bergman A. On the evolution of phenotypic plasticity in a spatially heterogeneous environment. Evolution. 1996;50:547–558. doi: 10.1111/j.1558-5646.1996.tb03867.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.