Abstract
Background and Aims
To address the issues associated with food security, environmental change and bioenergy in the context of crop plants, the production, identification and evaluation of novel plant phenotypes is fundamental. One of the major routes to this end will be wide hybridization and introgression breeding. The transfer of chromosomes and chromosome segments between related species (chromosome engineering or alien introgression) also provides an important resource for determining the genetic control of target traits. However, the realization of the full potential of chromosome engineering has previously been hampered by the inability to identify and characterize interspecific introgressions accurately.
Methods
Seven monosomic substitution lines have been generated comprising Festuca pratensis as the donor species and Lolium perenne as the recipient. Each of the seven lines has a different L. perenne chromosome replaced by the homoeologous F. pratensis chromosome (13 L. perenne + 1 F. pratensis chromosome). Molecular markers and genomic in situ hybridization (GISH) were used to assign the F. pratensis chromosomes introgressed in each of the monosomic substitutions to a specific linkage group. Cytological observations were also carried out on metaphase I of meiosis in each of the substitution lines.
Results
A significant level of synteny was found at the macro-level between L. perenne and F. pratensis. The observations at metaphase I revealed the presence of a low level of interspecific chromosomal translocations between these species.
Discussion
The isolation of the seven monosomic substitution lines provides a resource for dissecting the genetic control of important traits and for gene isolation. Parallels between the L. perenne/F. pratensis system and the Pooideae cereals such as wheat, barley, rye, oats and the model grass Brachypodium distachyon present opportunities for a comparison across the species in terms of genotype and phenotype.
Keywords: Lolium perenne, Festuca pratensis, chromosome engineering, molecular markers, genomic in situ hybridization, recombination, comparative genomics, Pooideae, genetics, introgression
INTRODUCTION
The identification and isolation of the genes via forward genetic approaches, i.e. genetic mapping, chromosome landing, etc., requires the presence of sufficient phenotypic variation for the trait in question. In inbreeding species such as wheat, very little genetic variation exists in adapted genotypes. However, wild hexaploid landraces and wild and distant relatives (alien species) (e.g. rye and Thinopyrum bessarabicum) provide a key source of variation for target traits. In outbreeding highly heterozygous crops such as forage grass, considerable variation still exists in elite plant material for many traits. Lolium perenne, the major source of forage in temperate regions of the world, shows considerable variation for quality traits such as digestibility, establishment, and spring and winter growth. However, L. perenne shows less variation for traits which are of importance for resistance to abiotic stresses, e.g. drought tolerance, winter hardiness, water use efficiency, root development and persistency (Kosmala et al., 2007). Considerable genetic variation for these traits does exist within closely related alien species such as Festuca pratensis, F. glaucescens, F. arundinacea, F. maire and F. atlantigena, with which Lolium species such as L. perenne and L. multiflorum can be readily hybridized (King et al., 2007b).
Thus wild and cultivated relatives (alien species) of crop species provide a wealth of genetic variation for potentially all characters of strategic and fundamental importance, e.g. yield, climate change and the environment, recombination frequency and distribution, and perenniality. This genetic variation from alien species can be exploited both directly, i.e. target genes can be transferred into a crop species leading to the development of superior varieties, and indirectly, i.e. genes from alien species can be used to determine the genetic control of target traits and the pathways involved in their expression (Armstead et al., 2006a, b, 2007; Sandve et al., 2008).
Lolium perenne and F. pratensis can be readily hybridized to form interspecific hybrids and are indeed found as natural hybrids through North West Europe (Borrill, 1975; Humphreys et al., 2003). At meiosis the chromosomes of these species recombine at high frequency, yielding progeny that contain L. perenne/F. pratensis recombinant chromosomes that can be characterized using genetic markers (King et al., 2002a, b, 2007a, b; Kopecky et al., 2009) and also genomic in situ hybridization (GISH; King et al., 1998a, b, 2007a). Here we report the isolation of seven L. perenne/F. pratensis monosomic substitution lines, i.e. in each of the lines one of the seven chromosomes of L. perenne has been replaced with its F. pratensis homoeologue (13 L. perenne + 1 F. pratensis chromosomes). The potential for the exploitation of these monosomic substitution lines is discussed.
MATERIALS AND METHODS
Monosomic substitution lines of Lolium perenne × Festuca pratensis (Fig. 1) were identified from an initial population of BC1 individuals using a combination of GISH, amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) screens, in a background of detailed karyotypic knowledge, as described by King et al. (1998a, 2002a, b). Briefly GISH was used to identify genotypes which contained whole chromosome introgressions. AFLP templates were then prepared from this core set of genotypes and these were screened with a series of selective primer pairs: (a) to confirm the F. pratensis introgression by identifying Festuca-specific AFLP bands segregating in the target genotypes and (b) to identify which introgressions were derived from the same and which from different F. pratensis chromosome introgressions. Genetically mapped and chromosome-specific RFLPs and, subsequently, sequence tagged site (STS) and simple sequence repeat (SSR) markers were used to associate the introgressed F. pratensis chromosomes with previously identified linkage groups (Table 1). An alternative strategy of screening BC1 genotypes with chromosome-specific markers prior to GISH evaluation was also used to identify the last two monosomic substitutions with an expanded BC1 family.
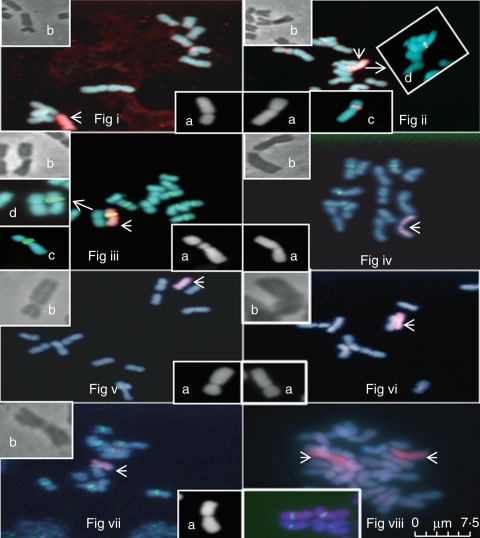
Fig. 1.
Mitosis in the seven monosomic substitution lines. (Fig i–vii) Monosomic lines 1–7; the substituted chromosome is shown by an arrow. Inset images: (a) 4′,6-diamidino-2-phenylindole (DAPI)-stained image of the relevant substitution line; (b) phase contrast image of the relevant substitution line. (Fig ii) Inset images: (c) and d) show the 5s site in monosomic line 2. (Fig iii) Inset images: (c) and (d) show monosomic line 3 carrying the pta71 ribosomal site. (Fig viii) A colchicine doubled line of monosomic line 3 (2n = 28) carrying a disomic substitution. Inset image: (a) shows the disomic chromosome substitutions for monosomic line 3 carrying the two pta71 sites. Scale bar = 7·5 µm.
Table 1.
Markers used to assign chromosome identities to F. pratensis whole-chromosome introgression lines
| Line 1 |
Line 2 |
Line 3 |
Line 4 |
Line 5 |
Line 6 |
Line 7 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | C* | S† | Marker | C | S | Marker | C | S | Marker | C | S | Marker | C | S | Marker | C | S | Marker | C | S |
| Rv0003 | 1 | A | Rv0062 | 2 | A,B | Rv0753 | 3 | A | Rv0161 | 4 | F | Rv0054 | 5 | A | Rv0067 | 6 | B | Rv0009 | 7 | A |
| Rv0033 | 1 | A,B | Rv0226 | 2 | A | Rv0774 | 3 | G | Rv0380 | 4 | A,B | Rv0184 | 5 | A | Rv0144 | 6 | A | Rv0011 | 7 | A |
| Rv0137 | 1 | A | Rv1008 | 2 | A | Rv0863 | 3 | A | Rv0454 | 4 | A,B | Rv0250 | 5 | A,B | Rv0297 | 6 | A | Rv0264 | 7 | A,B |
| Rv0165 | 1 | A | Rv1031 | 2 | A | Rv1046 | 3 | A,B | Rv0650 | 4 | A | Rv0342 | 5 | A | Rv0609 | 6 | A | Rv0333 | 7 | A |
| Rv0301 | 1 | A,B | Rv1068 | 2 | A | Rv1131 | 3 | A,B | Rv0922 | 4 | A | Rv0950 | 5 | A | Rv0641 | 6 | A,B | Rv0817 | 7 | A |
| Rv0367 | 1 | A | Rv1164 | 2 | A | Rv1172 | 3 | A | Rv0966 | 4 | A,B | Rv1112 | 5 | A,B | Rv0985 | 6 | A | Rv1171 | 7 | A,B |
| Rv0394 | 1 | A | Rv1239 | 2 | A | Rv1390 | 3 | F | Rv1053 | 4 | A | Rv1139 | 5 | A | Rv1036 | 6 | A | Rv1254 | 7 | A |
| Rv0624 | 1 | A | Rv1268 | 2 | A | Rv1439 | 3 | A | Rv1056 | 4 | A | Rv1307 | 5 | A | Rv1093 | 6 | F | Rv1408 | 7 | A |
| Rv1391 | 1 | A,B | Rv1269 | 2 | A | B3B8 | 3 | F | Rv1127 | 4 | G | CDO127 | 5 | H | Rv1208 | 6 | A,B | Rv1411 | 7 | A,B |
| 17ga1 | 1 | A | LpSSRH01A01 | 2 | C | Rv0677 | 3 | A | Rv1153 | 4 | A | Rv0157 | 3,7 | A | Rv1266 | 6 | A | 08ga1 | 7 | A |
| LpACT15H3 | 1 | E | Rv0188 | 2 | A,B | CDO345 | 3 | H,I,J | Rv1190 | 4 | A,B | Rv0752 | 4 | A | LpACT14C9 | 6 | E | LpACT13F9 | 7 | E |
| LpACA11H9 | 1 | F | CDO1417 | 2 | H | C949 | 3 | H,I | Rv1295 | 4 | A | Rv0809 | 4 | A | LpACA24B4 | 6 | F | LpACA11D4 | 7 | F |
| PR8 | 1 | F | CDO405 | 2 | H | C250 | 3 | H,I | LpACA8B9 | 4 | E | NFA048 | 6 | F | LpHCA17C6 | 7 | E | |||
| PR25 | 1 | F | CDO36 | 2 | H,J | WG889 | 3 | H,I | Rv0262 | 4 | A | Rye014 | 6 | B,C | B3C5 | 7 | F | |||
| PR37 | 1 | F | BCD855 | 2 | H | BCD828 | 3 | H,I | Rv0329 | 4 | A | Rye0739 | A | Rv0663 | 7 | A | ||||
| Rv1097 | 1 | A | Rv0447 | 3 | A | CDO455 | 3 | H,I | Rv0954 | 4 | A | Rv0908 | 7 | A,B | ||||||
| CDO98 | 1 | H | Rv0229 | 6 | G | PSR370 | 3 | H,I | Rv1089 | 4 | A | 06g10880 | 7 | D | ||||||
| CDO202 | 1 | H,J | PSR394 | 3 | H,I,J | Rv1302 | 4 | G | LpHCA17C6 | 7 | F | |||||||||
| Rv1154 | 2 | A | CDO920 | 3 | H,I,J | CDO122 | 4 | H,J | BCD147 | 7 | H | |||||||||
| R1613 | 3 | H,I | CDO20 | 4 | H | CDO545 | 7 | H,J | ||||||||||||
| CDO328 | 3 | H,I | Rv1087 | 1 | A | Rv0620 | 6 | A | ||||||||||||
| CDO460 | 3 | H,I,J | Rv0785 | 1 | A | |||||||||||||||
| C30 | 3 | H,I | Rv0810 | 1,7 | A | |||||||||||||||
| Rv0756 | 2 | G | Rv0103 | 2 | A | |||||||||||||||
| Rv0680 | 4 | A | Rv1063 | 3 | A | |||||||||||||||
| Rv0372 | 5 | G | ||||||||||||||||||
| Rv0551 | 7 | A | ||||||||||||||||||
* Chromosome to which the marker has been assigned by the mapping study referred to in column S.
† Mapping studies: A, Gill et al. (2006); B, Turner et al. (2008); C, Armstead et al. (2004); D, Armstead et al. (2008); E, King et al. (2008); F, J. King (unpubl. res.); G, G. Gill (Vialactia Biosciences, unpubl. res.); H, Armstead et al. (2002); J, Alm et al. (2003).
The seven monosomic substitution lines are perennial outbreeders and therefore have to be maintained as clones. Multiplying each genotype is done simply by tillering down and potting on. In this way it is possible to maintain the plants indefinitely.
Feulgen and GISH analyses were performed on pollen mother cells (PMCs) from each of the monosomic substitution lines at metaphase I of meiosis (Fig. 2) to observe the meiotic behaviour of the F. pratensis chromosomes (after King et al., 1998b).
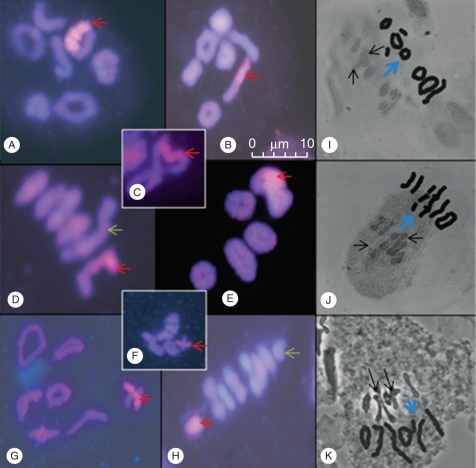
Fig. 2.
The analysis of chromosome pairing during meiotic metaphase I in L. perenne × F. pratensis monosomic substitution lines. (A–H) GISH images of meiotic metaphase I with F. pratensis (Fp) chromosome shown by a red arrow. (I–K) Phase images of pairing during meiotic metaphase I and drawn representations of the meiotic configuration shown by a blue arrow. (A) Monosomic line 3 showing a ring bivalent involving the Fp chromosome. (B) Monosomic line 5 showing a rod bivalent involving the Fp chromosome. (C) Monosomic line 4 showing the Fp chromosome as part of a quadrivalent. (D) Monosomic line 2 with a rod bivalent, but a quadrivalent is also present involving L. perenne (Lp) chromosomes shown by the green arrow. (E) Monosomic line 2 showing the Fp chromosome as part of a quadrivalent. (F) Proximal chiasma in monosomic line 1. (G) Proximal chiasma in monosomic line 5. (H) Monosomic line 2 showing the Fp chromosome as a univalent and also an Lp univalent (green arrow). (I) Monosomic line 3 showing two univalents. (J) Monosomic line 4 with one trivalent and three univalents. (K) Monosomic line 5 with one trivalent and one univalent.
RESULTS
Out of 161 BC1 genotypes screened with GISH, five genotypes appeared to carry a single, whole F. pratensis chromosome introgression (i.e. 1 F. pratensis + 13 L. perenne chromsomes). AFLP analysis identified unique F. pratensis-derived AFLP bands in all five genotypes containing the F. pratensis whole chromosome introgressions, indicating that all five genotypes probably contained different F. pratensis-derived chromsomes, i.e. five of the seven possible target chromosomes, a conclusion compatible with the karyotype observations (Fig. 3). Some overlapping F. pratensis-derived AFLP bands were identified between two of the genotypes. Repetition of the GISH analysis indicated that one of the genotypes had a whole chromosome plus a small terminal F. pratensis introgressed segment which had not been detected in the initial GISH images; this small introgressed segment was subsequently removed by a further round of backcrossing. Screening of the five different genotypes containing the whole chromosome introgressions with RFLP and SSR markers previously mapped to defined linkage groups (King et al., 2002a, 2008; J. King et al., unpubl. res.) identified the introgressed chromosomes as being derived from C1, C2, C3 (King et al., 2002a, b), C4 and C7 (Table 1). To isolate C5 and C6, a further 446 BC1 genotypes were generated and screened with C5- and C6-specific RFLP, STS and SSR markers to identify candidate genotypes containing at least partial C5 and C6 F. pratensis introgressions. GISH was then used to distinguish partial from whole chromosome introgressions, and this resulted in the identification of both C5 and C6 candidate monosomic substitutions. As with C1–C4 and C7, this was confirmed by karyotype evaluation (Fig. 3) and SSR screening (Table 1).
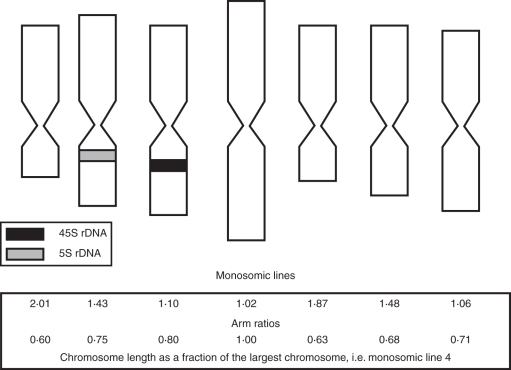
Fig. 3.
The individual chromosomes of F. pratensis assigned to their respective Triticeae linkage group. A combination of arm ratio, chromosome length, 5S rDNA and 45S rDNA allows identification of each of the F. pratensis chromosomes. The chromosomes are drawn to scale.
Meiotic analyses of Feulgen-stained PMCs from the diploid L. perenne genotype (the recurrent crossing parent) and each of the seven substitutions were analysed. A low frequency of univalents but no multivalents were recorded in the L. perenne genotype, but the meiotic analysis did reveal the presence of univalents and a low frequency of multivalent formation within the substitution lines (Table 2). The mean number of chiasmata per cell in each of the monosomic substitution lines did not vary significantly from the mean number recorded for L. perenne (χ2 = 0·03, 0·13, 0·03, 0·05, 0·003, 0·01 and 0·003 for substitution lines 1–7, d.f. = 1) at the 1 % level of probability. In order to determine if the introgressed F. pratensis chromosomes were involved in multivalent formation or the observed pairing failure, 25 PMCs per line were analysed at metaphase I of meiosis using GISH (Fig. 2). A very low frequency of multivalent formation involving the F. pratensis chromosome was observed, indicating the potential presence of interspecific chromosomal translocations (Table 3). The chiasma frequency within the homoeologous L. perenne/F. pratensis bivalent varied between 1·25 (substitution line 5) and 1·9 (substitution line 4). The chiasma frequency between Lolium/Lolium bivalents fell within this range, i.e. the chiasma frequency for the L. perenne/F. pratensis bivalent was slightly lower than the average chiasma frequency for all bivalents in substitution lines 1, 5 and 6, equal to it in substitution line 2 and higher in substitution lines 3, 4 and 7. The homoeologous L. perenne and F. pratensis chromosomes formed both rods (resulting from a single chiasmate exchange between two homoeologous chromosome arms) and rings (that resulted from two chiasmata between all four homoeologous chromosome arms; Fig. 2). Some differences were noted in the mean chiasmata per cell in Table 2 as compared with Table 3 although a χ2 test on these differences was not significant (χ2 = 1·79, d.f. = 6) at the 1 % level of probability.
Table 2.
Frequency of univalents, bivalents and multivalents in 50 Feulgen-stained pollen mother cells (PMCs) from L. perenne and each of the seven monosomic substitution lines
| Frequency of meiotic configurations in 50 PMCs* |
|||||
|---|---|---|---|---|---|
| Monosomic substitution line | Total no. of chiasmata* | I univalent | II bivalent | III trivalent | IV quadrivalent |
| 1 | 515 (10·30) | 4 (0·08) | 345 (6·90) | 2 (0·04) | 0 (0) |
| 2 | 542 (10·84) | 11 (0·22) | 340 (6·90) | 3 (0·06) | 2 (0·04) |
| 3 | 512 (10·24) | 16 (0·32) | 342 (6·84) | 0 (0) | 0 (0) |
| 4 | 502 (10·40) | 22 (0·44) | 312 (6·24) | 10 (0·20) | 6 (0·12) |
| 5 | 478 (9·56) | 33 (0·66) | 332 (6·64) | 1 (0·02) | 0 (0) |
| 6 | 498 (9·96) | 16 (0·32) | 340 (6·80) | 0 (0) | 1 (0·02) |
| 7 | 496 (9·92) | 54 (1·08) | 323 (6·46) | 0 (0) | 0 (0) |
| L. perenne (Liprior) Ba10890/8 (recurrent crossing parent) | 231 (9·24) | 4 (0·16) | 173 (6·92) | 0 (0) | 0 (0) |
*The means per cell are shown in parentheses.
Table 3.
GISH analysis of metaphase I of 25 pollen mother cells (PMCs) of each monosomic substitution line
| Frequency of meiotic configurations in 25 PMCs (%) |
Average chiasma frequency |
No. of Lp/Fp rods and rings |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Monosomic substitution line | Total no. of chiasma and mean per cell | I univalent | II bivalent | III trivalent | IV quadrivalent | For all bivalents, i.e. Lp/Lp and Lp/Fp | For Lp/Fp bivalents | Rods | Rings |
| 1 | 286 11·44 | 1 (0) 0·04 (0) | 173 (24) 6·92 (0·96) | 1 (1) 0·04 (0·04) | 0 0 | 1·64 | 1·33 | 16 | 8 |
| 2 | 308 12·32 | 3 (1) 0·12 (0·04) | 168 (22) 6·72 (0·88) | 1 (1) 0·04 (0·04) | 2 (1) 0·08 (0·04) | 1·77 | 1·86 | 3 | 19 |
| 3 | 293 11·72 | 2 (1) 0·08 (0·04) | 174 (24) 6·96 (0·96) | 0 0 | 0 0 | 1·67 | 1·84 | 2 | 22 |
| 4 | 283 11·32 | 2 (0) 0·08 (0) | 167 (21) 6·68 (0·84) | 2 (2) 0·08 (0·04) | 2 (2) 0·08 (0·04) | 1·62 | 1·90 | 2 | 19 |
| 5 | 292 11·68 | 2 (1) 0·08 (0·04) | 174 (24) 6·96 (0·96) | 0 0 | 0 0 | 1·67 | 1·25 | 18 | 6 |
| 6 | 309 12·36 | 0 0 | 175 (25) 7 (1·00) | 0 0 | 0 0 | 1·76 | 1·80 | 5 | 20 |
| 7 | 275 11·00 | 4 (2) 0·16 (0·08) | 173 (23) 6·92 (0·92) | 0 0 | 0 0 | 1·57 | 1·43 | 13 | 10 |
DISCUSSION
Table 1 lists the markers used for the association of the individual whole chromosome introgressions with particular chromosome designations (e.g. 1–7). While, in general, the syntenic relationships are consistent, 16 of the total 136 markers (approx. 12 %) identified different F. pratensis chromosomes from the one to which they had been mapped in L. perenne (Alm et al., 2003 also noted some limited, apparent breakdown in synteny between L. perenne and F. pratensis, with approx. 9 % of the common markers mapping to non-syntenic loci). There are a number of possible explanations for this. (a) While most SSR markers identify only a single locus, it is not infrequent that they can identify more than one locus on the same or on different linkage groups. Therefore, the locus tagged by a particular SSR in this introgression study may be different from the locus mapped in a different mapping population. (b) Small sequence differences between the SSR priming sites present in L. perenne and F. pratensis may lead to a particular SSR tagging different loci in the two species. (c) The conservation of synteny between L. perenne and F. pratensis chromosomes is not complete, i.e. there are some genomic rearrangements between the two species. The first two of these explanations really reflect limitations in marker technology, e.g. are the markers identifying the same loci? While this study cannot discriminate directly between these three explanations, work currently being undertaken on mapping populations produced from the monosomic substitution lines would suggest that any breakdown in synteny would be limited to disruptions to micro-colinearity rather than macro-colinearity. However, it is worth noting that for C4, approx. 25 % of the markers that tagged the F. pratensis-derived introgression had been mapped to different linkage groups in previous L. perenne mapping studies. This indicates that a genomic rearrangement might be more likely for this chromosome than the others. Further evidence for a genomic rearrangement comes from the observation that the group four F. pratensis chromosome is occasionally involved in multivalent formation. If this were the case, the nature of the genomic rearrangement would not appear to be a simple translocation, as the seven non-syntenous markers have been previously mapped to five different linkage groups (Table 1).
Univalents and rare multivalents have occasionally been observed in Feulgen-stained preparations of PMCs from some of the seven monosomic substitution lines (Table 2). In order to determine whether the F. pratensis chromosomes in these lines were involved in multivalent formation, or pairing failure resulting in univalent formation, GISH analysis was performed. Multivalents involving the F. pratensis chromosomes and the L. perenne genome would have indicated a breakdown in synteny between the two species at the macro-level. For example, in the absence of interspecific chromosomal rearrangements, only bivalents would be formed at metaphase I of meiosis, i.e. each Festuca chromosome shows complete synteny with its Lolium homoeologue. However, the involvement of a Festuca chromosome in a multivalent would indicate that it shows a syntenic relationship with more than one Lolium syntenic group. Multivalents were observed involving the Festuca chromosomes in substitution lines 1, 2 and 4. It therefore appears there maybe a breakdown in synteny between these chromosomes in Lolium and Festuca at the macro-level (especially as no multivalents were observed in a normal Lolium genotype; Table 2). However, the frequency of multivalent formation was very low, i.e. 0·04 %, indicating that the level of breakdown in synteny is low (Table 3). In addition, the presence of ring bivalents resulting from recombination involving both arms of the L. perenne and F. pratensis chromosomes indicates a relatively high level of conservation of synteny at the macro-level between the chromosomes of these species and thus confirms the results from the marker data. Thus any breakdown in synteny between L. perenne and F. pratensis that could lead to rare multivalent formation is likely to be complex, e.g. involve proximal translocations between the F. pratensis and L. perenne genomes.
Effective alien introgression strategies require high rates of homoeologous recombination between host and alien chromosomes (or very large populations and multiple generations) in order to minimize the transfer of an undesirable genetic load via linkage drag from the alien species. Recombination between homoeologous chromosomes within the forage grasses has previously been shown not to be compromised by differences in DNA content in terms of either total DNA content or repetitive sequences (Jenkins et al., 2000). Indeed, high rates of recombination between host L. perenne and alien F. pratensis chromosomes have already been observed for the chromosome 3 monosomic substitution line (King et al., 2007b). The cytogenetic observations confirm that all seven chromosomes are likely to recombine, though possibly with some limitations (the low numbers of univalents and multivalent observed). Work on a related diploid hybrid, L. temulentun × L. perenne, which also contained two sets of chromosomes which were genetically and structurally dissimilar, showed the hybrid to have a remarkable capacity to eliminate synaptonemal complex irregularities (which would interfere with pairing) and to produce homoeologous bivalents (Jenkins and White, 1990).
In this work we have described the transfer of the entire genome of F. pratensis into L. perenne through the development of seven monosomic substitution lines. This germplasm provides a unique source of genetic variation that can be exploited for the determination of the genetic control of target traits and the subsequent development of superior L. perenne varieties. Presently each of the seven substitutions are being screened for phenotypic variation for a range of traits that include tolerance to drought/cold, root morphology, nutrient use efficiency, disease resistance (Armstead et al., 2006b), etc. This will allow the identification of which of the seven F. pratensis chromosomes carries target genes for specific traits. Once a specific F. pratensis chromosome has been identified as carrying a gene controlling a target trait its exact position can be determined accurately via the exploitation of L. perenne/F. pratensis recombinant chromosome lines, i.e. a series of lines that carry small F. pratensis chromosome segments derived from the target monosomic substitution (J. King et al., unpubl. res.), thus providing a platform for the map-based cloning of the target gene(s) as previously demonstrated for the gene responsible for Mendel's I locus (Donnison et al., 2005; Moore et al., 2005; Armstead et al., 2006a, 2007; Ougham et al., 2008).
Within the Poaceae, the subfamily Pooideae contains ryegrasses, fescues, wheat, barley, rye oats and the model grass Brachypodium distachyon (purple false broom). The similarities and difference between these species (Table 4) present general opportunities for ‘comparing and contrasting’ across species in terms of general biology and, specifically, evolutionary and comparative genomics. For all the economically important grasses and cereals in this subfamily, alien introgression has been considered to be a useful route for introducing new traits (Humphreys et al., 2003; Hodgkin and Hajjar, 2008; Nevo and Chen, 2010). Particularly in wheat, the development of comprehensive collections of monosomic substitution and deletion lines has been a major target for both cytogeneticists and geneticists and, in parallel, wheat plant breeders have developed many alien introgression approaches for improving wheat germplasm. To these ends, a wide variety of wheat wild and cultivated relatives have been used in alien introgression programmes, e.g. rye, Triticum spp. Aegilops spp, Thinopyron spp. (Hohmann et al., 1996; King et al., 1997; Friebe et al., 1999; Ehdaie et al., 2003; Gupta et al., 2005; Jauhar et al., 2009; Motsnyi et al., 2009). In rye, barley and oats, while the range of species targeted for introgression is not so wide, considerable effort has still been expended in these directions (Rooney et al., 1994; Prieto et al., 2001; von Korff et al., 2004; Lukaszewski, 2006; Atienza et al., 2007; Schmalenbach et al., 2008, 2009; Falke et al., 2009; Fetch et al., 2009; Johnston et al., 2009; Schmalenbach and Pillen, 2009; Scholz et al., 2009). In contrast to the Pooideae crop species, B. distachyon is being developed as a model monocot with the understanding that its biology will be more analogous with the important cereals and grasses than that of Arabidopsis thaliana. However, within the genus Brachypodium there is also a wide variety of alien resources (Garvin et al., 2008; Ozdemir et al., 2008; Bakker et al., 2009) and alien introgression is likely to prove increasingly informative as a way of extending the (otherwise narrow) range of variation and thus increasing our capacity to understand and extrapolate from the biology of this model species, just has been the case with A. thaliana and its relatives (Ramos-Onsins et al., 2004; de Meaux et al., 2006; Frerot et al., 2010). The continuing development of next-generation sequencing (NGS) technologies has meant that the ‘art of the possible’ in genome analysis has changed to such an extent that detailed genomic characterizations of all these species are imminent (Varshney et al., 2009; Deschamps and Campbell, 2010). The implication of this is that the ability to develop DNA-based cross-references between these similar but also contrasting Pooideae genomes will become routine within the next few years. Genome-wide alien introgression is a tool for generating variation which is tractable to NGS-related analyses and, thus, these analyses provide the means for cataloguing the available variation at the level of molecular genomics. There remains the major challenge in associating genomic introgression with phenotype, though the development of dedicated, large-scale phenomics facilities might mitigate this. In addition, the implementation of parallel analyses at the levels of genotype and phenotype in the Pooideae species described in Table 4 should also help to leverage extra information and suggest further avenues of investigation. Hence the genome-wide alien introgression resource for L. perenne and F. pratensis described here will not only contribute to our knowledge of the biology of the ryegrasses and fescues and to our ability to manipulate their genomes for food security, but will also contribute to the generation of a model for understanding and exploiting the variation across the Pooideae genomes.
Table 4.
Characteristics of some Pooideae grasses and cereals
| Species | Brachypodium distchyon | Lolium perenne | Hordeum vulgare | Secale cereale | Avena sativa | Triticum aestivum |
|---|---|---|---|---|---|---|
| Common name | Purple false brome | Perennial ryegrass | Barley | Rye | Oats | Wheat |
| Tribe | Brachypodieae | Poeae | Triticeae | Triticeae | Aveneae | Triticeae |
| Chromosome no. (n) | 5 | 7 | 7 | 7 | 7 | 7 |
| Ploidy | 2n | 2n | 2n | 2n | 6n | 6n |
| Approx. genome size (Gb) | 0·35 | 2·3 | 5·5 | 8·1 | 13 | 17 |
| Agronomic status | Uncultivated | Cultivated | domesticated | Domesticated | Domesticated | Domesticated |
| Breeding system | Inbreeder | Outbreeder | Inbreeder | Outbreeder | Inbreeder | Inbreeder |
| Crop* | – | Forage/seed | Grain | Grain/forage | Grain | Grain |
| Life cycle | Annual | Perennial | Annual | Annual/perennial | Annual | Annual |
* Current major application(s) in the UK. Ryegrass also has many amenity uses and grain crops can be used as forages.
The germplasm developed is freely available upon request to the corresponding author.
ACKNOWLEDGEMENTS
The authors would like to thank the BBSRC and EMBO for funding this research.
LITERATURE CITED
- Alm V, Fang C, Busso CS, et al. A linkage map of meadow fescue (Festuca pratensis Huds.) and comparative mapping with other Poaceae. Theoretical and Applied Genetics. 2003;108:25–40. doi: 10.1007/s00122-003-1399-5. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Turner LB, King IP, Cairns AJ, Humphreys MO. Comparison and integration of genetic maps generated from F2 and BC1-type mapping populations in perennial ryegrass. Plant Breeding. 2002;121:501–507. [Google Scholar]
- Armstead IP, Turner LB, Farrell M, et al. Synteny between a major heading-date QTL in perennial ryegrass (Lolium perenne L.) and the Hd3 heading-date locus in rice. Theoretical and Applied Genetics. 2004;108:822–828. doi: 10.1007/s00122-003-1495-6. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Donnison IS, Aubry S, et al. From crop to model to crop: identifying the genetic basis of the stay-green mutation in the Lolium/Festuca forage and amenity grasses. New Phytologist. 2006a;172:592–597. doi: 10.1111/j.1469-8137.2006.01922.x. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Harper JA, Turner LB, et al. Introgression of crown rust (Puccinia coronata) resistance from meadow fescue (Festuca pratensis) into Italian ryegrass (Lolium multiflorum): genetic mapping and identification of associated molecular markers. Plant Pathology. 2006b;55:62–67. [Google Scholar]
- Armstead IP, Donnison IS, Aubry S, et al. Cross-species identification of Mendel's I locus. Science. 2007;315:73. doi: 10.1126/science.1132912. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Turner LB, Marshall AH, Humphreys MO, King IP, Thorogood D. Identifying genetic components controlling fertility in the outcrossing grass species perennial ryegrass (Lolium perenne) by quantitative trait loci analysis and comparative genetics. New Phytologist. 2008;178:559–571. doi: 10.1111/j.1469-8137.2008.02413.x. [DOI] [PubMed] [Google Scholar]
- Atienza SG, Martin AC, Martin A. Introgression of wheat chromosome 2D or 5D into tritordeum leads to free-threshing habit. Genome. 2007;50:994–1000. doi: 10.1139/g07-081. [DOI] [PubMed] [Google Scholar]
- Bakker EG, Montgomery B, Nguyen T. Strong population structure characterizes weediness gene evolution in the invasive grass species Brachypodium distachyon. Molecular Ecology. 2009;18:2588–2601. doi: 10.1111/j.1365-294X.2009.04225.x. [DOI] [PubMed] [Google Scholar]
- Borrill M. Festuca L. In: Stace CA, editor. Hybrization and the flora of the British Isles. London: Academic Press; 1975. pp. 543–547. [Google Scholar]
- Deschamps S, Campbell MA. Utilization of next-generation sequencing platforms in plant genomics and genetic variant discovery. Molecular Breeding. 2010;25:553–570. [Google Scholar]
- Donnison IS, O'Sullivan D, Thomas A, et al. Construction of a Festuca pratensis BAC library for map based cloning in Festulolium substitution lines. Theoretical and Applied Genetics. 2005;110:846–851. doi: 10.1007/s00122-004-1870-y. [DOI] [PubMed] [Google Scholar]
- Ehdaie B, Whitkus RW, Waines JG. Root biomass, water-use efficiency, and performance of wheat–rye translocations of chromosomes 1 and 2 in spring bread wheat ‘Pavon. Crop Science. 2003;43:710–717. [Google Scholar]
- Falke KC, Wilde P, Wortmann H, Geiger HH, Miedaner T. Identification of genomic regions carrying QTL for agronomic and quality traits in rye (Secale cereale) introgression libraries. Plant Breeding. 2009;128:615–623. [Google Scholar]
- Fetch T, Johnston PA, Pickering R. Chromosomal location and inheritance of stem rust resistance transferred from Hordeum bulbosum into cultivated barley (H. vulgare) Phytopathology. 2009;99:339–343. doi: 10.1094/PHYTO-99-4-0339. [DOI] [PubMed] [Google Scholar]
- Frerot H, Faucon MP, Willems G, et al. Genetic architecture of zinc hyperaccumulation in Arabidopsis halleri: the essential role of QTL × environment interactions. New Phytologist. 2010;187:355–367. doi: 10.1111/j.1469-8137.2010.03295.x. [DOI] [PubMed] [Google Scholar]
- Friebe B, Kynast RG, Hatchett JH, Sears RG, Wilson DL, Gill BS. Transfer of wheat–rye translocation chromosomes conferring resistance to Hessian fly from bread wheat into durum wheat. Crop Science. 1999;39:1692–1696. [Google Scholar]
- Garvin DF, Gu YQ, Hasterok R, et al. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Science. 2008;48:S69–S84. [Google Scholar]
- Gill GP, Wilcox PL, Arens P, et al. A framework linkage map of perennial ryegrass based on SSR markers. Genome. 2006;49:354–364. doi: 10.1139/g05-120. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Kulwal PL, Rustgi S. Wheat cytogenetics in the genomics era and its relevance to breeding. Cytogenetic and Genome Research. 2005;109:315–327. doi: 10.1159/000082415. [DOI] [PubMed] [Google Scholar]
- Hodgkin T, Hajjar R. Using crop wild relatives for crop improvement: trends and perspectives. In: Maxted N, Ford-Lloyd BV, Kell SP, Iriondo JM, Dulloo ME, Turok J, editors. Proceedings of the First International Conference on Crop Wild Relative Conservation and Use. 2008. pp. 535–548. Sicily, Italy, 14–17 September 2005. [Google Scholar]
- Hohmann U, Badaeva K, Busch W, Friebe B, Gill BS. Molecular cytogenetic analysis of Agropyron chromatin specifying resistance to barley yellow dwarf virus in wheat. Genome. 1996;39:336–347. doi: 10.1139/g96-044. [DOI] [PubMed] [Google Scholar]
- Humphreys MW, Canter PJ, Thomas HM. Advances in introgression technologies for precision breeding within the Lolium–Festuca complex. Annals of Applied Biology. 2003;143:1–10. [Google Scholar]
- Jauhar PP, Peterson TS, Xu SS. Cytogenetic and molecular characterization of a durum alien disomic addition line with enhanced tolerance to Fusarium head blight. Genome. 2009;52:467–483. doi: 10.1139/g09-014. [DOI] [PubMed] [Google Scholar]
- Jenkins G, White J. Elimination of synaptonemal complex irregularities in a Lolium hybrid. Heredity. 1990;64:45–53. [Google Scholar]
- Jenkins G, Head J, Foster JW. Probing meiosis in hybrids of Lolium (Poaceae) with a discriminatory repetitive genomic sequence. Chromosoma. 2000;109:280–286. doi: 10.1007/s004120000078. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Timmerman-Vaughan GM, Farnden KJF, Pickering R. Marker development and characterisation of Hordeum bulbosum introgression lines: a resource for barley improvement. Theoretical and Applied Genetics. 2009;118:1429–1437. doi: 10.1007/s00122-009-0992-7. [DOI] [PubMed] [Google Scholar]
- King IP, Forster BP, Law CC, et al. Introgression of salt-tolerance genes from Thinopyrum bessarabicum into wheat. New Phytologist. 1997;137:75–81. [Google Scholar]
- King IP, Morgan WG, Armstead IP, et al. Introgression mapping in the grasses. I. Introgression of Festuca pratensis chromosomes and chromosome segments into Lolium perenne. Heredity. 1998a;81:462–467. [Google Scholar]
- King IP, Morgan WG, Harper JA, Thomas HM. Introgression mapping in the grasses. II. Meiotic analysis of the Lolium perenne/Festuca pratensis triploid hybrid. Heredity. 1998b;82:107–112. [Google Scholar]
- King J, Armstead IP, Donnison IS, et al. Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics. 2002a;161:307–314. doi: 10.1093/genetics/161.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Roberts LA, Kearsey MJ, et al. A demonstration of a 1:1 correspondence between chiasma frequency and recombination using a Lolium perenne/Festuca pratensis substitution line. Genetics. 2002b;161:315–324. doi: 10.1093/genetics/161.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Armstead IP, Donnison IS, et al. Comparative analyses between Lolium/Festuca introgression lines and rice reveal the major fraction of functional annotated gene models are located in recombination poor/very poor regions of the genome. Genetics. 2007a;177:597–606. doi: 10.1534/genetics.107.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Armstead IP, Donnison IS, et al. Introgression mapping in the grasses. Chromosome Research. 2007b;15:105–113. doi: 10.1007/s10577-006-1103-0. [DOI] [PubMed] [Google Scholar]
- King J, Thorogood D, Edwards KJ, et al. Development of a genomic microsatellite library in perennial ryegrass (Lolium perenne L.) Annals of Botany. 2008;101:845–853. doi: 10.1093/aob/mcn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky D, Bartos J, Lukaszewski AJ, et al. Development and mapping of DArT markers within the Festuca–Lolium complex. BMC Genomics. 2009;10:473. doi: 10.1186/1471-2164-10-473. doi:10.1186/1471-2164-10-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Korff M, Wang H, Leon J, Pillen K. Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp spontaneum) as donor. Theoretical and Applied Genetics. 2004;109:1736–1745. doi: 10.1007/s00122-004-1818-2. [DOI] [PubMed] [Google Scholar]
- Kosmala A, Zwierzykowski Z, Zwierzykowska E, et al. Introgression mapping of genes for winter hardiness and frost tolerance from Festuca arundinacea into Lolium multiflorum. Journal of Heredity. 2007;98:311–316. doi: 10.1093/jhered/esm047. [DOI] [PubMed] [Google Scholar]
- Lukaszewski AJ. Cytogenetically engineered rye chromosomes 1R to improve bread-making quality of hexaploid triticale. Crop Science. 2006;46:2183–2194. [Google Scholar]
- de Meaux J, Pop A, Mitchell-Olds T. Cis-regulatory evolution of chalcone-synthase expression in the genus Arabidopsis. Genetics. 2006;174:2181–2202. doi: 10.1534/genetics.106.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BJ, Donnison IS, Harper JA, et al. Molecular tagging of a senescence gene by introgression mapping of a mutant stay-green locus from Festuca pratensis. New Phytologist. 2005;165:801–806. doi: 10.1111/j.1469-8137.2004.01269.x. [DOI] [PubMed] [Google Scholar]
- Motsnyi II, Fayt VI, Blagodarova EM. Identification and characteristics of the 1R (1B) substitution lines of bread wheat. Cytology and Genetics. 2009;43:169–176. [PubMed] [Google Scholar]
- Nevo E, Chen GX. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant, Cell and Environment. 2010;33:670–685. doi: 10.1111/j.1365-3040.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- Ougham H, Hörtensteiner S, Armstead I, et al. The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biology. 2008;10:4–14. doi: 10.1111/j.1438-8677.2008.00081.x. [DOI] [PubMed] [Google Scholar]
- Ozdemir BS, Hernandez P, Filiz E, Budak H. Brachypodium genomics. International Journal of Plant Genomics. 2008;2008:536104. doi: 10.1155/2008/536104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P, Ramirez MC, Ballesteros J, Cabrera A. Identification of intergenomic translocations involving wheat, Hordeum vulgare and Hordeum chilense chromosomes by FISH. Hereditas. 2001;135:171–174. doi: 10.1111/j.1601-5223.2001.t01-1-00171.x. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins SE, Stranger BE, Mitchell-Olds T, Aguade M. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics. 2004;166:373–388. doi: 10.1534/genetics.166.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapacz M, Gasior D, Zwierzykowski Z, Lesniewska-Bocianowska A, Humphreys MW, Gay AP. Changes in cold tolerance and the mechanisms of acclimation of photosystem II to cold hardening generated by anther culture of Festuca pratensis × Lolium multiflorum cultivars. New Phytologist. 2004;162:105–114. [Google Scholar]
- Rooney WL, Jellen EN, Phillips RL, Rines HW, Kianian SF. Identification of homoeologous chromosomes in hexaploid oat (A. byzantina cv Kanota) using monosomics and RFLP analysis. Theoretical and Applied Genetics. 1994;89:329–335. doi: 10.1007/BF00225163. [DOI] [PubMed] [Google Scholar]
- Sandve SR, Rudi H, Asp T, Rognli OA. Tracking the evolution of a cold stress associated gene family in cold tolerant grasses. BMC Evolutionary Biology. 2008;8:245. doi: 10.1186/1471-2148-8-245. doi:10.1186/1471-2148-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I, Pillen K. Detection and verification of malting quality QTLs using wild barley introgression lines. Theoretical and Applied Genetics. 2009;118:1411–1427. doi: 10.1007/s00122-009-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I, Korber N, Pillen K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theoretical and Applied Genetics. 2008;117:1093–1106. doi: 10.1007/s00122-008-0847-7. [DOI] [PubMed] [Google Scholar]
- Schmalenbach I, Leon J, Pillen K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theoretical and Applied Genetics. 2009;118:483–497. doi: 10.1007/s00122-008-0915-z. [DOI] [PubMed] [Google Scholar]
- Scholz M, Ruge-Wehling B, Habekuss A, et al. Ryd4Hb: a novel resistance gene introgressed from Hordeum bulbosum into barley and conferring complete and dominant resistance to the barley yellow dwarf virus. Theoretical and Applied Genetics. 2009;119:837–849. doi: 10.1007/s00122-009-1093-3. [DOI] [PubMed] [Google Scholar]
- Turner LB, Cairns AJ, Armstead IP, Thomas H, Humphreys MW, Humphreys MO. Does fructan have a functional role in physiological traits? Investigation by quantitative trait locus mapping. New Phytologist. 2008;179:765–775. doi: 10.1111/j.1469-8137.2008.02495.x. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Nayak SN, May GD, Jackson SA. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology. 2009;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]