Abstract
Recent published sequencing of fungal genomes has revealed that these microorganisms have a surprisingly large number of secondary metabolite pathways that can serve as potential sources for new and useful natural products. Most of the secondary metabolites and their biosynthesis pathways are currently unknown, possibly because they are produced in very small amounts and are thus difficult to detect or are produced only under specific conditions. Elucidating these fungal metabolites will require new molecular genetic tools, better understanding of the regulation of secondary metabolism, and state of the art analytical methods. This review describes recent strategies to mine the cryptic natural products and their biosynthetic pathways in fungi.
Keywords: natural products biosynthesis, genomic mining, polyketide synthase, nonribosomal peptide synthetase
Secondary metabolites are remarkable resources for medically useful compounds. Fungi live in complex ecosystems and must compete with other organisms, such as bacteria, algae, other fungi, protozoans and small metazoans. They have evolved the ability to produce secondary metabolites that kill, or inhibit the growth of, their competitors. Thus, it is not surprising that fungal secondary metabolites have included a number of important drugs such as the antibiotic penicillin (1), the immunosuppressant cyclosporine (2), and the anti-hypercholesterolemic agent lovastatin (3) (Figure 1) [1]. Recent genome sequencing, however, has revealed that genes involved in secondary metabolite biosynthesis are more abundant than anyone had anticipated. This suggests that there is still a vast number of compounds with new chemical structures that could be isolated from filamentous fungi [2].
Figure 1.
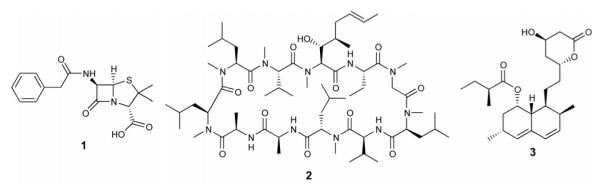
Chemical structures of medicinally important natural products: penicillin G (1), cyclosporine A (2), and lovastatin (3).
Fungi are known to produce several classes of secondary metabolites, including polyketides, non-ribosomal peptides, other amino acid derived compounds and terpenes [1]. Genomic analysis in Aspergillus nidulans, for example, identified 27 polyketide synthases (PKSs) and 14 non-ribosomal peptide synthetases (NRPSs), which are responsible for polyketide and non-ribosomal peptide biosynthesis, respectively [3]. Fungal polyketides are produced by multidomain type I PKSs which are iterative in nature and can be further grouped into nonreduced (NR), partially reduced (PR), and highly reduced (HR) PKSs by examining the existence of additional tailoring domains encoded in the gene [4,5]. An important feature that can facilitate genetic analysis of secondary metabolism biosynthesis pathways in fungi is the fact that the genes of individual secondary metabolite pathways are usually clustered together in the genome [6].
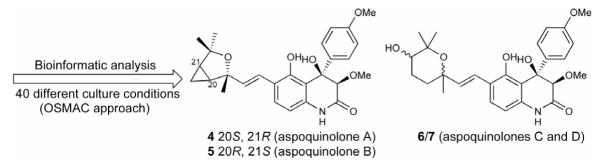
For reasons still not well understood, most biosynthetic gene clusters are either cryptic or expressed at levels too low to result in product detection in standard laboratory culture conditions. Systematically varying fermentation parameters, an approach termed OSMAC (One Strain-Many Compounds), has been successful in eliciting the production of some of these unknown compounds [7, 8]. An example of combining OSMAC and bioinformatic analysis is the discovery of aspoquinolones A – D (4 – 7, Figure 2) by Scherlach and Hertweck [9]. They recognized that the A. nidulans genome codes for multiple anthranilate synthases, which are responsible for quinoline or quinazoline alkaloid biosynthesis. They cultivated A. nidulans in numerous growth conditions and discovered aspoquinolones by UV and mass spectrometry screening.
Figure 2.
Compounds isolated from A. nidulans by combining bioinformatic analysis and the OSMAC approach.
Although the OSMAC approach allows to an extent the revelation of the hidden reservoir of chemical diversity, most biosynthetic gene clusters in fungi are still underexplored. It appears that some chemical or environmental signals for inducing secondary metabolite genes are missing in laboratory culture conditions. Genome mining thus offers a powerful tool for discovering cryptic natural products, especially when signaling pathways from external stimuli to gene expression are elucidated. The recently published genomes from A. nidulans, A. niger, and A. oryzae demonstrate that the majority of pathways identified in Aspergilli is neither redundant in different species nor duplicated within a given genome [2]. Excitingly, we now have genetic tools for gene-targeting to study these pathways with remarkable facility [10-12].
Manipulating expression of cluster-specific regulatory activators to discover novel secondary metabolites
The presence of cluster-specific regulatory activators in fungal biosynthetic gene clusters is a common feature, including tri6 for tricothecene biosynthesis [13], aflR for aflatoxin biosynthesis [14], and ctnA for citrinin biosynthesis [15], among others [16]. Yu et al. showed “proof-of-concept” that one could overexpress a pathway specific regulatory gene and generate a final metabolite [14]. Bergmann et al. first demonstrated that ectopic overexpression of apdR, a Zn2Cys6 regulator gene within a cryptic hybrid PKS-NRPS gene cluster, placed under the control of an inducible promoter results in the concerted activation of the gene cluster, allowing the identity of two new cytotoxic metabolites, aspyridones A (8) and B (9) (Figure 3) [17]. Using a similar approach, the asperfuranone (10) gene cluster was discovered in our laboratory by replacing the endogenous promoter of a regulatory activator in the asperfuranone gene cluster, afoA, with an inducible promoter (Figure 4) [18]. Deletion of afoD, a key hydroxylase in the asperfuranone biosynthesis, resulted in accumulation of the benzaldehyde derivative 11. This result established the function of two PKSs, one HR-PKS (AfoG) and one NR-PKS (AfoE), together involved in the 11 biosynthesis, prompting the proposed***** asperfuranone biosynthesis pathway. The advantage of this approach is that only a regulatory activator needs to be handled and in some cases, concerted expression of all pathway genes can be triggered. Because the large quantities of genomic sequence data from a wide variety of organisms are accessible and still exponentially growing in publicly databases, this approach clearly will provide a new avenue for drug discovery [19].
Figure 3.
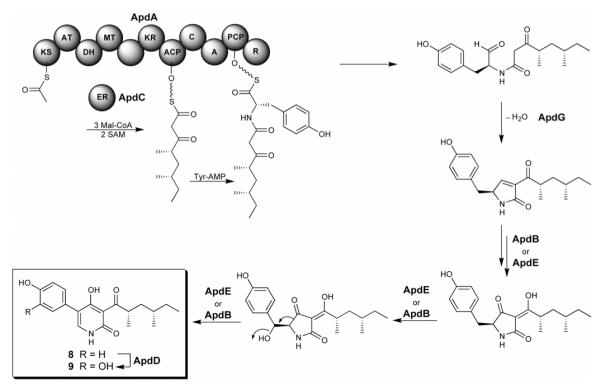
Unlocking a fungal cryptic PKS-NRPS via activation of a pathway-specific regulator to produce aspyridones A (8) and B (9).
Figure 4.
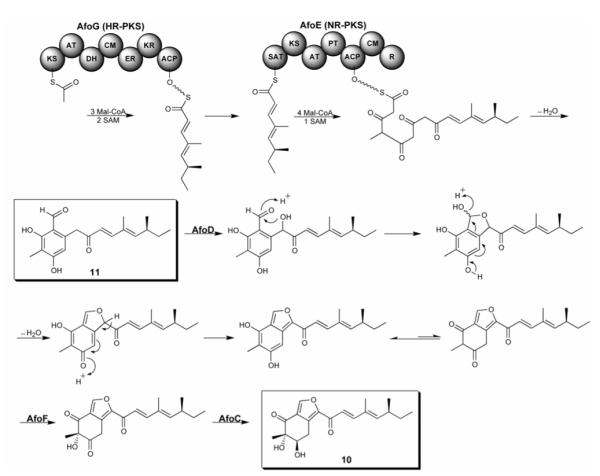
Unlocking fungal cryptic dual PKSs via activation of a pathway-specific regulator to produce asperfuranone (10).
Discovery of LaeA, a global regulator of secondary metabolism
The identification of LaeA (loss of aflR expression), a nuclear protein regulating secondary metabolite production in Aspergillus spp., leads to the hypothesis that LaeA functions as a global regulator of secondary metabolism in this genus [20]. Loss of LaeA decreased sterigmatocystin (12) and penicillin (1) production in A. nidulans and gliotoxin (13) production in A. fumigatus (Figure 5). Sequence analysis of LaeA indicated it encodes a methyltransferase with some sequence similarity to histone and arginine methyltransferases. The sequence similarity as well as the sub-telomeric locations of many of the targets of LaeA suggests that this protein acts via chromatin remodeling [1]. A whole-genome comparison of the transcriptional profile of wild-type, ΔlaeA, and complemented control strains of A. fumigatus showed that LaeA controls transcription of at least 9.5% of the genome, and 13 of 22 secondary metabolite gene clusters were positively regulated by LaeA. Seven of these regulated clusters are sub-telomeric, in the regions with a high degree of heterochromatin [21]. Microarray analysis of laeA deletion and overexpression A. nidulans strains, as proof-of-principle, led to the discovery of the terrequinone A (14) gene cluster (Figure 5) [22]. Thus, manipulation of laeA expression levels will be of benefit to identify previously unknown metabolites in fungi.
Figure 5.
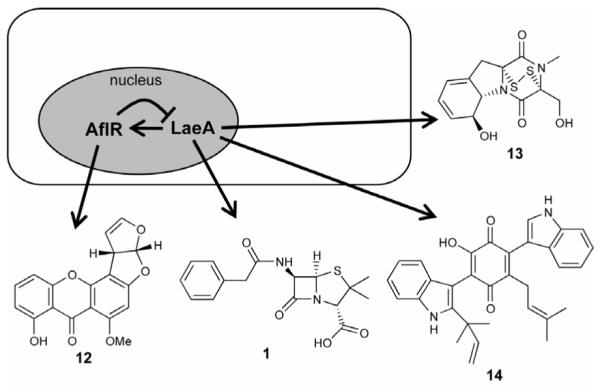
>Increasing secondary metabolite production via overexpression of the global regulator, LaeA.
Epigenetic regulation of fungal secondary metabolism
Efforts to uncover the mechanism of the global regulator LaeA revealed that some subtelomeric secondary metabolite clusters were located in heterochromatic regions of the genome. Importantly, gene transcription of telomere-proximal gene clusters and the level of the corresponding molecular generation are controlled by epigenetic regulation such as histone deacetylation. Deletion of hdaA (histone deacetylase) or treatment with a histone deacetylase (HDAC) inhibitor in Aspergillus nidulans resulted in the transcriptional activation of sterigmatocystin (12) and penicillin (1) gene clusters, both located in subtelomeres [23]. Cichewicz et al. rationally hypothesized that small-molecular epigenetic modifiers such as HDAC or DNA methyltransferase inhibitors could modulate secondary metabolite production. Treatment of Cladosporium cladosporioides with 5-azacytidine (a DNA methyltransferase inhibitor) stimulated the production of several oxylipins including (9Z,12Z)-11-hydroxyoctadeca-9,12-dienoic acid (15), its methyl ester (16), and a glycerol conjugate (17) (Figure 6). In contrast, administration of suberoylanilide hydroxamic acid (an HDAC inhibitor) yielded production of a complex series of perylenequinones including cladochromes (18 – 23) and calphostin B (24). Treatment of a Diatrype species with 5-azacytidine elicited the formation of lunalides A (25) and B (26) [24]. The Cichewicz group also isolated nygerone A (27) from Aspergillus niger when culturing with suberoylanilide hydroxamic acid [25]. These results demonstrate the potential of triggering cryptic metabolites through chemical epigenetic methodology.
Figure 6.
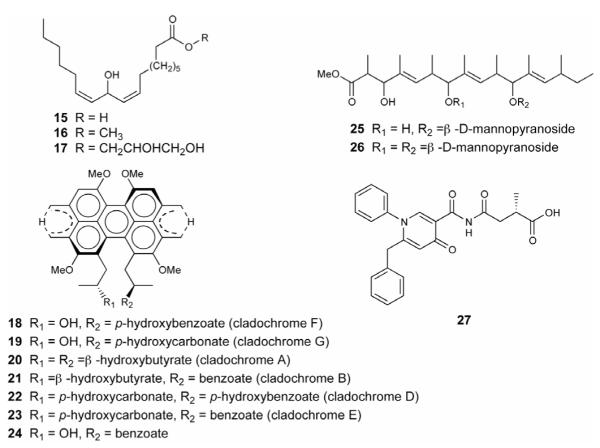
Unlocking fungal cryptic secondary metabolites via addition of DNA methyltransferase and histone deacetylase (HDAC) inhibitors to the culture medium.
Accruing evidence linking chromatin modifications with secondary metabolite cluster regulation led us to examine the hypothesis that additional chromatin modifying proteins were important in their regulation. We examined a member of COMPASS (complex associated with Set1) which methylates lysine 4 on histone 3 (H3K4) [26, 27]. The COMPASS complex consists of eight members, Set1, Bre2, Sdc1, Shg1, Spp1, Swd1, Swd2, and Swd3. Evidence from Set1 trimethyl-defective mutants in Saccharomyces cerevisiae suggest that mono- and/or dimethylation of H3K4 is important for cell growth, whereas trimethylation is required for silencing in telomeric regions [28]. CclA, an ortholog of bre2 in S. cerevisiae, was deleted in A. nidulans. Chemical profiling followed by genetic analysis led to the identification of two NR-PKS gene clusters, one cluster responsible for monodictyphenone (28), emodin (29) and emodin derivatives (30 – 33), and a second encoding the enzymes for F9775A (34) and B (35) (Figure 7) [29]. Interestingly, chromatin immunoprecipitation (ChIP) analysis of the two up-regulated genes in the monodictyphenone cluster of ΔcclA mutant showed that not only H3K4 but also H3K9 di- and trimethylation levels were suppressed. One non-activated gene nearby, however, was associated with only reduced levels of di- and trimethylation at H3K4 but not H3K9. Thus, strongly reduced levels of di- and trimethylation at H3K4 and H3K9 are required for de-repression during secondary metabolism. Since not all secondary metabolite gene clusters contain pathway specific activators, modification of the chromatin landscape provides another means to active cryptic gene clusters [29].
Figure 7.
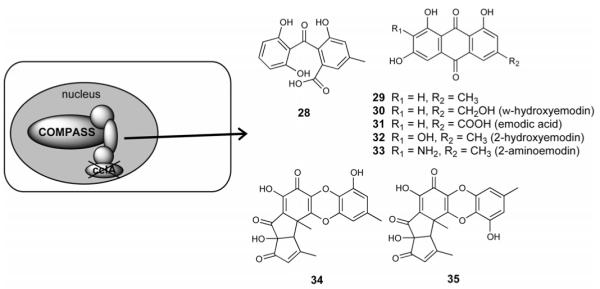
Unlocking fungal cryptic secondary metabolites via epigenetic regulation: Deletion of CclA, a member of the COMPASS complex.
Histone proteins are substrates for a wide array of modifications including acetylation, methylation, phosphorylation, and ubiquitination. SUMO is a small ubiquitin-like protein that is added posttranslationally to a number of proteins in the cell. In A. nidulans, there is a single SUMO gene, and the deletion of sumO causes only a slight inhibition of growth [30]. Secondary metabolite analysis showed that ΔsumO mutant decreased the production of austinol (36) and dehydroaustinol (37), dramatically increased the production of asperthecin (38), and did not alter the production of emericellamides (39 – 43) (Figure 8A). The aromatic nature of asperthecin (38) suggested an NR-PKS is responsible for its biosynthesis and the pathway was established through a series of knockout genetic analysis (Figure 8B) [31]. However, the mechanisms underlying SUMO regulation of secondary metabolism remains elusive; it may occur at different levels of regulation, including chromatin modification of the gene cluster, transcription factor modification, or the consequences of the effects on growth or primary metabolism.
Figure 8.
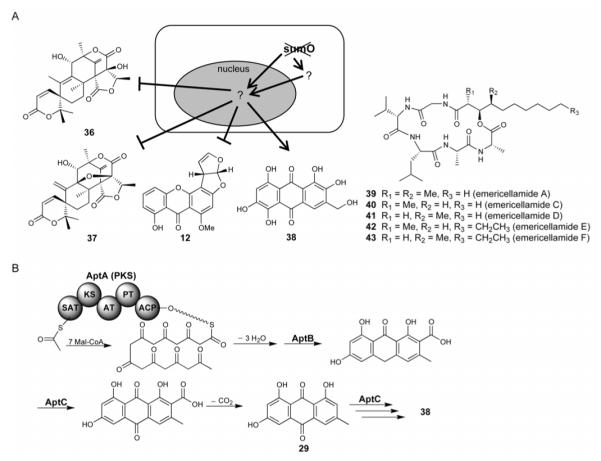
(A) Unlocking fungal cryptic secondary metabolites via deletion of SumO protein. (B) Proposed biosynthetic pathway of asperthecin (38) generated from genetic and chemical analysis.
Conclusion
With an ever-increasing number of complete genome sequences at hand, many novel gene clusters have now been identified. However, the corresponding secondary metabolites remain elusive, and researchers worldwide are working together to annotate gene function in the post-genomic era [32]. This review presents that, given exciting new approaches for identifying novel natural products, the next frontier of natural product discovery will be a synergistic combination of genomics, molecular genetics, biochemistry, and natural product chemistry to mine the fungal metabolome for useful natural products. Clearly, natural product chemistry and molecular genetics have a new interrelationship. Collaborations between mycologists, geneticists, biochemists and chemists are essential to facilitate the discovery of novel natural products and the genes involved in their biosynthesis, which in the end will benefit the commercial search of enzymatic reactions [33]. The OSMAC approach, co-culture experiments [25], specific regulator activation, and epigenetic regulation will continue to play crucial roles for discovering cryptic novel natural products. Although epigenetic approaches might have serious drawbacks, chiefly because targeting some histone-modifying complexes for loss-of-function could have negative effects on other cellular processes, continued development of genome mining tools will make these cryptic systems a potentially invaluable resource for the discovery of new chemical entities.
Acknowledgement
This review article was supported by Grant Number PO1GM084077 from the National Institute of General Medical Sciences to NK and CCCW.
References
- [1].Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nature Review Microbiology. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- [2].Schneider P, Misiek M, Hoffmeister D. In vivo and in vitro production options for fungal secondary metabolites. Molecular Pharmaceutics. 2008;5:234–242. doi: 10.1021/mp7001544. [DOI] [PubMed] [Google Scholar]
- [3].Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with. A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- [4].Sanchez JF, Chiang YM, Wang CCC. Diversity of polyketide synthases found in the Aspergillus and Streptomyces genomes. Molecular Pharmaceutics. 2008;5:226–233. doi: 10.1021/mp700139t. [DOI] [PubMed] [Google Scholar]
- [5].Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Organic & Biomolecular Chemistry. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- [6].Walton JD. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genetics and Biology. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- [7].Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [8].Gross H. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Applied Microbiology and Biotechnology. 2007;75:267–277. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]
- [9].Scherlach K, Hertweck C. Discovery of aspoquinolones A-D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Organic & Biomolecular Chemistry. 2006;4:3517–3520. doi: 10.1039/b607011f. [DOI] [PubMed] [Google Scholar]
- [10].Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proceedings of the National Academy of Science. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nature Protocol. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- [12].Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chemistry & Biology. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Proctor RH, Hohn TM, McCormick SP, Desjardins AE. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Applied and Environmental Microbiology. 1995;61:1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yu JH, Butchko RA, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Current Genetics. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
- [15].Shimizu T, Kinoshita H, Nihira T. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus. Applied and Environmental Microbiology. 2007;73:5097–5103. doi: 10.1128/AEM.01979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Natural Product Reports. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- [17].Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Natural Chemical Biology. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- [18].Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. Journal of the American Chemical Society. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brakhage AA, Schuemann J, Bergmann S, Scherlach K, Schroeckh V, Hertweck C. Activation of fungal silent gene clusters: a new avenue to drug discovery. Progress in Drug Research. 2008;66:3–12. doi: 10.1007/978-3-7643-8595-8_1. [DOI] [PubMed] [Google Scholar]
- [20].Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryotic Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLOS Pathogens. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chemistry & Biology. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- [23].Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryotic Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Organic & Biomolecular Chemistry. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- [25].Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Organic & Biomolecular Chemistry. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- [26].Sims RJ, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes & Development. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- [27].Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nature Chemical Biology. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wong KH, Todd RB, Oakley BR, Oakley CE, Hynes MJ, Davis MA. Sumoylation in Aspergillus nidulans: sumO inactivation, overexpression and live-cell imaging. Fungal Genetics and Biology. 2008;45:728–737. doi: 10.1016/j.fgb.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CCC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Applied and Environmental Microbiology. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, von Dohren H, Doonan J, Driessen AJ, Durek P, Espeso E, Fekete E, Flipphi M, Estrada CG, Geysens S, Goldman G, de Groot PW, Hansen K, Harris SD, Heinekamp T, Helmstaedt K, Henrissat B, Hofmann G, Homan T, Horio T, Horiuchi H, James S, Jones M, Karaffa L, Karanyi Z, Kato M, Keller N, Kelly DE, Kiel JA, Kim JM, van der Klei IJ, Klis FM, Kovalchuk A, Krasevec N, Kubicek CP, Liu B, Maccabe A, Meyer V, Mirabito P, Miskei M, Mos M, Mullins J, Nelson DR, Nielsen J, Oakley BR, Osmani SA, Pakula T, Paszewski A, Paulsen I, Pilsyk S, Pocsi I, Punt PJ, Ram AF, Ren Q, Robellet X, Robson G, Seiboth B, van Solingen P, Specht T, Sun J, Taheri-Talesh N, Takeshita N, Ussery D, vanKuyk PA, Visser H, van de Vondervoort PJ, de Vries RP, Walton J, Xiang X, Xiong Y, Zeng AP, Brandt BW, Cornell MJ, an den Hondel CA, Visser J, Oliver SG, Turner G. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genetics and Biology. 2009;46:S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fox EM, Howlett BJ. Secondary metabolism: regulation and role in fungal biology. Current Opinion in Microbiology. 2008;11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]