Abstract
Ribozymes offer a potentially important way to inactivate intracellular RNA from almost any gene whose nucleotide sequence is known. Recently, we found that hammerhead ribozymes directed against mRNA of tumour necrosis factor alpha (TNF alpha) and its derivatives, preferentially bind to a cellular protein(s). To better understand the effect of different 3'-terminal hairpins on ribozyme stability as well as their effect on the protein binding to the ribozyme, a mathematical treatment of the decay of three TNF alpha ribozymes that differed at their 3' ends was performed. One ribozyme contained a 3'-terminal hairpin derived from a transcription terminator of bacteriophage T7, another contained the same hairpin but modified to be highly enriched for G+C nucleotides, and a third lacked a hairpin. The TNF alpha ribozyme decay had two kinetic components. The slow component exhibited exponential decay with a half life of approximately 250 h in all cases. The 3'-terminal hairpin has no significant effect on this component. This slow phase accounted for 60-80% of ribozyme decay. The rapid phase also exhibited exponential decay. For this phase, a 3'-terminal hairpin roughly doubled the half-life (1.7-3.4). The slow phase of degradation was about three times faster for a ribozyme directed at the integrase mRNA of human immunodeficiency virus-1 than that seen with the TNF alpha ribozyme. Taken together, these results suggest that the ribozyme population is initially sensitive to degradation, with the presence of a hairpin provides some protection, and indicate that the addition of the hairpin to the ribozyme did not prevent the in vivo additional stabilizing effect of the protein(s).
Full text
PDF
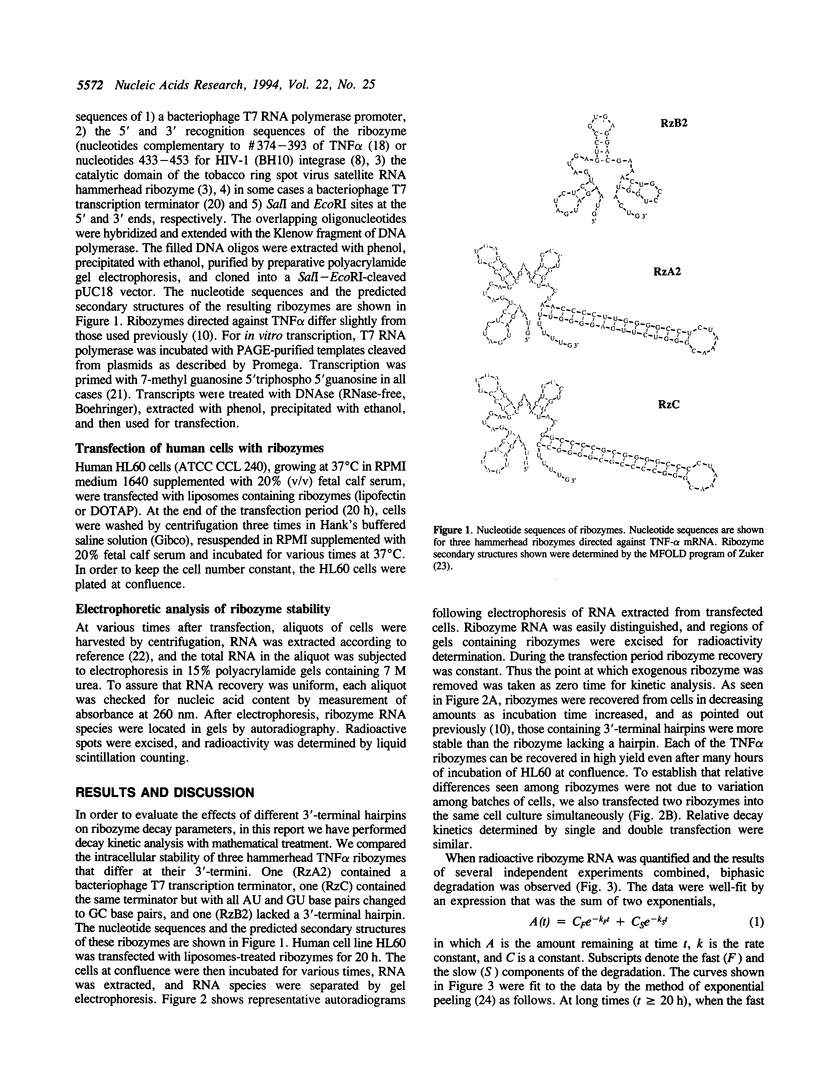
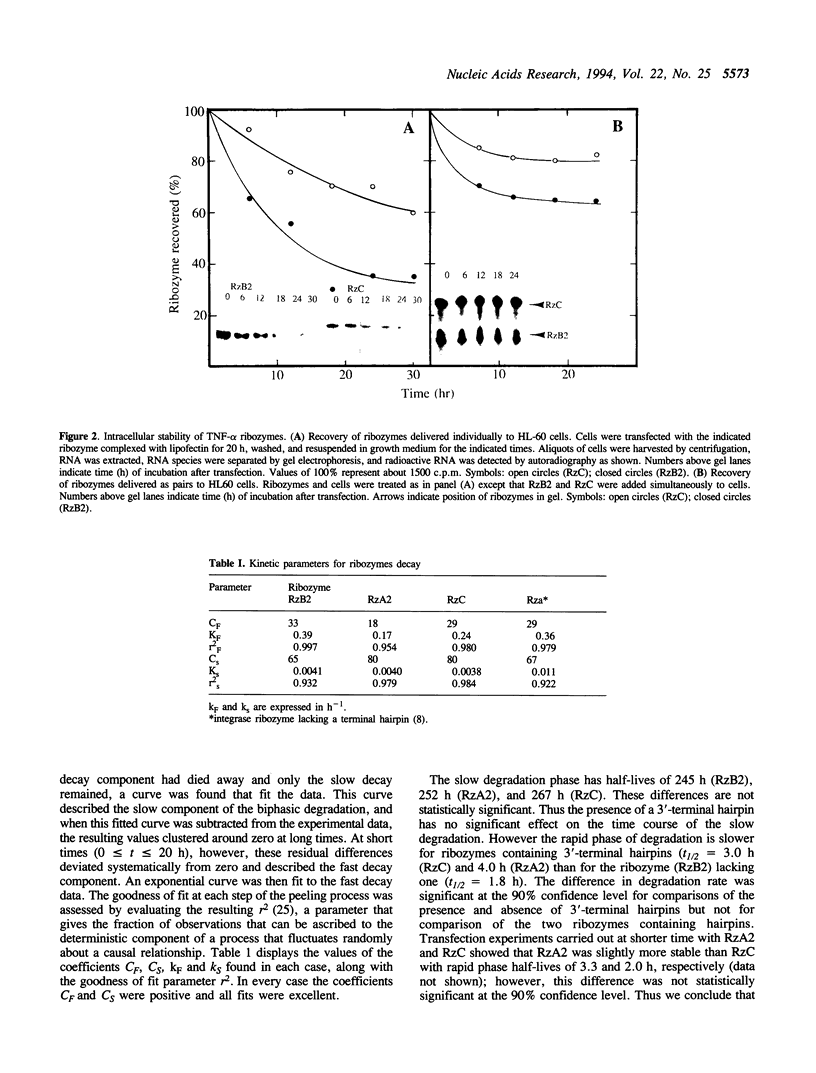
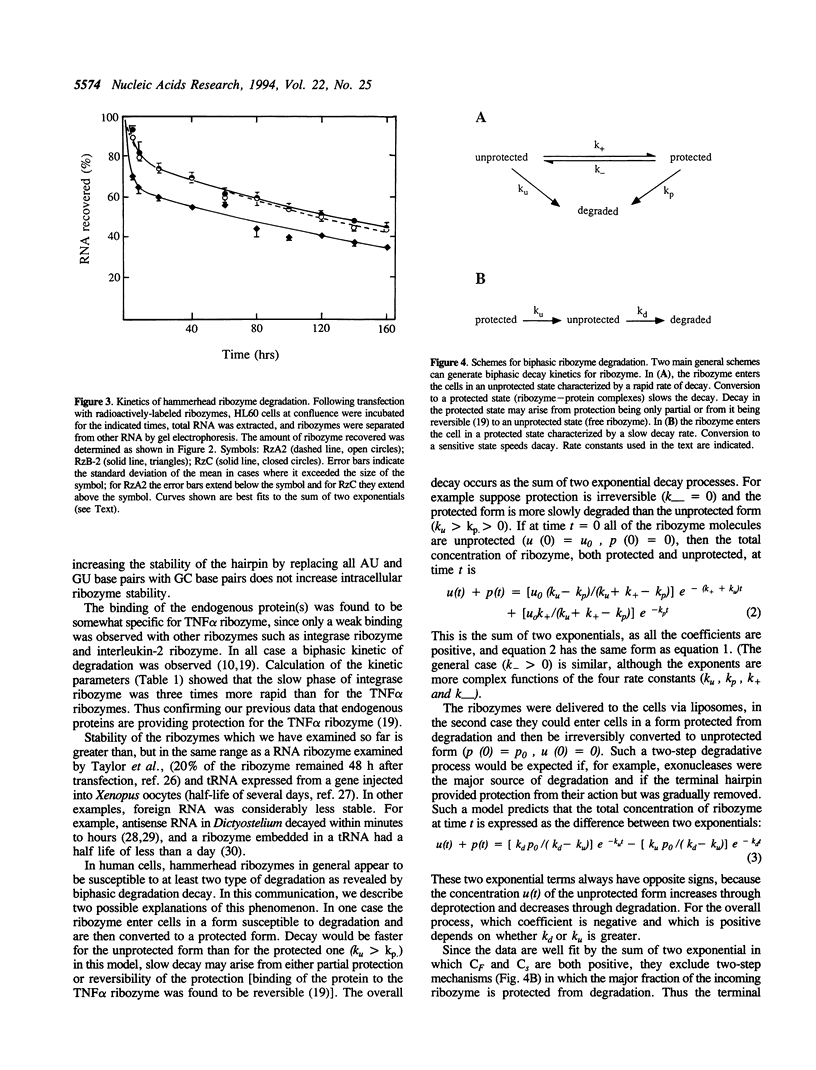

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branch A. D., Robertson H. D. Efficient trans cleavage and a common structural motif for the ribozymes of the human hepatitis delta agent. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10163–10167. doi: 10.1073/pnas.88.22.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Herschlag D., Piccirilli J. A., Pyle A. M. RNA catalysis by a group I ribozyme. Developing a model for transition state stabilization. J Biol Chem. 1992 Sep 5;267(25):17479–17482. [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cotten M., Birnstiel M. L. Ribozyme mediated destruction of RNA in vivo. EMBO J. 1989 Dec 1;8(12):3861–3866. doi: 10.1002/j.1460-2075.1989.tb08564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropulić B., Lin N. H., Martin M. A., Jeang K. T. Functional characterization of a U5 ribozyme: intracellular suppression of human immunodeficiency virus type 1 expression. J Virol. 1992 Mar;66(3):1432–1441. doi: 10.1128/jvi.66.3.1432-1441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Kashani-Sabet M., Funato T., Tone T., Jiao L., Wang W., Yoshida E., Kashfinn B. I., Shitara T., Wu A. M., Moreno J. G. Reversal of the malignant phenotype by an anti-ras ribozyme. Antisense Res Dev. 1992 Spring;2(1):3–15. doi: 10.1089/ard.1992.2.3. [DOI] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guerrier-Takada C., Altman S. Targeted cleavage of mRNA in vitro by RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3185–3189. doi: 10.1073/pnas.89.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K. M., Biasolo M. A., Dehni G., Palú G., Haseltine W. A. Inhibition of replication of HIV-1 by retroviral vectors expressing tat-antisense and anti-tat ribozyme RNA. Virology. 1992 Sep;190(1):176–183. doi: 10.1016/0042-6822(92)91203-7. [DOI] [PubMed] [Google Scholar]
- Loomis W. F., Fuller D. L. Antisense RNA inhibition of expression of a pair of tandemly repeated genes results in a delay in cell-cell adhesion in Dictyostelium. Antisense Res Dev. 1991 Fall;1(3):255–260. [PubMed] [Google Scholar]
- Nielsen D. A., Shapiro D. J. Preparation of capped RNA transcripts using T7 RNA polymerase. Nucleic Acids Res. 1986 Jul 25;14(14):5936–5936. doi: 10.1093/nar/14.14.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojwang J. O., Hampel A., Looney D. J., Wong-Staal F., Rappaport J. Inhibition of human immunodeficiency virus type 1 expression by a hairpin ribozyme. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D., Nedwin G. E., Hayflick J. S., Seeburg P. H., Derynck R., Palladino M. A., Kohr W. J., Aggarwal B. B., Goeddel D. V. Human tumour necrosis factor: precursor structure, expression and homology to lymphotoxin. Nature. 1984 Dec 20;312(5996):724–729. doi: 10.1038/312724a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sarver N., Cantin E. M., Chang P. S., Zaia J. A., Ladne P. A., Stephens D. A., Rossi J. J. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990 Mar 9;247(4947):1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- Scanlon K. J., Jiao L., Funato T., Wang W., Tone T., Rossi J. J., Kashani-Sabet M. Ribozyme-mediated cleavage of c-fos mRNA reduces gene expression of DNA synthesis enzymes and metallothionein. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10591–10595. doi: 10.1073/pnas.88.23.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M., Drlica K. Prevention of human immunodeficiency virus type 1 integrase expression in Escherichia coli by a ribozyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7303–7307. doi: 10.1073/pnas.88.16.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Interaction between tumour necrosis factor alpha ribozyme and cellular proteins. Involvement in ribozyme stability and activity. J Mol Biol. 1994 Oct 7;242(5):619–629. doi: 10.1006/jmbi.1994.1612. [DOI] [PubMed] [Google Scholar]
- Sioud M., Natvig J. B., Førre O. Preformed ribozyme destroys tumour necrosis factor mRNA in human cells. J Mol Biol. 1992 Feb 20;223(4):831–835. doi: 10.1016/0022-2836(92)90244-e. [DOI] [PubMed] [Google Scholar]
- Sun T. J., Van Haastert P. J., Devreotes P. N. Surface cAMP receptors mediate multiple responses during development in Dictyostelium: evidenced by antisense mutagenesis. J Cell Biol. 1990 May;110(5):1549–1554. doi: 10.1083/jcb.110.5.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. R., Kaplan B. E., Swiderski P., Li H., Rossi J. J. Chimeric DNA-RNA hammerhead ribozymes have enhanced in vitro catalytic efficiency and increased stability in vivo. Nucleic Acids Res. 1992 Sep 11;20(17):4559–4565. doi: 10.1093/nar/20.17.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe M., Liem S. E., Asad S., Read S. E., Joshi S. Resistance to human immunodeficiency virus type 1 (HIV-1) infection in human CD4+ lymphocyte-derived cell lines conferred by using retroviral vectors expressing an HIV-1 RNA-specific ribozyme. J Virol. 1991 Oct;65(10):5531–5534. doi: 10.1128/jvi.65.10.5531-5534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Ojwang J., Yamada O., Hampel A., Rapapport J., Looney D., Wong-Staal F. A hairpin ribozyme inhibits expression of diverse strains of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6340–6344. doi: 10.1073/pnas.90.13.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]



