Abstract
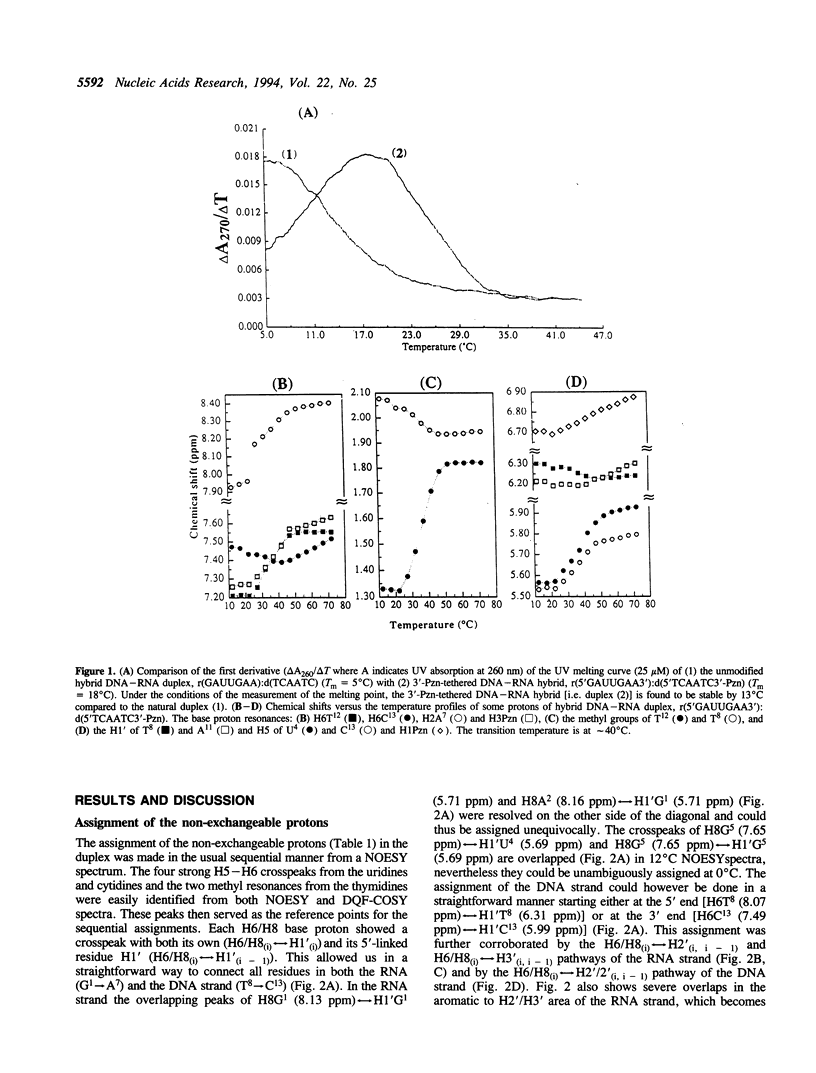
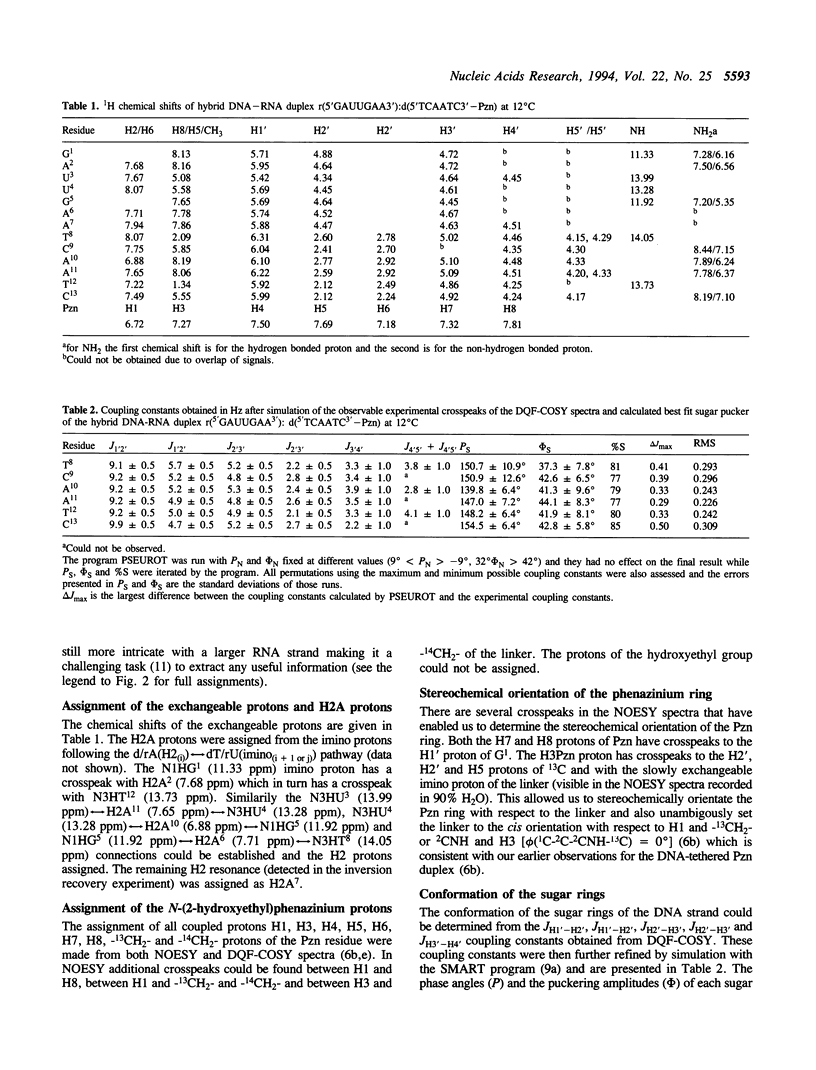
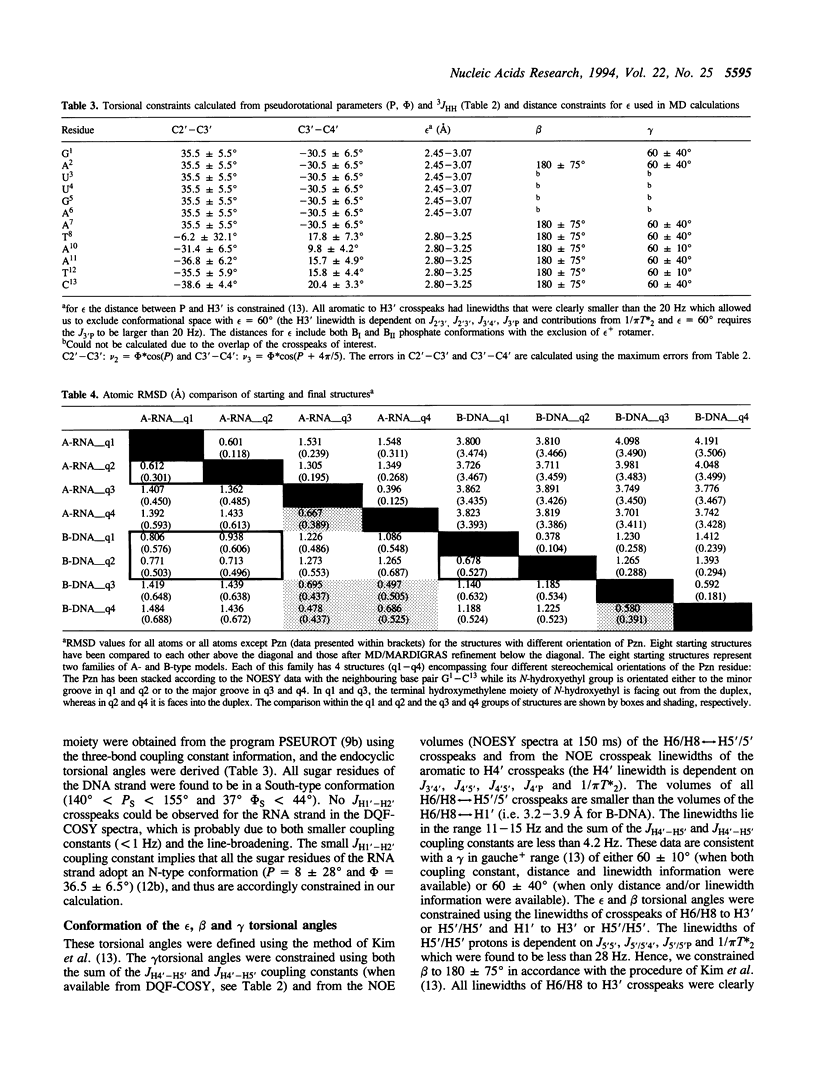
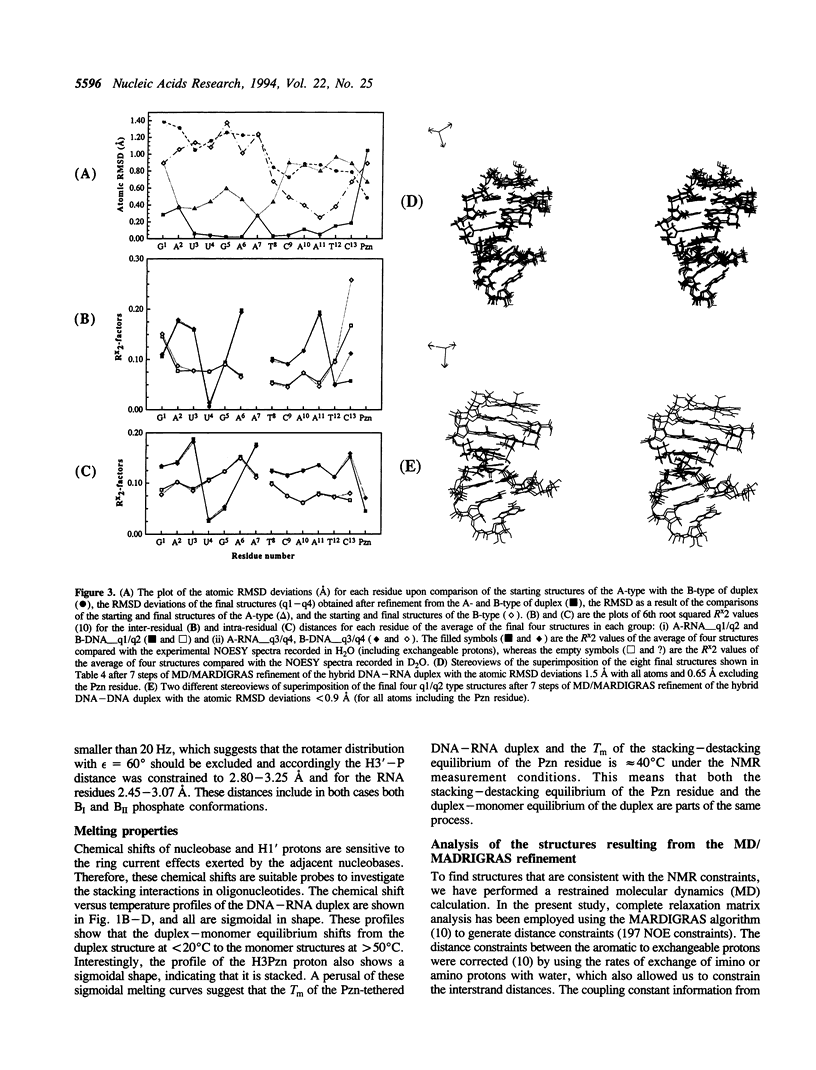
The 3'-Pzn group tethered to an oligo-DNA stabilizes a DNA-RNA hybrid duplex structure by 13 degrees C compared to the natural counterpart. This report constitutes the first full study of the conformational features of a hybrid DNA-RNA duplex, which has been possible because of the unique stabilization of this rather small duplex by the tethered 3'-Pzn moiety (Tm approximately 40 degrees C from NMR). In this study, a total of 252 inter- and intra-strand torsional and distance constraints along with the full NOE relaxation matrix, taking into account the exchange process of imino and amino protons with water, have been used. The 3'-Pzn-promoted stabilization of the DNA-RNA hybrid duplex results in detailed local conformational characteristics such as the torsion angles of the backbone and sugar moieties that are close to the features of the other natural DNA-RNA hybrids (i.e. sugars of the RNA strand are 3'-endo, but the sugars of the DNA strand are intermediate between A- and B-forms of DNA, 72 degrees < P < 180 degrees; note however, that the sugars of our DNA strand have a C1-exo conformation: 131 degrees < P < 154 degrees). This study suggests that 3'-Pzn-tethered smaller oligo-DNA should serve the same purpose as a larger oligo-DNA as a antisense inhibitor of the viral mRNA. Additionally, these types of tethered oligos have been found to be relatively more resistant to the cellular nuclease. Moreover, they are taken up quite readily through the cellular membrane (14) compared to the natural counterparts.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bichenkova E. V., Gorenshtein L. A., Vorob'ev Iu N., Tenne E. Iu, Zarytova V. F., Ivanova E. M., Mal'tseva T. V., Lebedev A. V. Issledovanie prostranstvennoi strukury dupleksa (Phn-NH(CH2)NH)pd(CCAAACA).pd(TGTTTGGC) s kovalentno prisodinennym 10-(2-gidroksiétil)fenaziniem v vodnom rastvore metodom 2M-(1)IaMR-spektroskopii i metodomogranichennoi molekuliarnoi mekhaniki. Bioorg Khim. 1992 Jul;18(7):901–910. [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. High-resolution NMR study of a synthetic DNA-RNA hybrid dodecamer containing the consensus pribnow promoter sequence: d(CGTTATAATGCG).r(CGCAUUAUAACG). Biochemistry. 1989 Mar 21;28(6):2435–2443. doi: 10.1021/bi00432a014. [DOI] [PubMed] [Google Scholar]
- Fritsch V., Wolf R. M. Molecular dynamics simulations of a r(GA12G).d(CT12C) hybrid duplex. J Biomol Struct Dyn. 1994 Jun;11(6):1161–1174. doi: 10.1080/07391102.1994.10508061. [DOI] [PubMed] [Google Scholar]
- González C., Stec W., Kobylanska A., Hogrefe R. I., Reynolds M., James T. L. Structural study of a DNA.RNA hybrid duplex with a chiral phosphorothioate moiety by NMR: extraction of distance and torsion angle constraints and imino proton exchange rates. Biochemistry. 1994 Sep 20;33(37):11062–11072. doi: 10.1021/bi00203a002. [DOI] [PubMed] [Google Scholar]
- Gryaznov S. M., Lloyd D. H. Modulation of oligonucleotide duplex and triplex stability via hydrophobic interactions. Nucleic Acids Res. 1993 Dec 25;21(25):5909–5915. doi: 10.1093/nar/21.25.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Lin L. J., Reid B. R. Determination of nucleic acid backbone conformation by 1H NMR. Biochemistry. 1992 Apr 14;31(14):3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- Lancelot G., Guesnet J. L., Vovelle F. Solution structure of the parallel-stranded duplex oligonucleotide alpha-d(TCTAAAC)-beta-d(AGATTTG) via complete relaxation matrix analysis of the NOE effects and molecular mechanics calculations. Biochemistry. 1989 Sep 19;28(19):7871–7878. doi: 10.1021/bi00445a049. [DOI] [PubMed] [Google Scholar]
- Lane A. N., Ebel S., Brown T. NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur J Biochem. 1993 Jul 15;215(2):297–306. doi: 10.1111/j.1432-1033.1993.tb18035.x. [DOI] [PubMed] [Google Scholar]
- Lokhov S. G., Podyminogin M. A., Sergeev D. S., Silnikov V. N., Kutyavin I. V., Shishkin G. V., Zarytova V. P. Synthesis and high stability of complementary complexes of N-(2-hydroxyethyl)phenazinium derivatives of oligonucleotides. Bioconjug Chem. 1992 Sep-Oct;3(5):414–419. doi: 10.1021/bc00017a010. [DOI] [PubMed] [Google Scholar]
- Lybrand T., Kollman P. Molecular mechanical calculations on the interaction of ethidium cation with double-helical DNA. Biopolymers. 1985 Oct;24(10):1863–1879. doi: 10.1002/bip.360241003. [DOI] [PubMed] [Google Scholar]
- Maltseva T. V., Agback P., Chattopadhyaya J. How much hydration is necessary for the stabilisation of DNA-duplex? Nucleic Acids Res. 1993 Sep 11;21(18):4246–4252. doi: 10.1093/nar/21.18.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny J. L., Boutorine A. S., Garestier T., Belloc F., Rougée M., Bulychev N. V., Koshkin A. A., Bourson J., Lebedev A. V., Valeur B. Fluorescence energy transfer as a probe for nucleic acid structures and sequences. Nucleic Acids Res. 1994 Mar 25;22(6):920–928. doi: 10.1093/nar/22.6.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel L. J., Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J Biomol Struct Dyn. 1987 Feb;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Sun J. S., François J. C., Montenay-Garestier T., Saison-Behmoaras T., Roig V., Thuong N. T., Hélène C. Sequence-specific intercalating agents: intercalation at specific sequences on duplex DNA via major groove recognition by oligonucleotide-intercalator conjugates. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9198–9202. doi: 10.1073/pnas.86.23.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ven'iaminova A. G., Gorn V. V., Zenkova M. A., Komarova N. I., Repkova M. N. Avtomaticheskii H-fosfonatnyi sintez oligoribonukleotidov s ispol'zovaniem 2'-O-tetragidropiranil'noi zashchitnoi gruppy. Bioorg Khim. 1990 Jul;16(7):941–950. [PubMed] [Google Scholar]
- Venyaminova A. G., Repkova M. N., Maltseva T. V. Preparative solid phase synthesis of a tetradecamer r(GAUUGAAAAUCCCC) by the H-phosphonate method using a 2'-O-tetrahydropyranyl protective group. Nucleic Acids Symp Ser. 1991;(24):263–263. [PubMed] [Google Scholar]
- Wang A. C., Kim S. G., Flynn P. F., Chou S. H., Orban J., Reid B. R. Errors in RNA NOESY distance measurements in chimeric and hybrid duplexes: differences in RNA and DNA proton relaxation. Biochemistry. 1992 Apr 28;31(16):3940–3946. doi: 10.1021/bi00131a008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J. DNA modification and cancer. Annu Rev Biochem. 1981;50:159–192. doi: 10.1146/annurev.bi.50.070181.001111. [DOI] [PubMed] [Google Scholar]
- Wilson W. D., Ratmeyer L., Zhao M., Strekowski L., Boykin D. The search for structure-specific nucleic acid-interactive drugs: effects of compound structure on RNA versus DNA interaction strength. Biochemistry. 1993 Apr 20;32(15):4098–4104. doi: 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]


