Abstract
The evolutionary relationships among known Chlamydophila abortus variant strains including the LLG and POS, previously identified as being highly distinct, were investigated based on rRNA secondary structure information. PCR-amplified overlapping fragments of the 16S, 16S-23S intergenic spacer (IS), and 23S domain I rRNAs were subjected to cloning and sequencing. Secondary structure analysis revealed the presence of transitional single nucleotide variations (SNVs), two of which occurred in loops, while seven in stem regions that did not result in compensatory substitutions. Notably, only two SNVs, in 16S and 23S, occurred within evolutionary variable regions. Maximum likelihood and Bayesian phylogeny reconstructions revealed that C. abortus strains could be regarded as representing two distinct lineages, one including the “classical” C. abortus strains and the other the “LLG/POS variant”, with the type strain B577T possibly representing an intermediate of the two lineages. The two C. abortus lineages shared three unique (apomorphic) characters in the 23S domain I and 16S-23S IS, but interestingly lacked synapomorphies in the 16S rRNA. The two lineages could be distinguished on the basis of eight positions; four of these comprised residues that appeared to be signature or unique for the “classical” lineage, while three were unique for the “LLG/POS variant”. The U277 (E. coli numbering) signature character, corresponding to a highly conserved residue of the 16S molecule, and the unique G681 residue, conserved in a functionally strategic region also of 16S, are the most pronounced attributes (autapomorphies) of the “classical” and the “LLG/POS variant” lineages, respectively. Both lineages were found to be descendants of a common ancestor with the Prk/Daruma C. psittaci variant. Compared with the “classical”, the “LLG/POS variant” lineage has retained more ancestral features. The current rRNA secondary structure-based analysis and phylogenetic inference reveal new insights into how these two C. abortus lineages have differentiated during their evolution.
Introduction
Chlamydophila abortus is an intracellular bacterium that is able to efficiently colonize the placenta of several mammals causing abortion and premature birth of stillborn or weak neonates [1]–[4]. This pathogen is endemic among small ruminants and represents a zoonotic pathogen. Pregnant women exposed to infected animals have the risk of spontaneous abortion or even a life-threatening disease [4]. Chlamydophila abortus is classified as a member of the family Chlamydiaceae which currently encompasses the two genera Chlamydia and Chlamydophila, subdivided into three (C. muridarum, C. suis, C. trachomatis) and six (C. abortus, C. caviae, C. felis, C. pecorum, C. pneumoniae, C. psittaci) species, respectively [2], [5]. Genetic analyses indicate that C. abortus has evolved from Chlamydophila psittaci, which also constitutes a zoonotic pathogen associated primarily with avian chlamydiosis [2], [6], [7].
Studies using different phenotypic and molecular approaches suggest that C. abortus is a homogeneous species and includes strains sharing distinctive inclusion morphology and antigenic profile, and nearly 100% sequence conservation in the ribosomal and ompA genes [2], [8]–[11]. However, two homologous strains, namely LLG and POS, isolated in Greece from an aborted goat and ewe, respectively [12], were considerably different among other C. abortus strains prevailing in the same area and were characterized as variants on the basis of unique inclusion morphology, differences in polypeptide profiles, non-reactivity with monoclonal antibodies against immunodominant C. abortus antigens, diversity of 23S domain I rRNA and ompA sequences, and different behavior in cell cultures and mouse model protection experiments [12]–[16]. In a recent study using multiple-locus variable number tandem repeat (VNTR) sequences, the LLG and POS strains were identified as the most divergent ones among other C. abortus strains, constituting a distinct genotype, in particular for the pmp5E and hctB loci involved in establishing the immunodominant and structural proteins, respectively [17]. Moreover, sequencing of the LLG RFLP-fragments of the plasticity zone, a region of extensive gene differences between Chlamydiaceae species, revealed considerable differences in the pseudogene content [18]. Similar variation in biological and/or genotypic characteristics, albeit to a lesser extent, has also been observed among other C. abortus strains [12]–[15], [17], [19].
The previous studies have raised novel questions regarding the actual evolutionary relationships of the variant C. abortus strains that share a common geographical origin. To this end, the information content of rRNA genes is especially useful for providing a solid framework for the assessment of evolutionary changes in lineages [20]–[24]. Moreover, rRNAs are functionally constrained structure mosaics ranging from highly conserved to more variable ones, with varying evolutionary rates among secondary structure elements [20], [25]–[29]. In the present study, PCR-amplified overlapping fragments of the “ribosomal operon” derived from C. abortus variant strains, including the LLG and POS, were subjected to cloning and sequencing. We firstly focused on the 16S rRNA and 16S-23S intergenic spacer (IS) genes since the 23S rRNA domain I gene sequences for the respective strains had been previously determined [12]. We aimed at investigating the pattern and distribution of signature or unique nucleotide residues in rRNA molecules among C. abortus variant strains as well as on inferring their phylogenetic relationships based on rRNA secondary structure. The information gained may contribute to a more thorough understanding of the mode of molecular evolution in C. abortus.
Materials and Methods
Chlamydial strains and DNA preparations
The C. abortus strains FAS, FAG, VPG, LLG and POS, all isolated in Greece from aborted sheep or goat fetuses [12], were used in the present study. All strains have been previously described on the basis of inclusion morphology, antigenic and molecular diversity [12], [15], and recently classified into three distinct VNTR genotypes [17]. Whole genomic DNAs were extracted (NucleoSpin tissue kit; Macherey-Nagel) from the second passage of the original isolates, propagated in yolk sac of embryonated chicken eggs, so as to represent fresh clinical isolates and not laboratory-adapted strains.
PCR amplification, cloning and sequencing of rDNAs
PCR amplifications resulting in four overlapping PCR-amplified rDNA fragments were conducted as previously described [2], [5], [30] with some modifications. Briefly, two PCR amplifications intended for amplifying the entire 16S rDNA were performed by using the primer pairs 16SFor/16SIGR and 16SF/16SR, yielding fragments encoding the 16S signature sequence and nearly the full-length 16S rRNA, respectively. Two additional PCR amplifications were performed by using the primer pairs 16SF2/23R and 16SF2/23SIGR for amplifying the 16S-23S IS flanked by 16S and 23S segments, including the 23S signature sequence (domain I). A schematic representation of the four overlapping PCR-amplified rDNA fragments as well as the primer pairs used for the PCR amplifications with the respective annealing temperatures are available as supporting material (Figure S1, Table S1).
The rDNA sequences were determined by both direct PCR sequencing and sequencing of cloned products. Initially, purified (NucleoSpin Extract II kit; Macherey-Nagel) PCR products from two separate PCR reactions for each fragment for each strain were sequenced (ABI 3730XL, Macrogen) on both strands using the respective PCR primers and an internally designated primer (Table S1). In addition, clone libraries of purified PCR products obtained from a separate series of PCR reactions were constructed by ligation into the pCR2.1 vector (TA cloning kit; Invitrogen) and transformation, by heat shock, into E. coli XL-1 Blue (Stratagene). Blue-white screening of transformants [31] was performed on LB agar containing ampicillin (100 µg/ml) and top spread with IPTG (0.5 mM) and X-Gal (80 µg/ml). From each clone library, ten white colonies were picked randomly and screened by PCR for the presence of rDNA inserts. Subsequently, four independent clones were selected and sequenced following extraction of the recombinant plasmid DNA (NucleoSpin Plasmid QuickPure kit; Macherey-Nagel). The T7 promoter and the M13R-pUC primer flanking the multiple cloning site of pCR2.1 DNA were used to sequence both DNA strands. On the basis of the four PCR-amplified rDNA fragments, which overlapped one another (Figure S1), as well as the PCR amplification and sequencing strategies, the corresponding sequences that were obtained each had a 6x up to 12x read coverage.
Secondary structure-based rDNA sequence analysis
The obtained sequences were initially compared with the public sequences using the BLAST program at NCBI (http://www.ncbi.nlm.nih.gov/). The overlapping rDNA sequences were initially aligned together using CLUSTAL X 1.83 [32], and then for each rDNA locus multiple sequence alignments with reference sequences downloaded from the NCBI database were computed. In order to construct sequence alignments on the basis of 16S and 23S rRNA secondary structure modelling, sequences were automatically aligned by SINA, as implemented in the SILVA SSU and LSU rRNA database project (http://www.arb-silva.de/; [33]). The 16S rDNA sequences were also aligned via the NAST aligner (http://greengenes.lbl.gov/; [34]). As a control for the effects of using secondary structure-based alignment algorithms, specific data for rRNA sequences were used from the Comparative RNA Web (CRW) relational database management system (RDBMS) (http://www.rna.ccbb.utexas.edu/; [35]). Helix numbering for the 16S and 23S rRNA secondary structures followed the respective reference sequence numbering system (E. coli GenBank acc. no. J01695) according to CRW [35]. Nucleotide frequency and conservation data were also derived from the CRW site.
Phylogeny reconstruction
Phylogeny reconstruction was performed using maximum likelihood (ML) and Bayesian Inference (BI) approaches. It has repeatedly been demonstrated [20], [23], [36], [37] that likelihood-based approaches (ML and BI) are able to recover the true tree more frequently than parsimony or distance-based (e.g., Neighbor-Joining) approaches. For ML-based analyses the most recent version 7.2.6 of RAxML (http://wwwkramer.in.tum.de/exelixis/software.html; [38]) was used as it has been shown to perform best among all other methods tested by Price et al. [37]. For BI-based analyses, MrBayes version 3.1.2 was used (http://mrbayes.csit.fsu.edu/; [39]).
RAxML under the GTR+Gamma substitution model [40] (see RAxML manual at http://icwww.epfl.ch/~stamatak/index-Dateien/Page443.htm) was used to infer 1,000 bootstrap replicates and to conduct 50 ML searches on the original alignment using 50 distinct randomized stepwise addition parsimony trees. The respective RAxML options were used to draw bootstrap support (BS) values onto the best-scoring ML tree obtained on the original alignment as well as to compute majority-rule consensus trees from the collections of bootstrap replicates and ML trees. Bayesian inference was also conducted under the GTR+Gamma model using two independent runs with four Metropolis-Coupled Markov Chains each. Ten million generations were performed for each region using default priors with trees sampled every 100th generation (burnin set to 10,000 generations) to obtain posterior probabilities.
Dendroscope (http://www-ab.informatik.uni-tuebingen.de/software/dendroscope/; [41]) and TreeGraph 2 (http://treegraph.bioinfweb.info/; [42]) were used for tree visualization and manipulation.
Results and Discussion
Nucleotide sequences of the rRNA molecules
Directly obtained sequences of the rDNA fragments produced by the two separate PCR reactions and the corresponding sequences of the separately produced independent clones (including all overlapping segments in each case) were 100% identical. This resulted in an unambiguous determination of the entire lengths of the 16S, 16S-23S IS, and 23S domain I rRNA genes for each C. abortus strain examined. Sequence identity among PCR-amplified rDNAs and cloned products for each strain indicated that only one gene from each rRNA locus is present in C. abortus, which is in agreement with previous findings [18].
However, the rDNA sequences obtained from the five C. abortus strains under investigation (GenBank accession numbers EF486853-EF486857) exhibited differences. Regarding the 16S rRNAs, the comparison revealed the presence of two sequence variants differing by four single nucleotide variations (SNVs). More precisely, the nucleotide sequences of LLG and POS strains were identical to each other but differed from those of the FAS, FAG, and VPG strains by the presence of nucleotides A, C, G, and G instead of G, U, A, and A at positions 80, 277, 396, and 681 (according to the E. coli numbering system) [35], respectively. Interestingly, at positions 277 and 396 the LLG and POS variants shared identical nucleotides with the C. abortus type strain B577T, while the sequences of FAS, FAG, and VPG strains were found to be identical to those of other classical (well-established) strains of this species (i.e., S26/3, EBA, EAE, and OEA).
The 16S-23S IS rRNA sequences obtained from the C. abortus strains were identical in all but one SNV. This variation, nucleotide U instead of C, located at position 79 (according to the C. abortus type strain B577T sequence; GenBank acc. no. U68445) was detected in the LLG and POS sequences.
The sequencing results of the 23S domain I region of the current study confirmed previous ones [12] regarding the presence of four SNVs. Three of them, namely A, C, and G instead of G, U, and A located at positions 152, [181-182], and 273 (E. coli numbering; square brackets indicate that E. coli lacks the corresponding position), respectively, were identified in the LLG and POS sequences, and one, U instead of C at position 547, was identified in the FAG and VPG sequences.
To evaluate the importance of these sequence differences (discussed below), alignments of 16S and 23S rRNA sequences of 71 and 67 strains, respectively, belonging to C. abortus and other related species were constructed on the basis of the corresponding secondary structure information. Alignment segments corresponding to the structural elements bearing the SNVs are available as supporting material (Figure S2A and S3A). An alignment of 16S-23S IS sequences of 57 chlamydial strains was constructed on the basis of the primary structure (Figure S4A), since the intergenic spacer is the most variable region throughout the rRNA operon and an analogous secondary structure-based reference numbering system does not exist. In Table 1 we provide the variant residues found in C. abortus compared with related taxa.
Table 1. Nucleotide variations in rRNA molecules of C. abortus strains compared with related taxa and respective nucleotide frequency data within the domain Bacteria.
| Organisms | Base or base pair in | ||||||||
| 16S rRNA | 16S-23S IS | 23S rRNA (domain I) | |||||||
| 80:89a | 247:277 | 45:396 | 681:709 | 79 | 152:174 | [181–182] | 273:364 | 547 | |
| Chlamydophila abortus B577T VR-656 (D85709, U68445) | G:U | G:C | U:G | A:U | C | G:U | U | A:U | C |
| C. abortus LLG (EF486856) | A:U | G:C | U:G | G:U | U | A:U | C | G:U | C |
| C. abortus POS (EF486857) | A:U | G:C | U:G | G:U | U | A:U | C | G:U | C |
| C. abortus FAS (EF486853) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | C |
| C. abortus FAG (EF486854) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | U |
| C. abortus VPG (EF486855) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | U |
| C. abortus S26/3 (CR848038) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | C |
| C. abortus EBA (U76710) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | C |
| C. abortus EAE (Z49871), A22 (U68444) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | C |
| C. abortus OEA (Z49872), OSP (U68446) | G:U | G:U | U:A | A:U | C | G:U | U | A:U | C |
| Chlamydophila psittaci 6BCT VR-125 (U68447)b | G:U | G:C | U:G | A/g:U | C | A:U | C | A:U | A/c |
| Chlamydophila caviae GPICT VR-813 (AE015925)b | A:U | G:C | U:G | A:U | C | A:U | C | A:U | C |
| Chlamydophila felis FP BakerT VR-120 (D85701, U68457)b | G:U | G:C | U:G | A:U | C | A:U | C | A:U | C |
| Chlamydophila pecorum E58T VR-628 (D88317, U68433) b | G:U | G:C | U:G | A:U | C | A:U | A | A:U | C |
| Chlamydophila pneumoniae TW-183T VR-2282 (L06108, U76711)b | G/a:U | G:C | U:G | A:U | C | A:U | C | A:U | C |
| C. pneumoniae N16 (U68426) | G:U | G:C | U:G | A:U | C | A:U | C | A:U | C |
| C. pneumoniae LPCoLN (FJ236984) | G:U | G:C | U:G | A:U | na | na | na | na | na |
| Chlamydia trachomatis A/Har-13T VR-571B (D89067, U68438)b | U:G/A | G:C | U:G | A:U | U | G:C | C | A/g:U | A |
| C. trachomatis L2/434/BU VR-902B (U68443, U68443) b | U:A | G:C | U:G | A:U | U | G:C | C | A:U | A |
| Chlamydia muridarum MoPnT VR-123 (D85718, U68436) b | U:G | G:C | U:G | A:U | C | G:C | C | G:U | A |
| Chlamydia suis S45T VR-1474 (U73110)b | U:A | G:C | U:G | A:U | C | G:C | C | A:U | A |
| Parachlamydiaceae sp. Bn9T VR-1476 (Y07556, AF193069, Y07555)b | − | G:C | U:G | A:U | C | U:A/C:G | U/C | G:C/A:U | U/A |
| Waddliaceae sp. WSU 86/1044T VR-1470 (AF042496)b | G:U | G:C | U:G | C/A:U | C | U:A | G | G:U/G:C | A |
| Simkaniaceae sp. ZT VR-1471 (U68460)b | A:U/− | G:C | U:G | A:U | A | C:G/U:A | U/− | G/A:U | U |
| E. coli (J016950) | A:U | G:C | G:C | A:U | A:U | − | G:C | A | |
| Domain Bacteria | |||||||||
| Base pair frequencies based on 16S & 23S rRNA modelsc | G:C 32.5 C:G 18.9 U:A 18.1 A:U 12.1 G:U 5.2 U:G 1.2 Gap 9.7 | G:C 99.2 C:G ---- U:A ---- A:U 0.1 G:U 0.4 U:G ---- Gap ---- | G:C 27.0 C:G 5.3 U:A 37.2 A:U 0.8 G:U 0.2 U:G 25.2 Gap ---- | G:C 10.5 C:G 16.7 U:A 40.6 A:U 31.2 G:U 0.1 U:G ---- Gap ---- | G:C 12.6 C:G 37.0 U:A 10.4 A:U 27.0 G:U 6.3 U:G 0.4 Gap 1.5 | G:C 28.2 C:G 19.5 U:A 21.7 A:U 7.2 G:U 4.7 U:G ---- Gap 15.6 | |||
| Single base frequencies based on 16S & 23S rRNA modelsc | A 14.05 G 37.93 C 19.75 U 19.75 | A 0.00 G 0.10 C 99.27 U 0.59 | A 37.42 G 30.29 C 31.05 U 1.20 | A 31.51 G 10.63 C 17.00 U 40.77 | A 27.78 G 19.63 C 37.78 U 13.70 | A 8.63 G 33.81 C 21.58 U 21.94 | A 53.76 G 9.32 C 16.13 U 20.79 | ||
Nucleotide positions of the 16S rRNA and 23S domain I rRNA are given according to E. coli J01695 secondary structure numbering system [35]; position of 16S-23S Intergenic Spacer based on primary structure is given according to the C. abortus type strain B577T sequence (U68445). Positions of the single nucleotide variations (SNVs) are indicated in boldface. The [181–182] position represents “insertion” position.
More than two or three accession numbers for each species or family including the type strain (T), were analyzed (the majority of analyzed strains and their accession numbers are given in the Figures S2, S3 and S4); lower-case letters denote residues found only in one of the examined strains; “na”, not available data; “−”, nucleotide gap in rRNA sequence comparison.
Dataset from http://www.rna.ccbb.utexas.edu/SAE/2A/nt_Frequency/
Secondary structure-based nucleotide analysis of the rRNA molecules
As an additional means of assessing genetic relatedness, and in order to check whether the observed SNVs are located within particular evolutionary variable or conserved regions of rRNAs potentially supporting phylogenetic groupings, we conducted comparisons with other available chlamydial sequences on the basis of their secondary structure.
16S rRNA analysis
All four SNVs corresponding to positions 80, 277, 396, and 681 occurred in stem regions (base-pairing regions) of the secondary structure helices H61(61-82/87-106), H240(240-259/267-286), H39(39-46/395-403), and H673(673-690/697-717) (helix numbering according to Comparative RNA Web (CRW) site; [35]), respectively. These SNVs did not result in a nucleotide substitution in the complementary position of the stem (Table 1, Figure S2A). Based on comparative sequence analyses (data available at CRW site; see Table 1), SNV at position 277 corresponded to a highly conserved residue throughout domain Bacteria (more than 98%), SNVs at positions 396 and 681 corresponded to less highly conserved residues (less than 80%), whereas SNV at position 80 was found to exist within an evolutionary variable region. Interestingly, at location 247∶277 the LLG/POS variant, as well as the C. abortus type strain B577T, exhibited the base pair G:C occurring in most members of the domain Bacteria (99.2%) and also shared by all Chlamydiales species, but not by the remaining C. abortus strains. The latter, possessed a G:U base pair rarely occurring at this location throughout domain Bacteria (0.4%) (Table 1). Similarly, at location 45∶396, the LLG/POS variant and the B577T strain presented the base pair U:G, shared with all Chlamydiales species but not the remaining C. abortus strains which presented the equally common U:A base pair. Notably, the feature G:U at location 681∶709 only found in the LLG/POS variant, was not present in the 16S rRNA molecule of any other species of the order Chlamydiales, with only one C. psittaci strain exception, and was also rarely found among bacteria (0.1%) (Table 1). Another attribute of the LLG/POS variant was the base pair A:U at location 80∶89, not present in the vast majority of the Chlamydiales (Table 1).
In each of the four 16S rRNA variations a transitional substitution was observed so that a G:U type base pairing, at no “dominant” G:U type sites [25], [43], was interchanged with canonical base pairing (A:U and G:C types) or vice versa (Table 1, Figure S2A), suggesting a strong selection for pyrimidine:purine base pairing. This sequence variation may not be necessarily involved in any obvious structural features that serve a specific binding mechanism; however, this depends on residue conservation [44] as well as on the functional significance of the structure element in which the variation occurs. Strikingly, a new phenotype mutant had been generated by a single C→U transition at the “universally conserved residue” G11:C23 (99.1%, three Phylogenetic domains [3P]) of the 5′ terminal pseudoknot H9 helix of 16S rRNA [45]. In the present case, the “conserved residue” G247:C277 (99.2%, domain Bacteria; 87.3%, 3P), flanked by an asymmetric internal loop inside the highly conserved H240 helix of the 5′ domain [46] (CRW site dataset; Figure S2A), could be selected as more stable than G:U (0.4%, domain Bacteria; 0.3%, 3P) in the context of a loop-closing base pair [25]. Interestingly, location 247∶277 and the region around it is important for recognition by the ribosomal protein S17, which strongly protects the region, binds, and stabilizes the H240 helix near the central junction [47]–[50]. The feature G247:U277 observed in most of C. abortus strains could function as a recognition signal for protein binding, which through stabilizing of the created “wobble” pair and alleviating what would otherwise be a deleterious condition could facilitate the evolutionary replacement of the base pair [28]. Additionally, it has been proven that G:U pairs could enhance stability as closing base pairs in specific contexts [43]. Some of the H240 positions also constitute parts of the S20 binding site [49], [50] and, genetic studies have shown that substitutions and deletions at these sites could abolish binding of S20 to 16S rRNA [51]. Generally, “conserved positions” throughout 3P represent the preservation of specific structural elements, which presumably act as scaffolds to provide the critical orientations of highly conserved residues in three-dimensional space [44]. Indeed, the “conserved position” 396 occurring in the interior of the H39 helix is flanked by a single bulge “universally conserved” adenosine residue (Figure S2A) [25], [46], [52], known as a binding site of the S4 protein, which is essential for the stability of the rRNA tertiary interactions [47], [49], [53], [54]. At position 396 (nucleotide 5′ of bulge), purines (A or G) are almost equally common (Table 1) suggesting that there is no particular pressure to favor one or the other base adjacent to the bulge. Structural reasons for the selection of U45:A396 instead of U45:G396 at this site are thus not apparent. The “conserved position” 681 is an intrahelical site of the helix H673 which is a functionally strategic region of the 16S rRNA [46], [55]. The location 681∶709 is found in the vicinity of “universally” or “highly conserved” residues known to be involved in heterodimer S6 and S18 protein bindings [47], [48], [56] or residues involved in S11 protein binding, which is essential for stabilizing the 16S rRNA central domain folding [47], [48], [57], [58] and specific E-site tRNA interactions [44], [59]. At this site, the high incidence of the canonical type base pairs (Table 1) could reflect its functional implication, whereas the rarity of the “wobble” G681:U709 pair could reflect a likely alteration in the respective interdependent interactions; the irregularity, probably caused by the G:U residue, may be a signal for specific protein bindings [28], [55], [60]. Remarkably, interdependencies of protein binding in the assembly of the central domain are similar but not identical among different microorganisms [57]. Finally, location 80∶89 is situated in a highly variable area corresponding to the “non-conserved region” (residues 79–100) of the H61 helix (Figure S2A), one of the most informative or discriminating regions for closely related organisms [20], [25], [44]. This area exhibits genetic-group specificity for the order Chlamydiales (variable region I of the 16S rRNA signature sequence) [2], [61] with intraspecific sequence variation also occurring within chlamydial species such as the equine-type (strain N16) of C. pneumoniae and the E, F, and L2 types of C. trachomatis (Figure S2A). The feature A80:U89 exhibited by the LLG and POS C. abortus variant strains also occurred in C. caviae as well as in C. pneumoniae and S. negevensis single strains, but without the corresponding variable region being entirely similar (Table 1, Figure S2A). Character homology in variable regions is not necessarily indicated by sequence identity or similarity [20] (discussed below).
23S domain I rRNA analysis
Two of three SNVs observed in the LLG/POS variant, corresponding to positions 152 and 273 (E. coli numbering), occurred in stem regions of the helices H150(150–158/168–176) and H271(271–297/341–366), respectively. Similarly to the 16S rRNA variations, none of these SNVs has resulted in a compensatory substitution, but G:U pairing was interchanged with canonical A:U pairing or vice versa (Table 1, Figure S3A). Both H150 and H271 helices comprise particularly variable regions that are phylogenetically informative for the identification and taxonomy of bacterial pathogens [26], [62], [63]. The SNV at position 152, corresponding to a conserved residue (less than 80%, domain Bacteria & 3P), was adjacent to a “non-conserved region” (residues 153–173) of the H150 helix, whereas the SNV at position 273 occurred within the “non-conserved region” (270–297/353–369) of the H271. At location 152∶174 the LLG/POS C. abortus variant exhibited the base pair A:U, also shared by all Chlamydophila species but not the remaining C. abortus strains. The latter, possessed a G:U base pair characterized by low frequency at this location throughout domain Bacteria (6.3%) (Table 1). In contrast, the base pair G:U at location 273∶364 found in the LLG/POS variant, was not present in any Chlamydophila species but only in isolated cases within the order Chlamydiales and with low frequencies among bacteria (4.7%) (Table 1). The third SNV of the LLG/POS variant, corresponding to position [181–182], occurred in an unpaired position immediately adjacent to the “universally conserved” adenosine residue (position 182) at the 3′ end of the multi-stem loop (Figure S3A) [52]. At this position, the LLG/POS variant presented the residue C shared by most Chlamydiales species while the remaining C. abortus strains presented the residue U. It is not apparent how the residue U prevailed and how it may affect the conformation of the AG opposition at the end of the H183 helix [64]. Finally, the SNV observed in 23S rRNA domain I of the FAG/VPG variant at position 547 occurred in the hairpin-loop of the H533 helix and corresponded to a conserved residue across domain Bacteria (less than 80%). It is worth noting that the sequence corresponding to residues 543–552 is a “non-conserved region” throughout 3P (data available at CRW site) and often presents remarkable intraspecific diversity within domain Bacteria [65]. The residue U found in the FAG/VPG variant, albeit frequently present among bacteria (20.79%), rarely occurs among Chlamydiales species (Table 1).
16S-23S IS analysis
The SNV detected in the LLG/POS variant, corresponding to position 79 (according to C. abortus type strain B577T sequence; acc. no. U68445, Figure S4A) was found to occur in a stem region which is predicted to be formed between the chlamydial spacer and a complementary segment of 16S promoter sequence [66]. In this context, the variant residue could be paired with a residue immediately adjacent to the 3′ end of the promoter P2-10 sequence (Figure S4B), a region of high functional stringency and conservation [66]–[68]. However, it is worth noting that residue U found in LLG/POS variant was also shared by C. trachomatis strains within the order Chlamydiales (Table 1).
Phylogenetic analysis
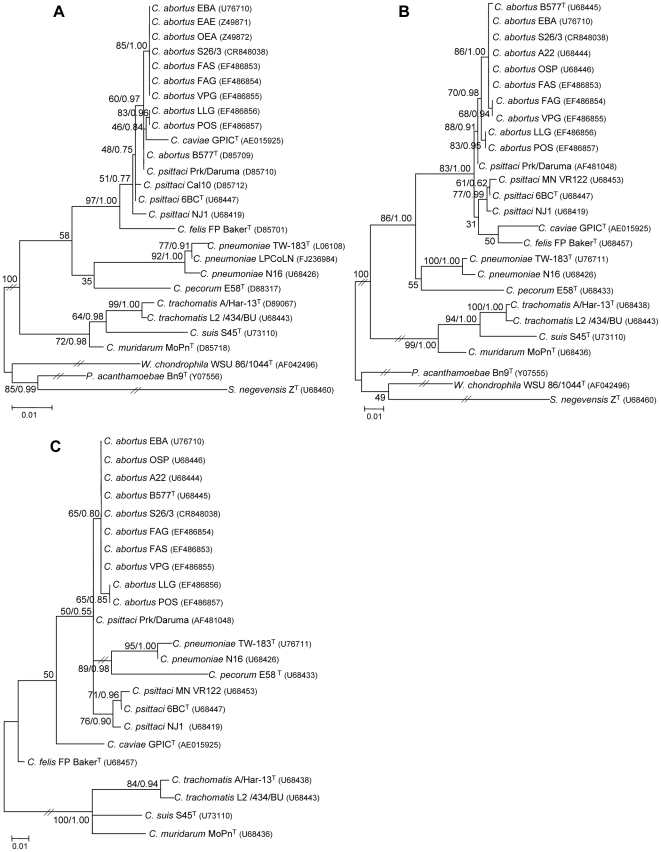
Phylogenetic trees were constructed using a subset of 27 or 26 full-length sequence alignments of the 16S, 23S domain I, and 16S-23S IS rRNAs. In particular, all available C. abortus sequences as well as representatives (type or reference strains) from all other hitherto defined species within Chlamydiaceae, including intraspecific variants, were employed to elucidate the evolutionary relationships of the C. abortus variants. The type strains of the families Parachlamydiaceae, Waddliaceae, and Simkaniaceae, phylogenetically positioned in the order Chlamydiales, were used as outgroups in the 16S and 23S rRNA trees. These outgroups were not used in 16S-23S IS tree due to their limited sequence identity with Chlamydiaceae and therefore the difficulty to align (Figure S4A). The best-scoring likelihood trees for 16S, 23S, and 16S-23S IS inferred by RAxML under the GTR+Gamma substitution model, are shown in Figure 1 (A, B and C). Branch support values (in congruent arrangements) for each of the two different approaches used (bootstrap support values from ML analyses, BS; Bayesian posterior probabilities, PP) are also indicated.
Figure 1. Best-scoring maximum likelihood trees based on 16S (A), 23S domain I (B), and 16S-23S IS (C) rRNA chlamydial sequences.
Full-length sequences of C. abortus variant strains and representatives from other Chlamydiaceae species were used. The type strains of other families within the order Chlamydiales were included as outgroups in the 16S and 23S rRNA trees. The trees were reconstructed using RAxML 7.2.6 [38]. The 16S and 23S rRNA trees were generated on the basis of secondary structure alignments created by SINA (SILVA SSU and LSU rRNA database project; [33]) while the 16S-23S IS tree was based on primary structure alignment computed using CLUSTAL X 1.83 [32]. Numbers on branches are support values to clusters on the right of them. Maximum likelihood bootstrap percentages and Bayesian posterior probabilities are included for clades that were consistently recovered using both phylogenetic methods (otherwise only bootstrap values are shown). Bayesian consensus trees are available as supporting material (Figure S5). Accession numbers for sequences retrieved from GenBank as well as for the sequences generated in this study are shown in parentheses. The mark//indicates that branches were shortened for visualization purposes.
16S rRNA analysis
The overall topology of the best-scoring ML tree inferred with RAxML was consistent with previously determined phylogenies using other algorithms [2], [6], [61] (Figure 1A). The tree constructed by the Bayesian approach differed in that it showed C. pneumoniae to form a distinct line of descent, separated from those of C. pecorum and other Chlamydiaceae taxa, resulting in an overall topology resembling the previously published phylogeny by Pettersson et al. [69] (Figure S5A). However, the Bayesian and ML trees were congruent with respect to the following characteristics:
A well-supported clade that contained C. felis, C. psittaci, C. caviae, and C. abortus was present in both analyses (BS, 97%; PP, 1.00).
Interestingly, both phylogenetic approaches recovered the C. caviae species (GPICT), usually positioned between the C. felis and C. psittaci clusters [2], [6], in the same group with LLG and POS C. abortus variant strains (Figure 1A, Figure S5A), even when filtering with Gblocks [70] was carried out. However, the ML tree (Figure 1A) indicated a relatively low BS support of 46%. Notably, a previous ML analysis recovered GPICT as a sister group to the C. abortus cluster [2], while in the present study exploratory Neighbor-Joining reconstruction (data not shown) led to a GPICT topology consistent with the currently accepted NJ-based phylogeny [2]. The position of GPICT will be discussed in more detail below.
The same close evolutionary relationship between C. psittaci and C. abortus was recovered by ML and Bayesian analyses. Chlamydophila abortus subclusters branched off from a common ancestor with the C. abortus type strain B577T and the C. psittaci variant strain Prk/Daruma, which, strikingly, shared identical 16S rRNA sequences (discussed below). The classical C. abortus strains were always grouped together in a subcluster (BS, 85%; PP, 1.00) and thereby separated from the LLG and POS C. abortus variant strains.
23S domain I rRNA analysis
The trees derived from both phylogenetic approaches (Figure 1B, Figure S5B) showed overall agreement with previously published topologies based on full-length or 23S domain I rRNA alignments [2], [6], [66]. In the ML tree (Figure 1B), C. psittaci strains (6BCT, MN-VR122, and NJ1) grouped with C. felis and C. caviae, though with poor statistical support (BS 31%), were separated from the C. psittaci variant Prk/Daruma lineage. The latter, in both analyses, was the closest relative (BS, 88%; PP, 0.91) to the C. abortus cluster (BS, 70%; PP, 0.98), which branched further into the LLG/POS C. abortus (BS, 83%; PP, 0.95) and the classical C. abortus (BS, 86%; PP, 1.00) subclusters. Within the latter, strains FAG and VPG formed a distinct clade (BS, 68%; PP, 0.94).
16S-23S IS analysis
The trees inferred from both phylogenetic approaches showed an almost identical overall topology (Figure 1C, Figure S5C), resembling a previously published 16S–23S IS tree [7]. The C. psittaci variant strain Prk/Daruma was again separated from the remaining C. psittaci strains. However, compared with the 16S and 23S trees, the evolutionary relationships of most Chlamydophila species were less well resolved. Nevertheless, a distinct C. abortus cluster is recovered again, with the LLG/POS C. abortus variant strains again forming a subcluster (BS, 65%; PP, 0.85).
Phylogeny and definition of the C. abortus cluster and subclusters
The current rRNA-based phylogenetic analyses provided strong evidence that C. abortus has evolved from C. psittaci, which is in agreement with previous findings based on other gene analyses [6], [7]. However, the C. abortus cluster and subclusters should be verified and defined by analysis of derived (autapomorphic or apomorphic) characters in the rRNA molecules such as signature or unique nucleotides [69], [71]–[73]. A signature nucleotide in this context is a nucleotide residue found explicitly in a certain position within the sequences of the particular cluster or group, where the base that is present differs from those found in the majority of other bacteria. A nucleotide residue at a certain position is said to be unique when present in all strains of a particular group or cluster and absent, with no or only a few exceptions, in the strains of any other chlamydial group or cluster. The characterization of unique nucleotide features was restricted to the Chlamydiales (autapomorphic characters) or to the Chlamydiaceae or Chlamydophila (apomorphic characters) taxa.
16S rRNA analysis
Among the five nucleotide differences observed between C. abortus and C. psittaci sequences (Table 2, Figure S2A and S2B), residue U277 represents a signature nucleotide for the classical C. abortus strains (see also Table 1). The subcluster of C. abortus classical strains is also supported by the residue A396 which is unique among all members of the order Chlamydiales. Perhaps the idiosyncratic U277 in the 16S molecule is the most pronounced attribute for classical C. abortus, since it corresponds to a highly conserved residue (discussed above). Interestingly, the C. abortus type strain B577T does not share the U277 and A396 residues, presenting an identical sequence with the C. psittaci variant Prk/Daruma strain. It should be noted that a type strain is not necessarily the most representative member of a species [74]. On the other hand, 16S rRNA sequence identity could not be indicative of species identity in some cases [75]. The LLG and POS C. abortus strains do not share residues U277 and A396 either (Tables 1, 2), however, the corresponding subcluster could be strongly supported by the residue G681 which is unique among all members of the order Chlamydiales (Table 1; discussed above). The variable residue A80 could also be informative for the LLG/POS subcluster, however this represents an ancestral and shared (symplesiomorphic) character occurring in the Simkaniaceae sp. ancestor and shared by C. caviae (strain GPICT) (see Table 1). At variable positions, identical residues are probably the result of multiple changes during the course of evolution, simulating an unchanged position (plesiomorphy). Such plesiomorphic-like sites may cause misleading branch attraction [20], like the one observed with C. caviae (strain GPICT) resulting in its grouping with LLG and POS strains in the 16S rRNA tree (Figure 1A, Figure S5A). Other plesiomorphic characters shared by C. abortus and C. caviae at positions where nucleotide differences between C. abortus and C. psittaci occur (Table 2) could further intensify the branch attraction and affect the tree topology.
Table 2. Nucleotide differences between C. abortus and C. psittaci rRNA molecules compared with related taxaa.
| Position inb | C. abortus | C. psittaci | C. caviae | C. felis | C. pecorum | C. pneumoniae | C. trachomatis | C. muridarum | C. suis | Parachlamydiaceae | Waddliaceae | Simkaniaceae | Domain Bacteria c |
| 16S rRNA | |||||||||||||
| 224 | A | G/A* | A | G | A | G | A | A | A | A | A | G/A | A 33.01; G 14.17; U 41.17; C 11.57 |
| 277 | U/C† | C | C | C | C | C | C | C | C | C | C | C | U 0.59; C 99.27; A 0.00; G 0.10 |
| 396 | A/G† | G | G | G | G | G | G | G | G | G | G | G | A 37.42; G 30.29; C 31.05; U 1.20 |
| 1267 | C | U/C* | C | U | U | U | C/U | C | U | U/C | C | A | C 66.45; U 30.80; A 0.73; G 0.26 |
| 1268 | A | G/A* | A | G | G | G | G | G | G | G | G | A | A 12.57; G 85.42; C 0.17; U 0.09 |
| 16S–23S Intergenic Spacer | |||||||||||||
| 49 | A | G/A* | G | A | A | A | G/A | A | G/A | U | G | – | |
| 55–56 | – | U/–* | A | A | – | – | U | U | C | G | – | U | |
| 185 | C | A/C* | C | A | C | C | U | U | U | U | A | G | |
| 192–193 | – | A/–* | – | A | – | A | A | A | A | A | A | C | |
| 198 | A | U/A* | A | A | A | A | A | A | A | G | A | G | |
| 204 | U | C | C | C | C | C | C | C | C | U | – | C | |
| 23S rRNA (domain I) | |||||||||||||
| 18 | U | C | C | C | C | C | C | C | C | C | C | C | U 18.73; C 80.88; A 0.40; G 0.00 |
| 132 | A | G/A* | G | G | A | A | G | G | G | A | A | G/A | A 17.23; G 46.82; C 33.71; U 0.75 |
| 147 | U | C/U* | C | C | U | U | C | C | C | U | U | C/U | U 37.78; C 25.19; G 34.44; A 1.11 |
| 152 | G/A† | A | A | A | A | A | G | G | G | U/C | U | C/U | G 19.63; A 27.78; U 13.70; C 37.78 |
| 157 | C | U/C* | C | C | C | U/C | C | C | C | C | C/U | C | C 62.22; U 11.11; G 15.93; A 2.96 |
| 181–182 | U/C† | C | C | C | A | C | C | C | C | U/C | G | U/– | |
| 240 | G | A/G* | A | G | G | G | G | G | G | A/U | G | U | G 39.71; A 22.38; U 21.30; C 16.61 |
| 297 | C | U/C* | U | U | U | U | U | U | U | C/A | C | A | C 23.38; U 29.86; A 25.90; G 20.50 |
| 547 | C/U ‡ | A/C* | C | C | C | C | A | A | A | U/A | A | U | C 16.13; U 20.79; A 53.76; G 9.32 |
More than three accession numbers for each species or family, including the type strain, were analyzed (the majority of analyzed strains and their accession numbers are given in the Figures S2, S3, and S4).
Nucleotide positions of the 16S and 23S domain I rRNA are given according to the E. coli J01695 secondary structure numbering system [35]; 16S–23S Intergenic Spacer positions based on primary structure is given according to the C. abortus type strain B577T sequence (U68445). Signature and unique residues for C. abortus strains and the corresponding positions are shown in bold; see text for details. Dashes indicate nucleotide gaps in rRNA sequence comparison.
Single base frequencies within domain Bacteria (dataset from http://www.rna.ccbb.utexas.edu/SAE/2A/nt_Frequency/).
Nucleotide in C. abortus LLG/POS variant;
Nucleotide in C. abortus FAG/VPG variant;
23S domain I rRNA analysis
Among the nine nucleotide differences observed between the C. abortus and C. psittaci sequences (Table 2, Figure S3A and S3B), residue U18 is unique for C. abortus strains, since it is not found in any other member of the order Chlamydiales, thereby defining the C. abortus cluster. Besides this, the residues in three other positions, namely 152, [181–182], and 297, could also be regarded as unique nucleotides. Residue C297, observed in all C. abortus strains but not among other Chlamydiaceae species, also could support the C. abortus cluster. Residues G152 and U[181–182] support the separation of the classical C. abortus subcluster, since they are not shared by other Chlamydophila or Chlamydiaceae species, respectively, as well as by the LLG/POS C. abortus variant (Tables 1, 2). The latter, LLG/POS variant subcluster, could be supported by the variable and informative residue G273 (Table 1). Finally, the group formed within C. abortus classical subcluster by the FAG and VPG strains is supported by the residue U547 which is not observed in other Chlamydiaceae members (Tables 1, 2).
16S–23S IS analysis
Another unique character of C. abortus cluster, also detected by Van Loock et al. [7], is the residue U204 in the IS sequence (according to C. abortus B577T sequence; acc. no. U68445) (Table 2). This residue is shared by both C. abortus subclusters (Table 2). Therefore, the LLG/POS C. abortus variant clade supported by U79 residue (Table 1) arises among other C. abortus strains (Figure 1C, Figure S5C). The topological difference of the “LLG/POS variant” in trees derived from the 16S–23S IS does not necessarily indicate a different path of evolution, since the IS region is more variable compared to 16S and 23S rRNAs.
Finally, as outlined in Table 2, at positions where differences between C. abortus and C. psittaci occur, the Prk/Daruma C. psittaci variant (including the Prk/Daruma, Prk46, Prk48, Prk49, 84/2334 and 1V avian strains) shared identical nucleotides with C. abortus strains. Nevertheless, this avian variant does not share the signature or unique C. abortus residues with only one exception, that of C297. The latter is particularly significant, since, based on the current rRNA phylogenetic analyses, the Prk/Daruma variant forms a distinct ancestral line for C. abortus supporting its intermediate position in the evolution of C. abortus from C. psittaci in agreement with previous reports [7], [61]. Recently, a multi-locus sequence typing scheme based on the partial sequences of seven housekeeping genes grouped the avian C. psittaci variant strain 84/2334 into C. abortus [76]. This does not contradict the above, as it is unlikely that independently evolving markers have preserved information on the same eras of evolutionary time [20].
Remarks on the evolutionary relationships among C. abortus variants
Based on rRNA secondary structure sequence data, we have investigated the evolutionary relationships among known Chlamydophila abortus variant strains originated from a common geographical region. Our results suggest that C. abortus strains could be regarded as representing two distinct phylogenetic lineages designated “classical” and “LLG/POS variant”. On the basis of maximum likelihood and Bayesian phylogenetic analyses these lineages were reliably recovered as subclusters supported by the presence of derived characters, with the C. abortus type strain B577T possibly representing an intermediate of the two lineages. The two C. abortus lineages, sharing three unique characters in the 23S domain I (residues U18 and C297) and 16S–23S IS (residue U204), but none in 16S (Table 2), could be distinguished on the basis of eight positions in the rRNA molecules (Tables 1, 2); four of these positions comprised nucleotides that appeared to be characteristic (signature or unique) of the “classical” lineage while three positions were unique for the “LLG/POS variant”. The U277 signature character, corresponding to a highly conserved residue of the 16S molecule, is the most pronounced attribute of the “classical” subcluster. Similarly, the unique G681 residue, conserved in a functionally strategic region also of the 16S molecule, is the most characteristic feature of the “LLG/POS variant”. Overall, the derived (signature or unique) C. abortus characters can serve as useful genetic markers for the identification of new strains before performing C. abortus-specific multilocus VNTR genotyping [17]. The rRNA-based phylogeny was consistent with the VNTR genotyping. In particular, the strains under investigation representing three different VNTR genotypes were also differentiated at least in one rRNA molecule (see also Table S2).
From an evolutionary perspective, both C. abortus lineages were found to be descendants of a common ancestor with the Prk/Daruma C. psittaci variant, and to have early diverged and separated during their evolution. Compared with the “classical” lineage, the “LLG/POS variant” has retained more ancestral features in the rRNA molecules as well as in other loci as gauged by the distinct similarity with C. psittaci-specific VNTR fragments [17]. The evolutionary events leading to rRNA sequence variations in both lineages have likely occurred once, as it is generally assumed for rRNA sequence evolution [20]. The observed rRNA sequence variations could possibly be explained by more rapid evolution due to a relatively recent shift to a host (ruminant), to which the C. psittaci variant ancestor had not been completely adapted. However, the FAG and VPG C. abortus strains, which represent the most common VNTR genotype of C. abortus in Greece and other countries [17], [77], have likely evolved from other “classical” strains, such as the FAS, following a gradual change (U547 residue) in the 23S domain I rRNA molecule.
Considering the relatively few sequence differences that lead to the classification of Chlamydiaceae into different species [2], [6] and based on the fact that most C. abortus derived characters corresponded to conserved residues, a subspecies status for each lineage may be applicable. The overall biological, biochemical and genotypic differentiation between LLG/POS variant and other C. abortus classical strains [13]–[18] in addition to their phylogenetic placement favor the delineation of these lineages as “subspecies”. Nevertheless, their systematics should be significantly aided by future sequence analyses of complete genomes [78].
In conclusion, the current rRNA secondary structure-based analysis and phylogenetic inference reveal new insights into how C. abortus variants of this study have differentiated during their evolution. The pattern and distribution of derived characters in functionally important regions of rRNA molecules could also make C. abortus a valuable model system for studies of molecular evolution in bacteria.
Supporting Information
Schematic representation of the 16S, 16S-23S intergenic spacer (IS) and 23S domain I rDNA showing the four overlapping PCR-amplified rDNA fragments as well as the relative positions of the primers used. The positions (a) of the primers are given according to the sequences determined in this study (GenBank accession numbers EF486853-EF486857). Numbers in parentheses are positions of the 16S (b) and 23S domain I (c) rRNA genes according to E. coli numbering system.
(DOC)
16S rRNA secondary structure-based alignment of Chlamydophila abortus and other Chlamydiales sp. sequences (71 strains), created with the SINA Webaligner (SILVA SSU reference alignment [33]). Alignment segments corresponding to the structural elements bearing SNVs (helices H61, H240, H39, and H673 in which the LLG/POS variant presents SNVs at positions 80, 277, 396, and 681, respectively) are shown in A. The positions in which C. abortus and C. psittaci species present nucleotide differences (positions 224/H122, 277/H240, 396/H39, and 1267&1268/H1241) are shown in A and B. Helix numbering and nucleotide positions are according to the E. coli numbering system (Comparative RNA Web, CRW site [35]). Relevant positions are indicated in boldface and shaded with their paired base positions; the latter appear in normal font. Loops and bulges are indicated with grey letters. Alignments were used to generate the Tables 1 and 2 of the paper.
(DOC)
23S domain I rRNA secondary structure-based alignment of Chlamydophila abortus and other Chlamydiales sp. sequences (67 strains), created with the SINA Webaligner (SILVA LSU reference alignment [33]). The helices and multistem-loop in which the LLG/POS variant presents SNVs (H150, ML between H150 & H183, and H271, at positions 152, [181–182], and 273, respectively), as well as the hairpin-loop in which the FAG/VPG variant presents a SNV (HL of the H533 at position 547) are shown in A. The positions in which C. abortus and C. psittaci species present nucleotide differences (positions 18/H15, 132/H131, 147/H131, 152/H150, 157/H150, [181–182]/ML177–182, 240/H235, 297/H271, and 547/HL545–548) are shown in A and B. Helix numbering and nucleotide positions are according to the E. coli numbering system (Comparative RNA Web, CRW site [35]). Relevant positions are indicated in boldface and shaded with their paired base positions; the latter appear in normal font. Loops and bulges are indicated with grey letters. Alignments were used to generate the Tables 1 and 2 of the paper.
(DOC)
A. 16S-23S rRNA intergenic spacer (IS) multiple sequence alignment of Chlamydophila abortus and other Chlamydiaceae sp. (57 strains), generated with CLUSTAL X (1.83) [32]. The IS sequences alignment of Parachlamydiaceae sp., Waddliaceae sp. and Simkaniaceae sp. strains, generated manually on the basis of the Chlamydiaceae sp. consensus sequence, is shown under the latter (see also [66]). The position in which the LLG/POS variant presents a SNV (position 79), and the positions in which C. abortus and C. psittaci species (shaded by yellow color) present interspecies differences (positions 49, 55–56, 185, 192–193, 198 and 204) are shaded. Relevant positions are indicated based on the 222 bp sequence of the C. abortus type strain B577T (U68445). Alignments were used to generate the Table 1 & 2 of the paper. B. Segment of stem region predicted to be formed between chlamydial 16S-23S rRNA IS and a complementary sequence of the 16S promoter [66]. The “-10 sequence” [68] is indicated with red letters.
(DOC)
Bayesian analysis (consensus trees) of 16S (A), 23S domain I (B), and 16S-23S IS (C) rRNA sequences of C. abortus strains and other Chlamydiales species. Numbers on branches indicate posterior probabilities. MrBayes version 3.1.2 was used [39]. The TreeGraph2 software [42] was used to display and manipulate the phylogenetic trees.
(DOC)
Primer pairs and internal primer used for rDNA amplification and direct sequencing.
(DOC)
Comparison of VNTR and rRNA genotypes.
(DOC)
Acknowledgments
Acknowledgement is due to Prof. George Mosialos for his most helpful advice.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was performed at the Laboratory of Microbiology and Infectious Diseases, Faculty of Veterinary Medicine of the Aristotle University of Thessaloniki. No external funding supported this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aitken ID. Chlamydial abortion. In: Martin WB, Aitken ID, editors. Diseases of Sheep. Oxford: Blackwell Science; 2000. pp. 81–86. [Google Scholar]
- 2.Everett KDE, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 3.Kerr K, Entrican G, McKeever D, Longbottom D. Immunopathology of Chlamydophila abortus infection in sheep and mice. Res Vet Sci. 2005;78:1–7. doi: 10.1016/j.rvsc.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Longbottom D, Coulter LJ. Animal chlamydioses and zoonotic implications. J Comp Pathol. 2003;128:217–244. doi: 10.1053/jcpa.2002.0629. [DOI] [PubMed] [Google Scholar]
- 5.Everett KDE. Chlamydia and Chlamydiales: more than meets the eye. Vet Microbiol. 2000;75:109–126. doi: 10.1016/s0378-1135(00)00213-3. [DOI] [PubMed] [Google Scholar]
- 6.Bush RM, Everett KDE. Molecular evolution of the Chlamydiaceae. Int J Syst Evol Microbiol. 2001;51:203–220. doi: 10.1099/00207713-51-1-203. [DOI] [PubMed] [Google Scholar]
- 7.Van Loock M, Vanrompay D, Herrmann B, Vander Stappen J, Volckaert G, et al. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int J Syst Evol Microbiol. 2003;53:761–770. doi: 10.1099/ijs.0.02329-0. [DOI] [PubMed] [Google Scholar]
- 8.Denamur E, Sayada C, Souriau A, Orfila J, Rodolakis A, et al. Restriction pattern of the major outer-membrane protein gene provides evidence for a homogeneous invasive group among ruminant isolates of Chlamydia psittaci. J Gen Microbiol. 1991;137:2525–2530. doi: 10.1099/00221287-137-11-2525. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths PC, Philips HL, Dawson M, Clarkson MJ. Antigenic and morphological differentiation of placental and intestinal isolates of Chlamydia psittaci of ovine origin. Vet Microbiol. 1992;30:165–177. doi: 10.1016/0378-1135(92)90111-6. [DOI] [PubMed] [Google Scholar]
- 10.Markey BK, McNulty MS, Todd D, Mackie DP. Comparison of ovine abortion and non-abortion isolates of Chlamydia psittaci using inclusion morphology, polyacrylamide gel electrophoresis, restriction endonuclease analysis and reactivity with monoclonal antibodies. Vet Microbiol. 1993;35:141–159. doi: 10.1016/0378-1135(93)90122-n. [DOI] [PubMed] [Google Scholar]
- 11.Salinas J, Souriau A, Cuello F, Rodolakis A. Antigenic diversity of ruminant Chlamydia psittaci strains demonstrated by the indirect microimmunofluorescence test with monoclonal antibodies. Vet Microbiol. 1995;43:219–226. doi: 10.1016/0378-1135(94)00100-b. [DOI] [PubMed] [Google Scholar]
- 12.Siarkou V, Lambropoulos AF, Chrisafi S, Kotsis A, Papadopoulos O. Subspecies variation in Greek strains of Chlamydophila abortus. Vet Microbiol. 2002;85:145–157. doi: 10.1016/s0378-1135(01)00506-5. [DOI] [PubMed] [Google Scholar]
- 13.Bouakane A, Benchaïeb I, Rodolakis A. Abortive potency of Chlamydophila abortus in pregnant mice is not directly correlated with placental and fetal colonization levels. Infect Immun. 2003;71:7219–7222. doi: 10.1128/IAI.71.12.7219-7222.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouakane A, Rekiki A, Rodolakis A. Protection of pregnant mice against placental and splenic infection by three strains of Chlamydophila abortus with a live 1B vaccine. Vet Rec. 2005;157:771–774. doi: 10.1136/vr.157.24.771. [DOI] [PubMed] [Google Scholar]
- 15.Vretou E, Loutrari H, Mariani L, Costelidou K, Eliades P, et al. Diversity among abortion strains of Chlamydia psittaci demonstrated by inclusion morphology, polypeptide profiles and monoclonal antibodies. Vet Microbiol. 1996;51:275–289. doi: 10.1016/0378-1135(96)00048-x. [DOI] [PubMed] [Google Scholar]
- 16.Vretou E, Psarrou E, Kaisar M, Vlisidou I, Salti-Montesanto V, et al. Identification of protective epitopes by sequencing of the major outer membrane protein gene of a variant strain of Chlamydia psittaci serotype 1 (Chlamydophila abortus). Infect Immun. 2001;69:607–612. doi: 10.1128/IAI.69.1.607-612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laroucau K, Vorimore F, Bertin C, Mohamad KY, Thierry S, et al. Genotyping of Chlamydophila abortus strains by multilocus VNTR analysis. Vet Microbiol. 2009;137:335–344. doi: 10.1016/j.vetmic.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 18.Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boumedine KS, Rodolakis A. AFLP allows the identification of genomic markers of ruminant Chlamydia psittaci strains useful for typing and epidemiological studies. Res Microbiol. 1998;149:735–744. doi: 10.1016/s0923-2508(99)80020-5. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig W, Klenk HP. Overview: A phylogenetic backbone and taxonomic framework for prokaryotic systematics. In: Boone DR, Castenholz RW, editors. Bergey's Manual of Systamatic Bacteriology. The Archaea and the Deeply Branching Phototropic Bacteria. New York: Springer-Verlag; 2001. pp. 49–65. [Google Scholar]
- 21.Ludwig W, Schleifer KH. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 22.Olsen GJ, Woese CR. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 23.Peplies J, Kottmann R, Ludwig W, Glöckner FO. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst Appl Microbiol. 2008;31:251–257. doi: 10.1016/j.syapm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Woese CR. Bacterial evolution. Microbial Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, et al. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smit S, Widmann J, Knight R. Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res. 2007;35:3339–3354. doi: 10.1093/nar/gkm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuyts J, Van de Peer Y, De Wachter R. Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res. 2001;29:5017–5028. doi: 10.1093/nar/29.24.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett KDE, Andersen AA. Identification of nine species of the Chlamydiaceae using PCR-RFLP. Int J Syst Bacteriol. 1999;49:803–813. doi: 10.1099/00207713-49-2-803. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. New York: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeSantis TZ, Hugenholtz P, Jr, Keller K, Brodie EL, Larsen N, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron and other RNAs. BMC Bioinformatics. 2002;3:1–31. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huelsenbeck JP. Systematic bias in phylogenetic analysis: is the Strepsiptera problem solved? Syst Biol. 1998;47:519–537. [PubMed] [Google Scholar]
- 37.Price MN, Dehal PS, Arkin AP. FastTree–2approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML-VI-HPC: maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 39.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. UK: Oxford University Press; 2006. Computational Molecular Evolution. Oxford Series in Ecology and Evolution. [Google Scholar]
- 41.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;22:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:1–9. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gautheret D, Konings D, Gutell RR. G.U base pairing motifs in ribosomal RNA. RNA. 1995;1:807–814. [PMC free article] [PubMed] [Google Scholar]
- 44.Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, et al. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 2002;321:215–234. doi: 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 45.Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 46.Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 48.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswamy P, Woodson SA. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20. J Mol Biol. 2009;392:666–677. doi: 10.1016/j.jmb.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988a;200:291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 51.Cormack RS, Mackie GA. Mapping ribosomal protein S20-16 S rRNA interactions by mutagenesis. J Biol Chem. 1991;266:18525–18529. [PubMed] [Google Scholar]
- 52.Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ. A story: unpaired adenosine bases in ribosomal RNAs. J Mol Biol. 2000;304:335–354. doi: 10.1006/jmbi.2000.4172. [DOI] [PubMed] [Google Scholar]
- 53.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 54.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 55.Mougel M, Philippe C, Ebel JP, Ehresmann B, Ehresmann C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 1988;16:2825–2839. doi: 10.1093/nar/16.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stern S, Powers T, Changchien LM, Noller HF. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J Mol Biol. 1988b;201:683–695. doi: 10.1016/0022-2836(88)90467-6. [DOI] [PubMed] [Google Scholar]
- 57.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Smith TF, Lee JC, Gutell RR, Hartman H. The origin and evolution of the ribosome. Biol Direct. 2008;3:1–13. doi: 10.1186/1745-6150-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 60.Allmang C, Mougel M, Westhof E, Ehresmann B, Ehresmann C. Role of conserved nucleotides in building the 16S rRNA binding site of E. coli ribosomal protein S8. Nucleic Acids Res. 1994;22:3708–3714. doi: 10.1093/nar/22.18.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pudjiatmoko, Fukushi H, Ochiai Y, Yamaguchi T, Hirai K. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1997;47:425–431. doi: 10.1099/00207713-47-2-425. [DOI] [PubMed] [Google Scholar]
- 62.Höpfl P, Ludwig W, Schleifer KH, Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989;185:355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- 63.Van Camp G, Chapelle S, De Wachter R. Amplification and sequencing of variable regions in bacterial 23S ribosomal RNA genes with conserved primer sequences. Curr Microbiol. 1993;27:147–151. doi: 10.1007/BF01576012. [DOI] [PubMed] [Google Scholar]
- 64.Elgavish T, Cannone JJ, Lee JC, Harvey SC, Gutell RR. A:A and A:G base-pairs at the ends of 16 S and 23 S rRNA helices. J Mol Biol. 2001;310:735–753. doi: 10.1006/jmbi.2001.4807. AA.AGhelix.ends: [DOI] [PubMed] [Google Scholar]
- 65.Antón AI, Martínez-Murcia AJ, Rodríguez-Valera F. Intraspecific diversity of the 23S rRNA gene and the spacer region downstream in Escherichia coli. J Bacteriol. 1999;181:2703–2709. doi: 10.1128/jb.181.9.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Everett KD, Andersen AA. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int J Syst Bacteriol. 1997;47:461–473. doi: 10.1099/00207713-47-2-461. [DOI] [PubMed] [Google Scholar]
- 67.Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. . J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 68.Engel JN, Ganem D. Chlamydial rRNA operons: gene organization and identification of putative tandem promoters. J Bacteriol. 1987;169:5678–5685. doi: 10.1128/jb.169.12.5678-5685.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersson B, Andersson A, Leitner T, Olsvik O, Uhlén M, et al. Evolutionary relationships among members of the genus Chlamydia based on 16S ribosomal DNA analysis. J Bacteriol. 1997;179:4195–4205. doi: 10.1128/jb.179.13.4195-4205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 71.Pettersson B, Tully JG, Bölske G, Johansson KE. Updated phylogenetic description of the Mycoplasma hominis cluster (Weisburg et al. 1989) based on 16S rDNA sequences. Int J Syst Evol Microbiol. 2000;50:291–301. doi: 10.1099/00207713-50-1-291. [DOI] [PubMed] [Google Scholar]
- 72.Pettersson B, Tully JG, Bölske G, Johansson KE. Re-evaluation of the classical Mycoplasma lipophilum cluster (Weisburg et al. 1989) and description of two new clusters in the hominis group based on 16S rDNA sequences. Int J Syst Evol Microbiol. 2001;51:633–643. doi: 10.1099/00207713-51-2-633. [DOI] [PubMed] [Google Scholar]
- 73.Weisburg WG, Tully JG, Rose DL, Petzel JP, Oyaizu H, et al. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989;171:6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young JM. Suggestions for avoiding on-going confusion from the Bacteriological Code. Int J Syst Evol Microbiol. 2000;50:1687–1689. doi: 10.1099/00207713-50-4-1687. [DOI] [PubMed] [Google Scholar]
- 75.Fox GE, Wisotzkey JD, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 76.Pannekoek Y, Dickx V, Beeckman DSA, Jolley KA, Keijzers WC, et al. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS One. 2010;5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laroucau K, Vorimore F, Sachse K, Vretou E, Siarkou VI, et al. Differential identification of Chlamydophila abortus live vaccine strain 1B and C. abortus field isolates by PCR-RFLP. Vaccine. 2010;28:5653–5656. doi: 10.1016/j.vaccine.2010.06.064. [DOI] [PubMed] [Google Scholar]
- 78.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the 16S, 16S-23S intergenic spacer (IS) and 23S domain I rDNA showing the four overlapping PCR-amplified rDNA fragments as well as the relative positions of the primers used. The positions (a) of the primers are given according to the sequences determined in this study (GenBank accession numbers EF486853-EF486857). Numbers in parentheses are positions of the 16S (b) and 23S domain I (c) rRNA genes according to E. coli numbering system.
(DOC)
16S rRNA secondary structure-based alignment of Chlamydophila abortus and other Chlamydiales sp. sequences (71 strains), created with the SINA Webaligner (SILVA SSU reference alignment [33]). Alignment segments corresponding to the structural elements bearing SNVs (helices H61, H240, H39, and H673 in which the LLG/POS variant presents SNVs at positions 80, 277, 396, and 681, respectively) are shown in A. The positions in which C. abortus and C. psittaci species present nucleotide differences (positions 224/H122, 277/H240, 396/H39, and 1267&1268/H1241) are shown in A and B. Helix numbering and nucleotide positions are according to the E. coli numbering system (Comparative RNA Web, CRW site [35]). Relevant positions are indicated in boldface and shaded with their paired base positions; the latter appear in normal font. Loops and bulges are indicated with grey letters. Alignments were used to generate the Tables 1 and 2 of the paper.
(DOC)
23S domain I rRNA secondary structure-based alignment of Chlamydophila abortus and other Chlamydiales sp. sequences (67 strains), created with the SINA Webaligner (SILVA LSU reference alignment [33]). The helices and multistem-loop in which the LLG/POS variant presents SNVs (H150, ML between H150 & H183, and H271, at positions 152, [181–182], and 273, respectively), as well as the hairpin-loop in which the FAG/VPG variant presents a SNV (HL of the H533 at position 547) are shown in A. The positions in which C. abortus and C. psittaci species present nucleotide differences (positions 18/H15, 132/H131, 147/H131, 152/H150, 157/H150, [181–182]/ML177–182, 240/H235, 297/H271, and 547/HL545–548) are shown in A and B. Helix numbering and nucleotide positions are according to the E. coli numbering system (Comparative RNA Web, CRW site [35]). Relevant positions are indicated in boldface and shaded with their paired base positions; the latter appear in normal font. Loops and bulges are indicated with grey letters. Alignments were used to generate the Tables 1 and 2 of the paper.
(DOC)
A. 16S-23S rRNA intergenic spacer (IS) multiple sequence alignment of Chlamydophila abortus and other Chlamydiaceae sp. (57 strains), generated with CLUSTAL X (1.83) [32]. The IS sequences alignment of Parachlamydiaceae sp., Waddliaceae sp. and Simkaniaceae sp. strains, generated manually on the basis of the Chlamydiaceae sp. consensus sequence, is shown under the latter (see also [66]). The position in which the LLG/POS variant presents a SNV (position 79), and the positions in which C. abortus and C. psittaci species (shaded by yellow color) present interspecies differences (positions 49, 55–56, 185, 192–193, 198 and 204) are shaded. Relevant positions are indicated based on the 222 bp sequence of the C. abortus type strain B577T (U68445). Alignments were used to generate the Table 1 & 2 of the paper. B. Segment of stem region predicted to be formed between chlamydial 16S-23S rRNA IS and a complementary sequence of the 16S promoter [66]. The “-10 sequence” [68] is indicated with red letters.
(DOC)
Bayesian analysis (consensus trees) of 16S (A), 23S domain I (B), and 16S-23S IS (C) rRNA sequences of C. abortus strains and other Chlamydiales species. Numbers on branches indicate posterior probabilities. MrBayes version 3.1.2 was used [39]. The TreeGraph2 software [42] was used to display and manipulate the phylogenetic trees.
(DOC)
Primer pairs and internal primer used for rDNA amplification and direct sequencing.
(DOC)
Comparison of VNTR and rRNA genotypes.
(DOC)