Abstract
In pigs, three different trichomonad species (Tritrichomonas foetus, Tetratrichomonas buttreyi and Tritrichomonas rotunda) have been described as commensals in the large intestine. The aim of this study was to gain further knowledge on the prevalence and pathogenicity of trichomonads in pigs by using a morphology-based approach. Chromogenic in situ hybridization (ISH) is a technique which allows direct localization of the protozoa in the intestinal tissue and correlation of the infection with pathologic changes. In the present study paraffin-wax embedded colon and ileum samples of 192 pigs were analyzed with this method. Using a probe specific for all known members of the order Trichomonadida (OT) 100 of the 192 pigs were tested positive. Thereof, about 10% showed moderate to high-grade parasitic load with trichomonads invading the lamina propria. Partial 18S ribosomal RNA gene sequencing of six of those animals showed a 100% sequence identity with T. foetus sequences. The majority of these animals were also tested positive for other enteropathogenic agents, such as Brachyspira sp., Lawsonia intracellularis, Escherichia coli, and porcine circovirus type 2. All OT-positive samples were further examined with another probe complementary to all known Tritrichomonas species sequences including T. foetus, T. augusta, T. mobilensis and T. nonconforma resulting in only 48 positives. These results suggest that T. foetus may not only be considered as an intestinal commensal but rather a facultative pathogen of pigs with a tendency for tissue invasion in the presence of other agents. Furthermore, the existence of other – yet to be identified – trichomonad species in the colon of pigs was shown.
Keywords: Pig, Trichomonads, Tritrichomonas foetus, Pathogenicity, Prevalence, Enteritis
1. Introduction
Infections with protozoal parasites of the order Trichomonadida (phylum Parabasala) are common in veterinary medicine. Trichomonads are anaerobic protists, possess hydrogenosomes instead of mitochondria, and can harbor up to six flagella. The order Trichomonadida comprises four families – Monocercomonadidae, Trichomonadidae, Tritrichomonadidae and Trichomitidae (Hampl et al., 2006). In pigs, a variety of trichomonads have been described to inhabit the intestinal tract (Rivera et al., 2008). Thereof, Tritrichomonas suis (Lun et al., 2005), which was shown to be the same species as Tritrichomonas foetus (Parsonson et al., 1976), has been most frequently found. T. foetus furthermore inhabits other host species like cats, where it elicits colitis (Levy et al., 2003). In pigs, T. foetus (Tachezy et al., 2002) is regarded a commensal mainly present in the colon and occasionally in the small intestine (Lun et al., 2005). Other trichomonads described in pigs are Tritrichomonas rotunda (Hibler et al., 1960) and Tetratrichomonas buttreyi (Rivera et al., 2008). In the case of T. rotunda no further literature exists, and there are no gene sequences of this species available. This species is not distinguishable by light microscopy from T. foetus and therefore may not be considered as an independent species. T. buttreyi has also been detected in the rumen and caecum of cattle and has been shown to be associated with diarrhea in heifers (Castella et al., 1997). However, T. buttreyi is still considered to be non-pathogenic in cattle (Cobo et al., 2004).
Currently all trichomonad species found in pigs are considered to be commensals and non-pathogenic. This might be due to the fact that in routine histopathological examination trichomonads are often overlooked or mistaken for cell debris or ingesta. To assess the prevalence and the pathogenicity of trichomonads a study was performed using chromogenic in situ hybridization (ISH) for the visualization of the parasites in porcine intestinal samples.
2. Materials and methods
2.1. Sampling
192 pigs suffering from chronic disease, the major symptoms of which were wasting, diarrhea, dyspnoea and skin lesions, were examined for infection with intestinal trichomonads. All pigs had already been tested for the presence of other pathogens such as Brachyspira sp., porcine circovirus type 2 (PCV-2), Lawsonia intracellularis and Escherichia coli (Komarek et al., 2009). During this former study all pigs were subjected to necropsy one to three hours after euthanasia, and amongst other samples, tissues from the first and the second colonic loop, as well as from the ileum were taken. The tissues were fixed in 7% neutral buffered formalin and embedded in paraffin wax for further examination.
2.2. In situ hybridization probes
Two runs of ISH were performed on paraffin embedded tissue sections including ileum and the first and the second colon loop. In the first run a probe able to detect all known representatives of the order Trichomonadida (OT probe) (5′-TTG CGG TCG TAG TTC CCC CAG AGC CCA AGA ACT-3′) was used (Mostegl et al., 2010). Positive samples were subjected to a second ISH run, during which ISH was performed on two consecutive tissue sections using the OT probe and a second probe able to detect all Tritrichomonas species (Tritri probe), respectively. The Tritri probe was newly designed after extensive homology studies of all 18S ribosomal RNA (rRNA) genes of different Tritrichomonas species (T. foetus, T. augusta, T. mobilensis, T. nonconforma) available in the gene bank, carried out with the Sci Ed Central (Scientific & Educational software, Cary, NC, USA) software package. A strongly homologous region (a maximum probe target mismatch of one nucleotide) complementary to a segment of the 18S rRNA gene of all Tritrichomonas species was chosen. The selected Tritri probe sequence was 5′-AGC TGT AGC CTT TTC AGG ACA GCA TCT CT-3′. This sequence was further analyzed using the Basic Local Alignment Search Tool (BLAST, www.ncbi.nlm.nih.gov/blast.cgi) to search against GenBank sequences and exclude unintentional cross-reactivity. Subsequently, the Tritri probe sequence was sent to Eurofins MWG Operon (Ebersberg, Germany) for probe synthesis and 3′ end labeling with digoxigenin. Afterwards the probe was tested positive on protozoal culture sections containing T. foetus or T. augusta, respectively. No positive signals were achieved with all other analyzed protozoal culture sections, including Histomonas meleagridis, Hypotrichomonas acosta, Monocercomonas colubrorum, Pentatrichomonas hominis, Tetratrichomonas gallinarum, Trichomonas gallinae and Trichomitus batrachorum (Mostegl et al., 2010). To rule out cross-reactivity the probe was tested negative on various embedded cultures or tissue samples including several species of other protozoan parasites, fungi, bacteria and viruses as listed in Mostegl et al. (2010).
2.3. In situ hybridization
Chromogenic ISH was performed according to a previously published protocol (Chvala et al., 2006). In brief, 3 μm paraffin wax embedded tissue sections were dewaxed and rehydrated. First, slides were treated with 2.5 μg/ml proteinase K (Roche, Basel, Switzerland) in Tris-buffered saline for 30 min at 37 °C for proteolysis. Afterwards the tissue sections were rinsed in distilled water to remove proteinase K and dehydrated in alcohol (95% and 100%), followed by air-drying. The slides were incubated over night at 40 °C with hybridization mixture, 100 μl of which was composed of 50 μl formamide, 20 μl 20× standard saline citrate buffer (SSC), 10 μl dextran sulfate (50%, w/v), 12 μl distilled water, 5 μl boiled herring sperm DNA (50 mg/ml), 2 μl Denhardt's solution and 1 μl OT or Tritri probe, respectively, at a concentration of 20 ng/ml. The next day, the slides were washed in decreasing concentrations of SSC (2× SSC, 1× SSC and 0.1× SSC; 10 min each), for removal of unbound probe. Afterwards the sections were incubated with anti-digoxigenin-AP Fab fragments (Roche) (1:200) for 1 h at room temperature. The hybridized probe was visualized subsequent to an additional washing step using the color substrates 5-bromo-4-chloro-3-inodyl phosphate (BCIP) and 4-nitro blue tetrazolium chloride (NBT) (Roche). After 2 h of incubation the color development was stopped using TE buffer (pH 8.0). Subsequently, the slides were counterstained with haematoxylin and mounted under coverslips using Aquatex (VWR International, Vienna, Austria).
2.4. Collection of data
Each tissue section was evaluated for two criteria. The first parameter was the parasitic load, which ranged from scattered (s) (0–3 parasites per high power field (hpf)), low- (l) (4–15 parasites per hpf), moderate- (m) (16–40 parasites per hpf), to high-grade (h) (>40 parasites per hpf). As second parameter the localization of the trichomonads in the intestine was assessed, ranging from parasites found in the intestinal lumen (lu), or inside crypt lumina (c) to emigrating into the lamina propria mucosae (e). In animals displaying m or h grade infections, the pathological changes were evaluated according to the following four parameters: (1) crypt hyperplasia, (2) moderate to severe non suppurative inflammation of the propria, (3) crypt abscesses and (4) presence of dilated, mucus-filled crypts.
2.5. PCR and gene sequence analysis
To classify the found trichomonads, a PCR followed by gene sequence analysis was carried out on tissue samples of sections with h/e, m/e and l/e infection grades. Two samples of each grade were sequenced. For this reason, GenBank sequences of the 18S rRNA gene from T. foetus, T. augusta, Tritrichomonas muris and T. buttreyi were aligned, and primers flanking a region containing at least one nucleotide mismatch in-between the species were designed. Subsequently, the primers were subjected to BLAST. BLAST analysis showed similar sequences only with protozoa from the phylum Parabasala, but not with other relevant microorganisms like bacteria, fungi or viruses. The designed sequencing primers: 1064F: 5′-AAC TTA CCA GGA CCA GAT GT-3′ and 1298R: 5′-CAC GGA CCT GTT ATT GCT AC-3′ flanked a region of 234 bp (accession no. M81842).
DNA extraction from three 10 μm thick sections from formalin fixed and paraffin embedded material was performed prior to PCR. The sections were dewaxed in xylene, washed in ethanol and air-dried. Subsequently, DNA was extracted using the nexttec Clean Column kit (Nexttec, Leverkusen, Germany) according to the manufacturer's protocol.
For DNA amplification the PCR master mixture reactions consisted of 10 μl HotMasterMix (5Prime, Eppendorf, Hamburg, Germany), 0.4 μM of each primer, 2 μl template DNA and distilled water to a total volume of 25 μl. PCR started with a first heat denaturation step at 94 °C for 2 min, followed by 40 cycles of heat denaturation at 94 °C for 30 s, primer annealing at 60 °C for 30 s and DNA elongation at 72 °C for 1 min. PCR was completed with a final elongation step at 72 °C for 10 min.
PCR was carried out after the efficacy of DNA extraction from paraffin-embedded tissue and the integrity of DNA had been tested using primers specific for the β-actin gene (Weissenböck et al., 2003). The negative control was a PCR mixture containing distilled water instead of template DNA. 10 μl of the PCR reaction was analyzed on a 2% Tris acetate–EDTA–agarose gel. Subsequent to staining with ethidium bromide the agarose gel was visualized with a BioSens SC-Series 710 gel documentation system using the BioSens gel imaging system software (GenXpress, Wiener Neudorf, Austria). PCR products showing the expected size were sequenced according to Bakonyi et al. (2004), except that DNA purification after amplification was carried out using the DyeEx 2.0 spin kit (QIAGEN, Hilden, Germany) instead of ethanol precipitation. The obtained nucleotide sequences were subjected to BLAST to search against the GenBank sequences.
3. Results
3.1. In situ hybridization with OT probe
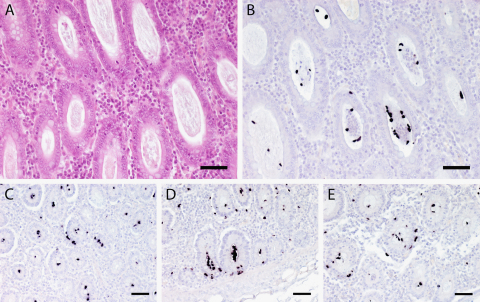
Of the 192 pigs investigated 100 (52.1%) were assessed as positive in at least one of the examined intestinal sections with the OT ISH probe (Fig. 1). In 55 cases scattered protozoal organisms, in 26 pigs low numbers of trichomonads, in 10 animals moderate, and in 9 cases high amounts of specifically stained parasites were found (Table 1). In one case only the ileum section revealed s/lu infection with trichomonads, in seven cases both, ileum and colon were positive, while in all other positive cases only the colon was affected.
Fig. 1.
HE staining and ISH using the OT probe of the colon of a pig. (A) HE staining shows dilated mucus-filled crypts in which presence of trichomonads can barely be recognized. Bar = 80 μm. (B) At ISH the trichomonads are easily discernible by their purple to black staining within the mucus-filled dilated crypts. Additionally, mild crypt abscesses are present. Bar = 80 μm. (C–E) ISH shows numerous trichomonads within crypt lumina but a considerable percentage of them is clearly localized in the crypt surroundings indicating emigration into the lamina propria mucosae. Bar = 80 μm.
Table 1.
Overview of the quantities and locations of trichomonads in the pigs which were positive after staining with the OT probe (1st run). Trichomonad quantity reached from scattered (s), low (l), and moderate (m) to high-grade (h). Location of trichomonads was assessed as: intestinal lumen (lu), crypts (c) or emigration into the lamina propria mucosae (e).
| Lumen (lu) | Crypts (c) | Emigration (e) | Total | |
|---|---|---|---|---|
| Scattered (s) | 39 | 9 | 7 | 55 |
| Low (l) | 12 | 8 | 6 | 26 |
| Moderate (m) | 3 | 1 | 6 | 10 |
| High (h) | 1 | 0 | 8 | 9 |
| 55 | 18 | 27 | 100 | |
Evaluation score: s: 0–3 parasites per high power field (hpf); l: 4–15 parasites per hpf; m: 16–40 parasites per hpf; h: >40 parasites per hpf.
3.2. Association of trichomonad presence with pathologic lesions
In total, nine cases of infected pigs (eight: h/e; one: h/lu) revealed high parasitic load. All animals with a h/e infestation grade showed pathological lesions, and in the majority of them multiple changes were present. Of the 10 pigs with moderate trichomonad infection (six: m/e, three: m/lu, one: m/c) only one was found positive for three assessed parameters (all except crypt hyperplasia) and one had crypt abscesses and mucus-filled dilated crypts. In five animals only one of the pathological changes was present and three pigs displayed no histological lesions at all (data of cases with high and moderate grades of infections are summarized in Table 2).
Table 2.
Total numbers of pigs displaying moderate (m) or high-grade (h) parasitic load assessed using the OT probe (1st run), categorized for different pathological lesions found. For each tissue section the quantity of trichomonads (m or h) was evaluated in context with their location (either within the intestinal lumen (lu), within crypts (c) or emigrating into the lamina propria mucosae (e)). Multiple entries are included and not shown separately.
| m/lu | m/c | m/e | h/lu | h/e | |
|---|---|---|---|---|---|
| Crypt hyperplasia | 4 | ||||
| ++–+++ non suppurative inflammation | 1 | 2 | 1 | 6 | |
| Crypt abscesses | 1 | 2 | 8 | ||
| Mucus-filled dilated crypts | 1 | 3 | 2 | ||
| No lesions | 2 | 1 |
Evaluation score: s: 0–3 parasites per high power field (hpf); l: 4–15 parasites per hpf; m: 16–40 parasites per hpf; h: >40 parasites per hpf.
3.3. Association of in situ hybridization with other pathogens
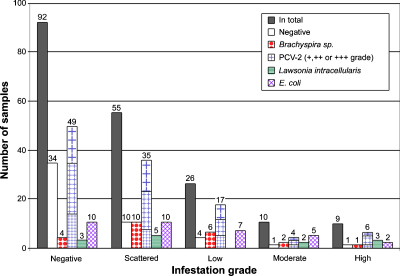
The presence of intestinal trichomonads with infection of other pathogens such as Brachyspira sp., porcine circovirus type 2, L. intracellularis and E. coli were correlated (data summarized in Fig. 2). In general, the majority of cases with presence of trichomonads also had an infection with other intestinal pathogens. In both, m and h infestation grades only one case out of ten or nine animals, respectively, was negative for all tested pathogens. One case each of both infestation grades, however, was only slightly positive in the PCV-2 ISH, a result which is considered of minor clinical importance.
Fig. 2.
Total numbers of pigs positive for trichomonads and other pathogens. The X-axis shows trichomonad infections in different infestation degrees (scattered, low, moderate, high or not infected (Neg)); the bars stand for total number, not infected with one of the tested pathogens (negative), positive for Brachyspira sp., PCV-2, L. intracellularis or E. coli, respectively. Multiple infections are included in the amount of samples and not shown separately.
3.4. Gene sequencing analysis
All six achieved sequences from tissue sections of samples having different infestation degrees of trichomonads emigrating into intestinal tissues showed 100% sequence identity with T. foetus sequences accessible in the GenBank.
3.5. In situ hybridization with Tritri probe
Of 100 pigs tested positive in the first ISH run with the OT probe, the positive result could be reproduced in 91 cases in the second run, whereas only 48 of 91 animals revealed positive signals when tested with the Tritri probe (in detail see Table 3). Generally, animals which had moderate or high-grade trichomonad infestation showed positive signals with both probes.
Table 3.
Results of comparative testing of the samples positive after the 1st run of staining with the OT probe. Two consecutive tissue sections were tested with the OT and the Tritri probe, respectively. Displayed are the different infestation degrees reaching from scattered (s), low (l), moderate (m) to high-grade (h) in context with the location of trichomonads (within the intestinal lumen (lu), within crypts (c) or emigration into the lamina propria mucosae (e)), achieved with each probe. Tissue samples negative with the OT probe in this run compared to the first run are indicated as negative.
| Lumen (lu) |
Crypts (c) |
Emigration (e) |
Total |
|||||
|---|---|---|---|---|---|---|---|---|
| Probe | OT | Tritri | OT | Tritri | OT | Tritri | OT | Tritri |
| Scattered (s) | 31 | 5 | 12 | 11 | 7 | 4 | 50 | 20 |
| Low (l) | 10 | 1 | 9 | 8 | 5 | 3 | 24 | 12 |
| Moderate (m) | 1 | 0 | 1 | 1 | 8 | 8 | 10 | 9 |
| High (h) | 0 | 0 | 0 | 0 | 7 | 7 | 7 | 7 |
| In total | 42 | 6 | 22 | 20 | 27 | 22 | 91 | 48 |
| Negative | 9 | |||||||
| 100 | ||||||||
Evaluation score: s: 0–3 parasites per high power field (hpf); l: 4–15 parasites per hpf; m: 16–40 parasites per hpf; h: >40 parasites per hpf.
4. Discussion
In this work the pathogenicity and prevalence of intestinal trichomonads in pigs should be investigated. Due to the authors’ knowledge this is the first time that trichomonads in pigs are not only detected in faecal samples but were directly visualized in the intestinal tissue. For assessment of the pathogenicity the parasitic load together with the localization of the found trichomonads and the associated pathological lesions were taken into account. Especially multiplication within the crypt lumina to large numbers and invasion into the lamina propria mucosae are considered to elicit tissue changes and resulting clinical signs. In 27 pigs trichomonads were found in the lamina propria mucosae (Fig. 1), of which 14 displayed a moderate to high-grade protozoal load. Due to the rather low number of such cases a statistically correct estimation whether there is an association with other enteropathogenic agents (Brachyspira sp., PCV-2, L. intracellularis, E. coli) seems to be difficult. However, h/e cases with concomitant histological lesions have been found only exceptionally in the absence of other enteropathogenic agents which suggests that trichomonads act rather as secondary than as primary pathogens. Considering cases with low grade PCV-2 positivity as not significantly impaired increases the cases of trichomonas infection in the absence of another enteropathogenic pathogen to more than 20%. This aspect may be considered a further argument for the potential pathogenicity of trichomonads in pigs. In any case, it is very likely that the excessive multiplication of trichomonads even in cases with presence of other pathogens is responsible for additional pathogenic effects and aggravation of clinical disease. The gene sequencing analysis to further classify the emigrating trichomonads identified them as T. foetus. Taken together, these data suggest that intestinal trichomonads in pigs namely T. foetus may no longer be considered as apathogenic and irrelevant commensals.
In only 91 of the 100 pigs the positive ISH result with the OT probe could be repeated in the second ISH run. This variability might be due to the fact that at repeated cutting some tissue material is lost before a suitable paraffin wax tissue section can be achieved. To be able to better compare the two probes used in the second run (OT and Tritri probe) and minimize the detection variability, the second run was performed on two consecutive tissue sections.
Only 48 pigs were positive using the Tritri probe compared to 91 positive cases with the OT probe. Taking into account that 3 μm thin sections were used and that T. foetus is about 11–15 μm long and 3–4 μm wide (Tachezy et al., 2002), the majority of the trichomonads should be present in both tissue sections. Most of the pigs positive with the OT probe and negative with the Tritri probe displayed scattered to low infection grades and in the majority of animals trichomonads were found in the intestinal lumen. Therefore, the presence of other apathogenic trichomonads than T. foetus has to be considered, which is currently further studied.
In summary, approximately 10% of the tested pigs displayed moderate to high infection grades with trichomonads; in most of these cases there were marked pathological lesions and approximately 80% of them had an accompanying infection with other clinically relevant enteropathogenic agents. All sequenced samples with low, moderate or high load of trichomonads emigrating into the lamina propria mucosae showed 100% identity with a T. foetus strain. Therefore, the authors propose that T. foetus may not longer be considered as a mere commensal of the intestine in pigs, but rather as a possible facultative pathogenic agent with the ability for mucosa invasion. Additionally, the collected data suggest the common presence of other most likely apathogenic, commensalic, not yet described, intestinal trichomonads.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
The authors wish to thank Karin Fragner and Klaus Bittermann for their excellent technical support. This work was funded by the Austrian Science Fund (FWF) grant P20926.
References
- Bakonyi T., Gould E.A., Kolodziejek J., Weissenböck H., Nowotny N. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: comparison with the South African strain SAAR-1776 and other flaviviruses. Virology. 2004;328:301–310. doi: 10.1016/j.virol.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Castella J., Muńoz E., Ferrer D., Gutiérrez J.F. Isolation of the trichomonad Tetratrichomonas buttreyi (Hibler et al., 1960) Honigberg, 1963 in bovine diarrhoeic faeces. Vet. Parasitol. 1997;70:41–45. doi: 10.1016/s0304-4017(96)01140-5. [DOI] [PubMed] [Google Scholar]
- Chvala S., Fragner K., Hackl R., Hess M., Weissenböck H. Cryptosporidium infection in domestic geese (Anser anser f. domestica) detected by in-situ hybridization. J. Comp. Pathol. 2006;134:211–218. doi: 10.1016/j.jcpa.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Cobo E.R., Cantón G., Morrell E., Cano D., Campero C.M. Failure to establish infection with Tetratrichomonas sp. in the reproductive tracts of heifers and bulls. Vet. Parasitol. 2004;120:145–150. doi: 10.1016/j.vetpar.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Hampl V., Vrlík M., Cepicka I., Pecka Z., Kulda J., Tachezy J. Affiliation of Cochlosoma to trichomonads confirmed by phylogenetic analysis of the small-subunit rRNA gene and a new family concept of the order Trichomonadida. Int. J. Syst. Evol. Microbiol. 2006;56:305–312. doi: 10.1099/ijs.0.63754-0. [DOI] [PubMed] [Google Scholar]
- Hibler C.P., Hammmond D.M., Caskey F.H., Johnson A.E., Fitzgerald P.R. The morphology and incidence of the trichomonads in swine, Tritrichomonas suis (Gruby & Delafond), Tritrichomonas rotunda, n. sp. and Trichomonas buttreyi, n. sp. J. Protozool. 1960;7:159–171. [Google Scholar]
- Komarek V., Maderner A., Spergser J., Weissenböck H. Infections with weakly haemolytic Brachyspira species in pigs with miscellaneous chronic diseases. Vet. Microbiol. 2009;134:311–317. doi: 10.1016/j.vetmic.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Levy M.G., Gookin J.L., Poore M., Birkenheuer A.J., Dykstra M.J., Litaker R.W. Tritrichomonas foetus and not Pentatrichomonas hominis is the etiologic agent of feline trichomonal diarrhea. J. Parasitol. 2003;89:99–104. doi: 10.1645/0022-3395(2003)089[0099:TFANPH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lun Z.-R., Chen X.-G., Zhu X.-Q., Li X.-R., Xie M.-Q. Are Tritrichomonas foetus and Tritrichomonas suis synonyms? Trends Parasitol. 2005;21:122–125. doi: 10.1016/j.pt.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Mostegl M.M., Richter B., Nedorost N., Maderner A., Dinhopl N., Kulda J., Liebhart D., Hess M., Weissenböck H. Design and validation of an oligonucleotide probe for the detection of protozoa from the order Trichomonadida using chromogenic in situ hybridization. Vet. Parasitol. 2010;171:1–6. doi: 10.1016/j.vetpar.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonson I.M., Clark B.L., Dufty J.H. Early pathogenesis and pathology of Tritrichomonas foetus infection in virgin heifers. J. Comp. Pathol. 1976;86:59–66. doi: 10.1016/0021-9975(76)90028-1. [DOI] [PubMed] [Google Scholar]
- Rivera W.L., Lupisan A.J.B., Baking J.M.P. Ultrastructural study of a tetratrichomonad isolated from pig fecal samples. Parasitol. Res. 2008;103:1311–1316. doi: 10.1007/s00436-008-1134-x. [DOI] [PubMed] [Google Scholar]
- Tachezy J., Tachezy R., Hampl V., Šedinová M., Vanaácová S., Vrlík M., Van Ranst M., Flegr J., Kulda J. Cattle pathogen Tritrichomonas foetus (Riedmüller, 1928) and pig commensal Tritrichomonas suis (Gruby & Delafond, 1843) belong to the same species. J. Eukaryot. Microbiol. 2002;49:154–163. doi: 10.1111/j.1550-7408.2002.tb00360.x. [DOI] [PubMed] [Google Scholar]
- Weissenböck H., Hubálek Z., Halouzka J., Pichlmair A., Maderner A., Fragner K., Kolodziejek J., Loupal G., Kölbl S., Nowotny N. Screening for West Nile virus infections of susceptible animal species in Austria. Epidemiol. Infect. 2003;131:1023–1027. doi: 10.1017/s0950268803001031. [DOI] [PMC free article] [PubMed] [Google Scholar]