Abstract
A clustered DNA lesion, also known as a multiply damaged site, is defined as ≥ 2 damages in the DNA within 1–2 helical turns. Only ionizing radiation and certain chemicals introduce DNA damage in the genome in this non-random way. What is now clear is that the lethality of a damaging agent is not just related to the types of DNA lesions introduced, but also to how the damage is distributed in the DNA. Clustered DNA lesions were first hypothesized to exist in the 1990’s, and work has progressed where these complex lesions have been characterized and measured in irradiated as well as in non-irradiated cells. A clustered lesion can consist of single as well as double strand breaks, base damage and abasic sites, and the damages can be situated on the same strand or opposing strands. They include tandem lesions, double strand break (DSB) clusters and non-DSB clusters, and base excision repair as well as the DSB repair pathways can be required to remove these complex lesions. Due to the plethora of oxidative damage induced by ionizing radiation, and the repair proteins involved in their removal from the DNA, it has been necessary to study how repair systems handle these lesions using synthetic DNA damage. This review focuses on the repair process and mutagenic consequences of clustered lesions in yeast and mammalian cells. By examining the studies on synthetic clustered lesions, and the effects of low vs high LET radiation on mammalian cells or tissues, it is possible to extrapolate the potential biological relevance of these clustered lesions to the killing of tumor cells by radiotherapy and chemotherapy, and to the risk of cancer in non-tumor cells, and this will be discussed.
Keywords: clustered DNA lesions, double strand breaks, DNA repair, mutation induction, ionizing radiation
1. Introduction
DNA damage and death due to ineffective repair are the basis of many cancer treatments. The DNA lesions produced by radiotherapy and certain chemotherapies include modifications to thymine, adenine, guanine and cytosine, sites of base loss (AP sites), single strand breaks (SSBs) and double strand breaks (DSBs) [1]. These types of damages are very similar to those produced by endogenous reactive oxygen species and chemicals such as hydrogen peroxide, yet ionizing radiation and bleomycin sulfate require fewer DNA lesions to produce a lethal event (the dose at which 63% of the cells are killed, according to a Poisson distribution) in a mammalian cell than hydrogen peroxide. For a lethal event 1000 SSBs are produced by ionizing radiation, 150 SSBs by bleomycin sulfate, but 400,000 SSBs are required following treatment with hydrogen peroxide [2]. So why are ionizing radiation and bleomycin sulfate more lethal than hydrogen peroxide? This was the question being considered by scientists in the late 1980’s and early 1990’s. At this time the DSB was considered the lethal lesion, with base damage only important in mutagenesis and not cell death. However, in 1988, John Ward put forward the idea of locally multiply damaged sites (LMDS) and proposed that the difference in lethality among DNA damaging agents was due to the spatial distribution of damage and the inability of the cell to repair these clusters of lesions in the DNA molecule [2,3].
The initial evidence for the existence of ionizing radiation-induced clustered DNA lesions was provided by the biophysical modelers. Although simulations of irradiated cells predicted that clusters of damage were unlikely to be produced by a direct ionization or primary radical [4], the clusters of ionizations generated from low secondary electrons from X- and γ-radiation tracks had the potential to produce multiple hydroxyl radicals from the water molecules closely associated with the DNA. The clusters of hydroxyl radicals were predicted to generate multiple lesions within a helical turn of the DNA from clinically relevant doses of X- and γ-radiation [4, 5]. Bleomycin sulfate was also found to produce multiple lesions. Bleomycin reacts preferentially with double-stranded DNA and modifies the bases in the order of T > C > A > G [6; 7]. One part of the molecule is thought to intercalate into the DNA, while another section is capable of binding metal ions such as iron. Reactive oxygen species formed from the reaction of the bleomycin with iron and oxygen have been implicated in the damaging mechanism [8]. Incubation of bleomycin-treated DNA in vitro with endonuclease III or putrescine to cleave at AP sites resulted in an increased production of DSBs [9]. This demonstrated that bleomycin forms clustered lesions consisting of SSBs and AP sites. Neocarcinostatin, another radiomimetic compound, also produces clustered lesions that consist of AP sites and SSBs two base pairs apart, and the breaks have 3' phosphate and 5' aldehyde termini [10]. Hydrogen peroxide, however, reacts with a metal ion in a Fenton reaction to produce hydroxyl radicals and so single lesions are produced at the sites of bound metal ions randomly distributed in the DNA [2]. Hence it was postulated that single lesions were easier to repair than the clustered lesions and agents generating clustered lesions would be more lethal to a mammalian cell. Etoposide, another cancer drug treatment, has also since been found to introduce multiple DSBs into single chromatin domains [11] and hence clustered lesions may also be involved in the lethality of this drug. John Ward defined a clustered lesion (multiply damaged site, MDS, or LMDS as they were originally known) as ≥2 damages within a 20 bp region [12]. Since ionizing radiation produces a greater yield of base damage than SSBs or DSBs [2], the individual types of lesions within a cluster could potentially be base damage and AP sites as well as strand breaks, and this started to change the thinking about the relevance of base damage to cell death.
2. Cluster designation and their detection
There are many possible permutations of clustered lesions that include clusters with damages on one strand (which includes tandem lesions) as well as those with damage in opposing strands (bistranded lesions), and as discussed in this review, they have different biological outcomes in terms of mutagenesis and repair inhibition.
Tandem lesions are two damages situated immediately next to each other in the same DNA strand. Unlike the bistranded clusters, these lesions can be generated from one radical creating a single damage with a reactive intermediate that reacts with a neighboring nucleoside 5’ or 3’ to the initial damage site, creating two damages [13]. Liquid chromatography-mass spectrometry is used to detect tandem lesions: this technique identified adjacent lesions of oxidized guanine and a formamido-derivative of a pyrimidine in X-irradiated calf thymus DNA [14–17]. A similar type of lesion was also detected in mouse cells exposed to oxidative stress [18]. Double lesions, which are also induced by radiation from one radical, consist of two damages that are not necessarily in adjacent nucleosides. In fact, two damages can even occur on one nucleoside. An example of this is the 8, 5’-cyclo-purine damage, which involves a covalent link between the base and the carbohydrate moiety of one nucleoside. Since the 8, 5’-cyclo-purine involves only the generation of one damaged nucleoside in the DNA it will not be discussed in this review. A second type of double lesion detected in irradiated DNA is a base damage (frequently guanine) situated in close proximity to a SSB on the same strand. Again this lesion can be generated from a single radical originating from the base, and the SSB is induced at the adjacent nucleoside or within a few nucleosides of the base damage [19, 20]. The mechanism of formation of these double lesions occurs by electron transfer originating at a nucleobase radical intermediate. Box et al [20] postulated that this process may also generate bistranded double lesions. Recently, a new clustered lesion was identified. It comprises of a SSB and an adjacent interstrand crosslink with a cytosine on the opposite strand. This lesion results from a single sugar radical generated by ionizing radiation or bleomycin [21].
Bistranded clusters have been designated into two groups: non-DSB which consist of base damage, AP sites and/or SSBs, and DSB clusters. The DSB clusters do have the possibility of being complex DSBs that have oxidative damage situated near the DSB, and these are predicted to exist in irradiated DNA especially as the linear energy transfer (LET) of the radiation increases [22]. The higher the LET, the denser the ionization track and so the greater the probability of complex lesions being produced in the DNA. The non-DSB clusters are also designated into oxypurine (purine base damage), oxypyrimidine (pyrimidine base damage) and abasic site clusters (containing AP sites). The simplest form of a clustered lesion is actually a DSB, generated from two closely opposed SSBs, and it was Rydberg [23] that detected the production of short (0.1–2 kb) DNA fragments following irradiation of human diploid cells, indicating the introduction of two DSBs within a short distance. This confirmed that clustered lesions involving strand breakage were being formed in the cells. The fragments increased linearly with dose and were produced more efficiently by high LET radiation, as predicted by track structure simulations [4]. Studies measuring DSBs post-irradiation in mammalian cells [24] and bacteria [25] demonstrated that an increase in DSBs was detected if cells were allowed time to repair DNA damage. These really were the first experiments to hint at the potential biological relevance of clustered lesions; they showed that repair of radiation damage could actually increase the level of what was considered the most lethal lesion, the DSB. We now recognize that this increase in breakage is likely the conversion of bistranded clusters to DSBs.
The breakthrough in detection, measurement and to some extent characterization of bistranded clustered lesions was the development of the technique combining quantitative gel electrophoresis with the treatment of irradiated DNA with base excision DNA repair enzymes (for review see [26]). Oxidative lesions such as base damage and AP sites are repaired predominantly by base excision repair (BER; [27]). The DNA N-glycosylases involved in removal of oxidative base damage and the AP endonucleases that cleave at AP sites introduce a SSB into the DNA. Therefore treatment of irradiated DNA containing bistranded clusters of base damage or AP sites with these repair enzymes increases fragmentation of the DNA, and the quantitation of the increased fragmentation is used to determine the level of clusters. This technique did provide the experimental evidence that clustered lesions contained base damage and AP sites, as well as strand breaks. Bistranded clusters of all types were shown to be generated not only in irradiated DNA [28, 29], but also in irradiated cells [30, 31]. In X-irradiated cells, less than 30% of the total clustered lesions were DSBs, while 70% were non-DSB clusters: for each DSB there was 1 oxypurine cluster, 0.9 oxypyrimidine cluster and 0.75 abasic site cluster [31]. Unexpectedly, when this technique was used to examine the effect of radiation LET on the induction of clusters in DNA, it was found that all types of non-DSB clusters decreased with increasing LET [32]. One limitation of this gel electrophoresis assay is the detection of small DNA fragments. DSBs generated from oxidative clusters situated within a few kilobases could go undetected by the assay. In fact simulations of proton and alpha particles predict short fragment production (0–23 kb) from DSBs but these could not be detected by experimental methods [33]. Other explanations for the decrease in non-DSB clusters include the presence of complex DSBs or inhibition of the repair enzymes in the assay by near-by damage. Complex DSBs “hide” the base damage, as a complex DSB is indistinguishable from a DSB without associated damage using this technique. If the enzymes in the assay are unable to cleave the oxidative damage, non-DSB clusters will not be revealed as DSBs and remain undetected. All of these reasons would underestimate the clustered lesion damage induced by high LET radiation. We therefore have to rely upon the computational simulations that indicate lesions are more complex and increase with increasing LET [22, 34, 35] until new assays are available to quantify all the oxidative damage induced by higher LET radiation.
This gel electrophoresis technique has proved capable of detecting clustered lesions in cells generated by endogenous reactive oxygen species. Bennett et al [36] measured clustered lesions at the level of a few non-DSB clusters per Giga bases in human skin and in hematopoietic cells from people. Interestingly, smokers were found to have a higher level of clusters in their cells than non-smokers [37]. It has even been possible to use this technique to detect persistent clustered lesions in mouse skin 20 weeks after irradiation of the animals [38]. More recently, malignant tumors were found to have a higher level of oxidative damage clusters compared to non-malignant tissue (50–400/Giga base pair compared to 100–1500/ Giga base pair in tumors [39]) and this may be due to a higher metabolic demand of malignant cells, or due to compromised mitochondria and a higher production of reactive oxygen species. Interestingly, tumor-bearing mice were also found to have elevated non-DSB clusters in distant tissues such as the gastrointestinal tract, and this was explained by a chronic inflammatory condition in these mice [40].
The neutral comet assay has also been useful for detecting oxidative clustered lesions. In fact it was used to detect an increase in DSB formation during repair in irradiated bacteria [41] and mammalian cells [42; 43]. The neutral comet assay has also been adapted to measure clusters consisting of opposing oxidative damage by treating the permeabilized cells with base excision repair enzymes prior to electrophoresis [44]. The combination of the repair enzymes and gel electrophoresis has substantially increased our knowledge about clustered lesions, their existence, persistence and repair.
3. Potential biological consequences of clustered lesions
As mentioned above, oxidative DNA damage is predominantly repaired by the BER pathway, whereas DSBs are repaired by non-homologous end-joining (NHEJ) throughout the cell cycle and homologous recombination (HR) in late S and G2/M. Even though the repair of the individual lesions within clusters share common late steps of BER, the proteins involved in damage recognition for oxidative base damage, AP sites and SSBs differ. Due to the complexity of damage introduced by ionizing radiation and the variety of proteins that are involved in repair initiation, it is not possible to dissect how each lesion is repaired, or how repair is inhibited at the lesions within radiation clusters in cells. This problem has been addressed by using synthetic lesions in either deoxyoligonucleotides (oligonucleotides) or plasmid DNA. A great deal of work has been performed using purified bacterial and mammalian DNA repair enzymes and mammalian cell extracts. This work has been extensively reviewed by Eccles and colleagues in this current issue and so will not be reviewed here. However, to aid in the discussion of the work performed in eukaryotic cells and to understand the potential biological consequences a general overview of the work is provided.
The majority of the work in vitro has been performed on non-DSB bistranded clusters consisting of two lesions. Prior to the hypothesis of clustered lesions, DNA repair was always thought to be beneficial to cell survival, but DNA repair of clustered damage can convert two potentially mutagenic lesions into a potentially lethal DSB. In general, initiation of repair at two opposing lesions situated ≥ 3 bps apart results in two SSB-repair intermediates in vitro generating a DSB. However, clusters containing two opposing lesions situated less than 3 bp apart were found in vitro not to form a DSB. Instead, the initiation of repair at one of the lesions was found to create a SSB-repair intermediate that inhibited either the binding of the repair enzyme to the damage in the opposing strand or inhibited the activity of the repair enzyme. In fact two opposing oxidative DNA lesions in plasmid in bacteria were found to enhance the mutagenicity of the individual lesions [45–48]. The delay of repair at one lesion due to the SSB-repair intermediate on the opposing strand provided enough time for replication to occur and introduce a mutation at the remaining oxidative damage. Work in vitro has also demonstrated that the repair at such a SSB-repair intermediate within a cluster can be inhibited at the insertion of the missing nucleotide [49] as well as the ligation step [50].
Tandem lesions or double lesions on the same strand have the potential of enhancing mutagenesis. Again the initiation of repair at one damage was found to inhibit repair at the near-by lesion [51]. It is also possible that replication through the individual lesions could be inhibited by the near-by second lesion. This could result in persistent SSBs due to collapse of the replication fork, or the use of a translesion polymerase to replicate through this damage that would likely generate mutations.
An increase in the complexity of the damage can consist of tandem or double lesions in opposition to damage in the opposing strand. Computer simulations using Monte Carlo models [34, 35] predict that high LET radiation can generate clusters consisting of 10–25 lesions over a 100–200 bp DNA region. Low LET gamma rays is also predicted to produce clusters with 20% containing three lesions, 6% containing four lesions and 2% containing five or more lesions (personal communication, Dr. R.D. Stewart, Purdue University, IN). Limited studies have been performed in vitro with clusters consisting of ≥ 3 lesions [52–54], but in general the increase in complexity tended to inhibit the formation of DSBs.
DSBs in combination with oxidative base damage generate complex DSBs. Work on these lesions has increased recently but there are few published studies. The presence of a SSB reduces repair of the near-by oxidative damage, as well as the oxidative damage reducing the repair of the SSB [55, 56], and ligation of a DSB by T4 DNA ligase was reduced by a near-by 7,8-dihydro-8oxoguanine (8-oxoG; [57]).
Studies in vitro have greatly increased our understanding of clustered lesion repair. However, it is important to remember that repair in a physiological setting inside a cell can be regulated at many levels and so work has progressed to examine how defined clustered lesions are processed in eukaryotes and prokaryotes. The remainder of this review will discuss the advances in clustered lesion repair in eukaryote cells.
4. Repair and mutagenesis at synthetic clustered DNA lesions in yeast
The synthetic clustered lesions used in biochemical studies have been inserted into non-replicative vectors (to analyze repair) or into replicative vectors (to explore repair and mutagenesis), and transfected into cells. Saccharomyces cerevisiae yeast cells are of particular interest for such investigations, because of the series of DNA repair isogenic mutants that are available. In addition, the DNA repair mechanisms are well conserved between S. cerevisiae and human cells, even though some differences exist. As stated above, the repair process of clustered lesions may convert potentially mutagenic lesions into potentially lethal DSBs. The yeast transformation assay is well suited to investigate DSB formation at clustered lesions, since it is a straight forward assay and easy to perform. This assay compares the transformation efficiencies of plasmids carrrying various clustered lesions to that of a control plasmid carrying an unmodified oligonucleotide. In other words, plasmid viabilities are compared and the plasmids do replicate in this assay. Notably, a low-copy number plasmid linearized by a DSB is not repaired, is rapidly degraded and harbors a very low ability to “survive” and be replicated. The Sage lab have inserted clusters of increasing complexity into vectors: two bistranded oxidized bases (8-oxoG and 5-hydroxyuracil separated by 3, 6 or 8 bp, three base damages (bistranded 8-oxoG and 5-hydroxyuracil plus 8-oxoadenine; 8-oxoA), or MDS of 4–5 lesions, carrying the last three base damages plus a 5-formyluracil and a 1 nt gap [58] (Figure 1A). Bistranded clustered lesions composed of two uracil residues or AP sites separated by 6 bp were also inserted into plasmid. It was observed that none of the vectors carrying oxidative base damage lost viability in comparison with the vector containing the undamaged oligonucleotide, in repair proficient yeast cells as well as in cells deficient in BER, nucleotide excision repair (NER), NER plus BER, translesion synthesis (TLS) or in HR [58]. Surprisingly, DSBs were also not produced at the most complex MDS that comprised of oxidative damaged bases and a gap. In contrast DSBs were formed, as monitored by loss of plasmid survival, when harboring opposing uracils or AP sites, in wild type cells. The formation of DSBs at clustered uracils was recently confirmed, using an elegant S. cerevisiae–based system which allowed integration of a DNA fragment carrying lesions into a selectable gene on one of two chromosome XV in a diploid strain [59]. Furthermore, the PCR-based analysis of the transformants confirmed that DSBs were generated and that they were efficiently repaired by HR. Incidently, this study showed that in S. cerevisiae, chromosomal integration of the desired DNA fragment occurred before repair of the inserted lesions, and that repair of clustered lesions can thus be investigated at the chromosomal level in a chromatin environment. Interestingly even when a non-replicative, integrative vector, carrying a complex MDS with base damage and a gap (oG/hU/oA/fU/gap in Figure 1A) was examined, there was no evidence of DSB formation [58]. It is possible that the different outcomes in these assays of clusters containing oxidative base damage or AP sites and uracils, which are readily converted to AP sites, may be explained by how the replication machinery is affected by the clusters: the majority of oxidative base damage is by-passed by replication, while an AP site can block replication. Replication fork collapse at opposing AP sites could result in a DSB. However, a recent study in wild-type yeast demonstrated that single AP sites could be by-passed up to 73% and REV1 was the major translesion polymerase [60]. This suggests that DSB formation in yeast from AP sites and uracils is likely due to initiation of repair and SSB-repair intermediates. With yeast extracts from wild type cells, it was further demonstrated that cleavage of a cluster of an opposing uracil and AP site occurred within a few minutes, whereas incision rate at an oxidized base was much lower. In addition, excision at 5-hydroxyuracil occurs first and prevents excision at 8-oxoG on the opposite strand. These observations led the authors to propose that the kinetics of the initial repair steps at clustered lesions is a major parameter that affects the conversion of clustered lesions into DSBs in cells. Similar data were obtained in bacteria (Eccles et al in this issue). It is well established now that repair of 8-oxoG is slow, while that of oxidized pyrimidines is more rapid, and that of uracil and AP sites is fast in yeast and bacteria [48, 52, 53, 61–65]. Indeed, 8-oxoG opposite dihydrothymine, thymine glycol or another 8-oxoG protects from the formation of a DSB, whereas the fast excision/incision of bistranded uracils generates DSBs as repair intermediate in bacteria and yeast [45, 47, 58, 59, 63, 65–67].
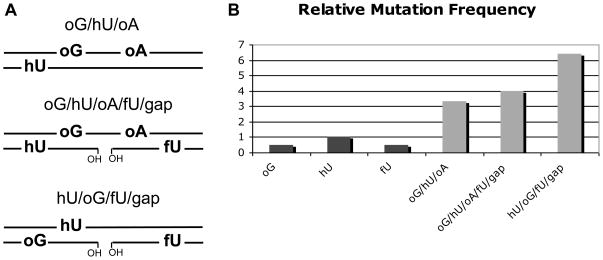
Figure 1. Relative mutation frequencies at synthetic MDS in yeast.
a) Duplexes carrying damaged sites were inserted into yeast centromeric plasmid, then used to transform yeast cells. oG, 8-oxoguanine; oA, 8-oxoA; hU, 5-hydroxyuracil; fU, 5-formyluracil. 8-oxoG and hU are separated by 3 bp, oG and oA by 7 nucleotides. The gap is at 7 nucleotide from oG or hU and fU on the same strand. b) relative mutation frequency at single oG, hU, fU and at the three MDS.
The mutation frequencies and the type of mutations at three complex MDS (Figure 1A) have also been investigated in wild type cells and compared to that of single lesions, all in the same sequence context (Kozmin & Sage, unpublished data). Due to the absence of selection and the direct sequencing of recovered plasmids, all mutagenic events can be detected. Mutations are mostly base substitutions and -1 deletions, targeted at base damage. Targeted mutations on both strands also occur. Interestingly, at such complex MDS carrying 3–4 oxidized bases with or without a gap that simulates a SSB, no large deletions are recovered, as in bacteria for bistranded oxidative clusters [46, 48, 67]. It can be anticipated that in yeast and bacterial chromosomes, point mutations at MDS carrying 8-oxoG prevail over loss of genetic material. Formation of a DSB or replication block at clusters carrying uracil, AP sites or thymine glycol at the chromosomal level (the last two lesions having the capacity to induce collapse of replication forks) may stimulate repair by HR or single strand annealing, resulting in accurate repair or deletions, whereas in mammalian cells repair by NHEJ is likely involved (see section 5).
A second important message is illustrated by work from the Sage lab in Figure 1B: the mutation frequencies at complex MDS in wild type yeast cells is drastically increased compared to single lesions. In S. cerevisiae, the mutation frequencies (MF) at single 8-oxoG, 5-hydroxyuracil and 5-formyluracil range between 4–10% and 8-oxoadenine is not mutagenic. The mutation frequencies at the studied MDS ranges between 35 and 70%. The synergistic effect of lesion clustering on mutation induction indicates that repair at clusters is strongly impaired. Indeed, the mutation induction at 8-oxoG is increased 5–6.5 times in oG/hU/oA and in oG/hU/oA/fU/gap compared to that at a single 8-oxoG. A more than 6-fold increase in mutation frequency at 5-hydroxyuracil is observed with hU/oG/fU/gap in comparison with a single lesion. In bacteria, mutation increases of similar orders were observed at 8-oxoG for bistranded clusters, depending on interlesion spacing [45–48, 67].
Mutation spectra in repair proficient and repair deficient cells provide information on repair processes. Interestingly, at oG/hU/oA/fU/gap the mutation frequency at 8-oxoG is increased 6.5 fold, whereas the mutation frequency at 5-hydroxyuracil remains at that of the single lesion in wild type cells. As observed with yeast extracts [58], the oxidized pyrimidine is thus cleaved first, preventing excision at 8-oxoG which persists and leads to mutation during replication. Mutation spectra for oG/hU/oA/fU/gap were also performed in strains inactivated for all the known DNA-N-glycosylases of BER, in ntg1 ntg2 strain inactivated for the major DNA-N-glycosylases in charge of removing oxidized pyrimidines, and in the triple mutant rad14 ntg1 ntg2 also deficient in NER. Only the mutations at 5-hydroxyuracil increase significantly (by a factor 2) in these cells. This is in accord with the above suggestion that in this particular context 5-hydroxyuracil is repaired in wild type cells, whereas 8-oxoG is not. In contrast, inverting 8-oxoG and 5-hydroxyuracil positions with regards to the gap (hU/oG/fU/gap, see Figure 1A) leads to an absence of mutation induction at 8-oxoG whereas mutations at 5-hydroxyuracil drastically increase. In this new context, 8-oxoG may be well repaired [53], and 5-hydroxyuracil may not, even though it is excised by yeast extract [58]. This demonstrates that the distribution of lesions within a cluster is of major importance for the repair and mutagenic processes. Noteworthy, the mutation spectra at oG/hU/oA/fU/gap in ntg1 ntg2 and in rad14 ntg1 ntg2 cells are superimposable. This demonstrates that NER is not involved in the repair of complex MDS such as those examined. The types of mutations at 8-oxoG are G to T and -1 deletion, while 5-hydroxyuracil is changed for a T. Moreover, the type of mutations at 8-oxoG partly differs from that at a single 8-oxoG, which may suggest that some mutations are introduced during repair by a non-replicative DNA polymerase.
Collectively, these data show that repair of lesions within bistranded clusters or more complex MDS carrying oxidative base damage and SSB is compromised in yeast. This drastically increases point mutations at the modified bases and multiple mutations occur, but large deletions do not, at least in yeast using a plasmid assay. On the other hand, clusters comprising of opposing uracils or AP sites separated by more than 3 bp generate DSBs, which either leads to cell lethality if left unrepaired or to large deletions if misrepaired.
5. Repair of clustered lesions in mammalian cells
5.1 Tandem lesions
Work in vitro [51] and E.coli [68, 69] has demonstrated that tandem lesions are difficult to repair and can be either a block to replication or result in mutagenesis. The result depends upon the particular type of damage in the tandem lesion. An example is thymine glycol 3’ to an oxidized AP site or a tetrahydrofuran (furan), which is a stable AP site analog. This tandem lesion in vitro inhibited endonuclease IV cleavage at the AP site and endonuclease III cleavage at the thymine glycol, although the human AP endonuclease (Ape1) was able to cleave at the AP site and initiate long patch BER [70]. This lesion was also found to be a substrate for UvrABC, the bacterial nucleotide excision repair enzyme. These results therefore have interesting implications for the possible pathways involved in repair of tandem lesions in mammalian cells. Recently, it was shown that ~ 50% of the 8-oxo-purine damage (8-oxoG and 8-oxoA) produced by γ-irradiation of DNA forms tandem lesions and >40% cannot be removed by DNA N-glycosylases [17]. Therefore tandem damage is highly relevant to mammalian cells but very little work has been performed in mammalian cells. One type of lesion consisting of an 8-oxoG in tandem with a formylamine lesion has been studied in vitro and in mammalian cells. The 8-oxoG in this lesion can be removed in vitro by formamidopyrimidine DNA N-glycosylase (Fpg), which also excised a small percentage of the formylamine residues in the tandem lesions [71]. To examine by-pass and mutagenesis in mammalian cells, the 8-oxoG/formylamine lesion was positioned in a single stranded vector and introduced into simian COS7 cells [72]. Even though 70% of single 8oxoGs could be by-passed during replication, only 45% of single formylamine residues were replicated through with an adenine inserted opposite the formylamine residue. Formylamine damage originates from a pyrimidine and so this lesion could be mutagenic if formed from a cytosine. By-pass of the tandem lesion decreased to 17%, with adenine inserted opposite the formylamine and cytosine opposite the 8-oxoG. The low level of by-pass indicates this tandem lesion, which has been shown to be generated by ionizing radiation [14], could be cytotoxic to mammalian cells. This study demonstrates the importance of tandem lesions in survival and mutagenesis, and much more work examining the biological consequences of tandem lesions in mammalian cells is required.
5.2 Bistranded clusters
Most of the work on clustered lesions in mammalian cells has focused on bistranded clusters due to the potential for conversion of these clusters to DSBs or complex DSBs. The use of gel electrophoresis in combination with treatment of DNA with BER enzymes has been useful in examining repair as well as induction of bistranded non-DSB clusters. In fact, ~10% of non-DSB clusters introduced by γ-radiation were found to be converted to DSBs within 30 minutes in NHEJ-defective Chinese hamster cells [73]. Using this technique it was also found that although some abasic site clusters are processed post-irradiation, there is the potential in a replicating cell for bistranded clusters to be “split”, which would result in lesions only on one strand [74].
Mammalian cells over-expressing repair enzymes have also been used to demonstrate that non-DSB clusters can be converted to lethal lesions: expression of E.coli endonuclease III in NHEJ-deficient cells sensitized the cells to bleomycin sulfate [75], and the over-expression of either hNTH1 (a human pyrimidine DNA N-glycosylase) or hOGG1 (a human purine DNA N-glycosylase) in TK6 cells was found to radiosensitize the cells to killing and mutagenesis, and increase DSB formation [42]. An increase in the Hprt mutation frequency post-γ-irradiation was also observed when hOGG1 was over-expressed in CHO cells [53]. In fact use of siRNA to reduce hOGG1 in the TK6 cells resulted in increased survival and fewer DSBs post-γ-irradiation [43]. The expression of E.coli Fpg in Chinese hamster ovary (CHO) cells was also found to decrease endogenous oxypurine clusters, as well as the accumulation of Hprt mutations [76]. The cells repair capacity has a significant impact on the biological consequences of clustered lesions. The limitation of these cell studies is that the specific types of lesions being “repaired” are unknown since the DNA N-glycosylases have a broad spectrum of substrate recognition and they can also cleave at AP sites. It is highly likely that E.coli endonuclease III expressed in mammalian cells cleaved at AP sites in clustered lesions generated by bleomycin sulfate [75]. However, these studies do support the idea that manipulating the repair capacity of a tumor could be a potential complementary treatment to radiotherapy or chemotherapy.
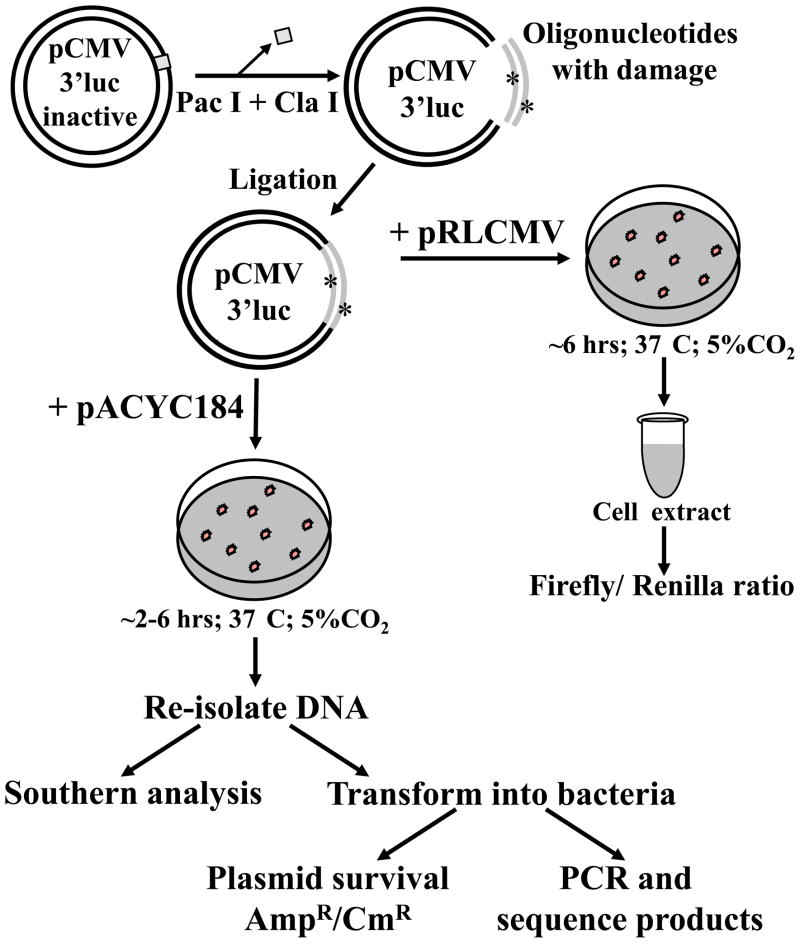
To understand the potential biological consequences of clustered damage in cells, it is necessary to determine what happens to clustered lesions in cells with physiological levels of repair enzymes. To examine this, the Harrison lab have designed an assay where the clustered lesion is situated in the firefly luciferase reporter coding sequence in a vector that cannot replicate in mammalian cells (Figure 2). Following transfection, time is allowed to repair the DNA and firefly luciferase activity is measured. Loss of firefly luciferase activity for a clustered lesion compared to undamaged DNA indicates the formation of a DSB. Re-isolated plasmid DNA can be examined by Southern analysis or analyzed for deletions by PCR, and repair junctions can be sequenced. Using Southern analysis it was determined that the luciferase activity correlated with the amount of re-isolated plasmid at 2 hrs post-transfection [77]. Single lesions are examined to ensure activity is not lost due to transcriptional mutagenesis [78]. Bistranded two lesion clusters examined to date in mammalian cells using this assay include two closely opposed uracils [79], two closely opposed 8-oxoG (unpublished data), two closely opposed furans [80] and a furan opposite an 8-oxoG [77]. In mammalian cells, the only clustered lesions found to convert to a DSB contained two opposing furans, and using siRNA it was demonstrated that the major class II AP endonuclease (Apex1 in mouse cells) cleaves at these lesions [77, 80]. Similar to the in vitro studies, cleavage increased as the distance between the two furans increased from 2 to 5 bp apart, suggesting that Apex1 was inhibited by a SSB-repair intermediate situated < 3 bps from the second furan. As the lesions were increased to 12 bp apart DSB formation decreased. This was likely due to the hydrogen bonding of the 12 bp overhang holding the structure together, allowing the lesions to be repaired as single lesions rather than two interacting lesions in a cluster; a similar situation was found in bacteria [64].
Figure 2. Assay to examine DSB formation of synthetic clustered lesions in mammalian cells.
pCMV3’luc inactive is ampicillin (AmpR) and does not express active firefly luciferase. Linearization with Pac I and Cla I removes the inactivating sequence from the luciferase open reading frame and the vector will express active firefly luciferase upon insertion of a 45 bp double stranded oligonucleotide. The initial DNA carrying the damage is generated by ligation of linearized pCMV3’luc inactive with a double-stranded oligonucleotide containing no damage, a single lesion or a clustered lesion. The ligation products are used in the assay, hence circular DNA as well as background linear DNA is introduced into the cells. A control ligation without the oligonucleotide is always used and results in negligible firefly luciferase activity in cells. To examine DSB formation indirectly using the reporter activity, the ligation is co-transfected using a Nucleofector® with pRLCMV, which expresses renilla luciferase. After 6 hrs, cell extracts are prepared and the firefly/ renilla luciferase activity ratio is determined. A decrease in this ratio compared to undamaged sequence indicates DSB formation of the clustered damage. To examine the products of the repair, the initial ligation products are co-transfected with pACYC184, which encodes chloramphenicol resistance (CmR). After transfection (2–6 hrs) DNA is re-isolated and used for Southern analysis, or transformed into bacteria to determine whether the clustered lesion decreased plasmid survival. The plasmid DNA from the colonies can be analyzed by PCR for the presence of deletions or insertions, and can be sequenced to examine the junctions of repair. For full experimental details see Malyarchuk et al [77, 80].
The in vitro studies of bistranded clusters predict DSB formation from two opposing lesions situated ≥ 3 bp apart. In mammalian cells, clusters of base damage or an 8-oxoG opposite a furan did not convert to a DSB according to the firefly luciferase activity assay, and the lack of DSB formation from a cluster with two uracils, or an 8-oxoG opposite a furan was confirmed by Southern analysis [77,79]. In bacteria, clusters of 8-oxoG opposite a uracil or clusters of two 8-oxoG also do not form a DSB. The lack of breakage of these lesions in bacteria, yeast and mammalian cells could be due to the levels of enzyme expressed in the cell as mentioned above (section 4) or to the inhibition of the DNA N-glycosylase by an opposing SSB-repair intermediate. Studies over-expressing hNth1 and Ogg1 do support the idea that the amount of enzyme available in the cell may be key to converting base damage/AP site clusters to DSBs [42]. Using nuclear extracts from mouse fibroblasts, cleavage of a furan was readily detected using 200 ng of nuclear extract, while the same amount of extract was unable to cleave at an 8-oxoG in a 1 hour time period [77]. To examine 8-oxoG cleavage in vitro other groups have used 2.5–10 μg of nuclear extracts from Chinese Hamster cells [81]. In fact, the repair efficiency of 8-oxoG was previously found to be ~10 times less than that of natural AP sites and only partly dependent on the cellular level of initiating enzymes [61,62]. This therefore supports the idea that a furan in a cluster in mammalian cells would be more readily converted to a SSB than an 8-oxoG. Inhibition of Ogg1 by the SSB-repair intermediate at the furan could allow complete repair of the furan prior to cleavage of the 8-oxoG and would explain the lack of DSB formation.
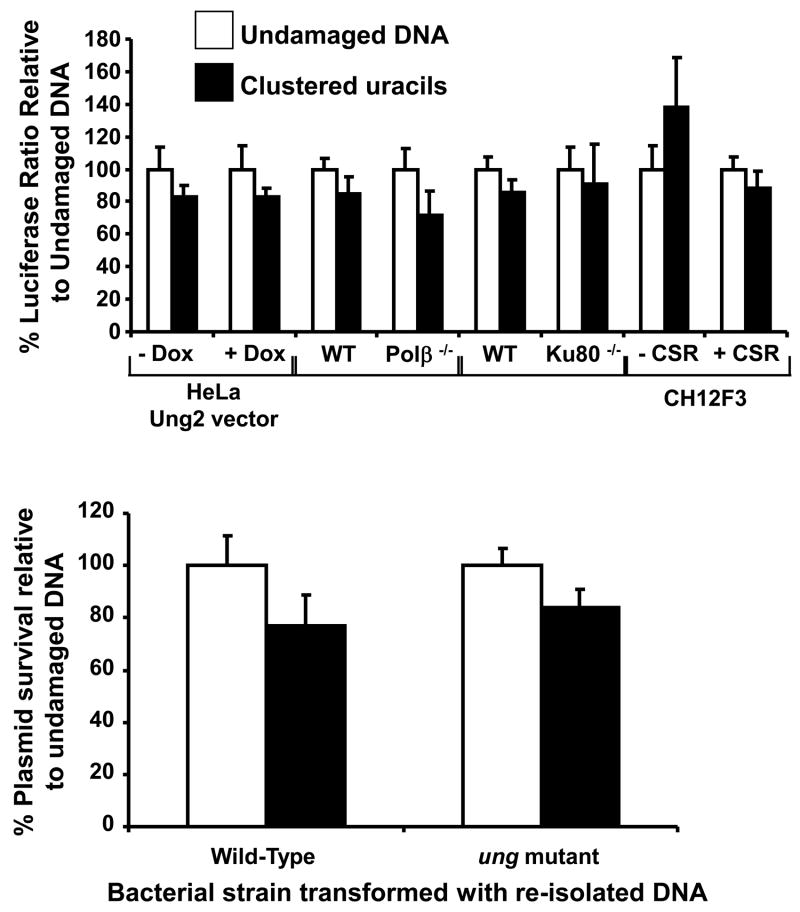
Uracil is not an oxidative DNA damage but is useful as a model substrate for short patch BER. Studies in bacteria [63, 66] and yeast (see section 4) have demonstrated that two closely opposed uracils form DSBs in cells, and that this was dependent on uracil DNA glycosylase initiating repair. As mentioned above (section 4) clusters of uracils or AP sites could be converted to DSBs due to replication fork collapse. However, in the bacterial studies by D’souza et al [63], DNA replication was prevented during the assay by the addition of novobiocin to inhibit DNA gyrase and breakage did still occur. This data and the yeast studies of AP site replication by-pass [60] suggest that DSB formation is not dependent on replication. However, in mammalian cells in the non-replicating system, it was determined that two closely opposed uracils do not result in a decrease in luciferase activity and do not result in loss of DNA [79]. To try to understand this difference in mammalian cells, the Harrison lab performed the luciferase assay (Figure 2) with different types of clusters in a number of different types of mammalian cells. Two uracils situated 5 bps apart and 5’ to each other with the uracil base paired to an adenine or a guanine have been examined as well as two uracils 3 bps apart and 3’ to each other. These different clusters gave similar results and showed no evidence of DSB formation in cells. The cell types used include mouse fibroblasts deficient in Ku80 to determine whether NHEJ was promoting accurate repair of the DSB, mouse fibroblasts deficient in DNA polymerase β to see if repair was altered by using the δ or ε DNA polymerases, and HeLa cells that can be induced to over-express uracil DNA glycosylase to make sure the first step of repair was not limiting. As can be seen from Figure 3A, no significant decrease in luciferase activity was detected in any of these cell lines. To make sure the uracil was being removed in the mammalian cells, the re-isolated DNA was transformed into wild-type or Ung-deficient E.coli. If the uracil cluster was still present in the plasmid DNA, Ung and BER would convert the opposing uracils to a DSB and plasmid survival would substantially decrease only in the wild-type bacteria [63]. No difference in plasmid survival was seen between wild-type and Ung-deficient bacteria (Figure 3B). This suggests the uracil was either completely removed or one of the uracils in the cluster was removed. It is possible that repair of transient breaks introduced at each lesion was fast enough to prevent a DSB forming in mammalian cells, or that one lesion was completely repaired before initiation of repair at the opposing lesion. Ku70/80 has also been implicated in the prevention of DSB formation from an opposing base damage and SSB in vitro, not by involvement of NHEJ but by inhibiting the removal of the base damage by binding the SSB [82]. If one uracil was converted to a SSB then DSB formation could have been prevented by Ku. The one cell type that is known to convert opposing uracils to a DSB to initiate class switching of antibodies is the B cell [83]. The mechanism involves deamination of cytosine to uracil by activation-induced cytosine deaminase. Multiple uracils are inserted at hot spots in the Ig-heavy chain locus. Removal of the uracils by uracil DNA glycosylase and conversion of the AP sites to SSBs is required for class switch recombination. Generation of SSBs could occur by replication fork collapse at the AP sites, Mre-11-NBS-Rad50 cleaving at AP sites in single-stranded DNA during replication, or Ape1 and 2 cleaving at the AP sites [83, 84]. A murine B cell lymphoma line (CH12F3) was therefore treated with TGF-β1, anti-CD40 antibodies and IL-4 to induce class-switching [85, 86]. After 48 hrs, ~42% of cells expressed IgA antibodies. Cells were transfected with the vector containing two opposing uracils and firefly luciferase activity did not substantially decrease indicating no DSB formation (Figure 3A). The Harrison lab has therefore been unable to detect DSB formation from two opposing uracils in mammalian cells. It is possible that DNA replication is required in the class switching B cells to generate a DSB from the opposing uracils, or that multiple uracils are needed to overwhelm BER. It has been postulated that the steps in BER are tightly regulated to prevent repair intermediates causing deleterious effects to the cell. This “passing the baton” idea [87] involves the interaction of the enzymes in this multi-stage pathway, e.g. the second enzyme in the pathway would interact with the first enzyme to prevent release of repair intermediates. Evidence for this includes Ape1 displacing thymine DNA glycosylase from AP sites [88], the stimulation of uracil DNA glycosylase [89] and Ogg1 by Ape1 [90], the interaction of Ape1 and DNA polymerase β [91], and the interaction of DNA polymerase β with XRCC1 [92,93] and DNA ligase I [94]. Mammalian cell studies examining clusters of uracil or oxidative base damage support this idea. So why then do we detect DSB formation at opposing furans if the AP endonuclease is linked to DNA polymerase β? A furan cannot be repaired by short patch repair and it is possible that after AP endonuclease cleavage, the switch to long patch repair to remove the blocking lesion may be key to forming the DSB from two furans. Oxidized abasic sites are generated by ionizing radiation and these lesions also require long patch repair. It is possible that these lesions form non-DSB clusters that are converted to DSBs post-irradiation. The repair efficiency of uracils by short and long patch BER has been found to be ~10 times lower than a natural AP site in mammalian cells [61, 62]. Thus obviating the need for the first initiating step of a DNA N-glycosylase may allow the class II AP endonuclease in the mammalian cell to rapidly cleave opposing AP sites. If the DNA is damaged extensively following irradiation, short patch repair may be delayed by limiting amounts of DNA polymerase β, resulting in an attempt at repair by long patch repair or DSB formation due to the link between AP endonuclease and DNA polymerase β being compromised. Interruption of “passing the baton” may also explain why under or over-expressing DNA glycosylases alters TK6 cell survival and DSB production following ionizing radiation [43]. Following irradiation, cellular DNA contains a high amount of different oxidative damages, which once repair initiates funnels into short and possibly long-patch BER. Under normal glycosylase expression levels it is possible that the link between enzymes in the BER pathway is interrupted by the availability of DNA polymerases to complete repair of all the SSB-repair intermediates. Decreased expression of the glycosylases would therefore result in decreased DSB production, while increased expression would further upset the balance of BER, increasing DSB production. The assays performed in mammalian cells using synthetic damage (Figure 2) may need to test multiple base damages each separated > 3 bp apart in opposing strands to detect DSB formation. These are points that warrant further study to understand the mechanism of cell killing by clustered lesions.
Figure 3. Two opposing uracil damages are not converted to a DSB in mammalian cells.
The assay was performed as shown in Figure 2 to assess the DSB formation from two uracils positioned 5 bps apart on opposing strands. The uracil was base-paired with an adenine. The firefly/renilla ratio was measured in (A) for Hela cells induced to over-express Ung2 using doxycyline, for a comparison between mouse cells wild-type (WT) and knocked-out for DNA polymerase β(polβ−/−) or Ku80 (Ku80−/−), and for CH3F12 unstimulated or stimulated to perform class switch recombination (CSR) with IL-4, TGF-β and anti-CD40 for 48 hours. Activity is shown for the undamaged sequence as well as the clustered uracils, and the average percentage of activity compared to the undamaged sequence, and the standard deviation was calculated as described in Malyarchuk et al [77, 80]. The over-expression of Ung2, the loss of DNA polymerase β or Ku80, and the stimulation of the B lymphoma cell line to perform class switch recombination did not significantly alter the firefly luciferase activity and hence DSB formation from the clustered uracils. To make sure that the uracil cluster was removed from the DNA while in the mammalian cells, the assay was also performed as in Figure 2 and the plasmid DNA re-isolated from wild-type mouse fibroblasts (B). Three separate DNA samples were transformed in duplicate into either wild-type E.coli or ung mutants. The average and standard error are shown. No significant difference was found in plasmid survival between the wild-type and ung mutant bacteria.
In summary, four factors may determine whether the clustered damage is converted to a DSB: the expression level of the initiating enzyme, the efficiency of repair of the lesion, the amount of damage induced in the DNA and whether short or long patch BER is involved in repair. The difference between the yeast and mammalian cells for clustered uracil lesions may well be due to the efficiency of uracil removal, since in yeast, removal of a uracil is as efficient as removal of an AP site [58].
Studies examining the mutagenic potential of synthetic bistranded oxidative DNA damage clusters in mammalian cells have not yet been published. This is because it is difficult to generate purified double-stranded vectors carrying the opposing damage. Use of ligations could produce confusing results due to the background of deletions that will be found from NHEJ of the input linear DNA. It will not be possible to distinguish the background deletions from the actual deletions generated by breakage and replication unless the deletions occur only in the oligonucleotide sequence ligated into the vector.
5.3 Complex DSBs
Complex DSBs are predicted to increase as the LET of the radiation increases, and even following low LET radiation, modeling studies predict that 30–40% of DSB are complex DSBs [22]. These complex DSBs could also be generated from non-DSB clusters consisting of base damage and AP sites. It has been demonstrated that bistranded clusters consisting of three furans, two furans with an 8-oxoG, and three furans with an 8-oxoG can be converted to DSBs by Apex1 in mouse fibroblasts, albeit at a reduced level compared to a cluster of two furans [77]. Work in vitro with nuclear extracts indicates that these complex non-DSB clusters would generate DSBs with near-by oxidative damage [77] even though the oxidative damage did reduce the activity of pure human Ape1 and Apex1 in the mouse extracts. This work suggests that complex DSBs can be generated by abortive BER of non-DSB clusters and previous studies examining repair of clusters with two opposing furans in Ku80-deficient cells implicated NHEJ and alternative NHEJ in the repair of DSBs generated from non-DSB clusters [80]. An important question to be answered is how the DSB repair pathways handle complex DSBs. In vitro studies examining BER of oxidative damage at or close to break termini have demonstrated that the presence of the DSB does compromise removal of the oxidative damage, and a recent study implicated DNA PKcs and NHEJ in the repair of the complex DSB prior to removal of the clustered oxidative base damage situated near the break [95]. The effect of oxidative base damage on NHEJ is being examined by a number of groups. In vitro studies using oligonucleotides have shown that the presence of an 8-oxoG within 3 bases of a 3’ terminus, or 6 bases of a 5’ terminus can delay rejoining by T4 DNA ligase and DNA ligase IV/XRCC4. An AP site also decreased the efficiency of ligation by T4 DNA ligase [57]. One published study [96] using mammalian cells has examined survival of a linear plasmid with a DSB containing an AP site in the 5’ overhang. The presence of the AP site severely compromised repair of the plasmid. From the plasmid that did survive, only 16% of repair products were accurate: small (42%) and large deletions (11%) as well as insertions (29%) were found. The products suggested that DNA polymerases skipped the lesion to complete repair, there was also evidence of translesion synthesis of the AP site and deletions frequently occurred at regions of microhomology (1–4 bases). Repair of the DSBs required XRCC4 and complex DSB repair also involved Artemis. This study clearly demonstrates that NHEJ can be compromised by oxidative damage near the DSB, and mutagenic repair occurs to complete repair of these complex DSBs.
6. Biological significance of clustered DNA lesions for high vs low LET radiotherapy
The extensive studies using synthetic clustered DNA lesions have largely contributed to our understanding of the biological consequences of clustered lesions in cells or tissues. Clustered damage sites are of large diversity, they are processed differently depending on the nature of the base modification, the interlesion spacing, the presence or not of strand breaks. The expected biological consequences range from point mutations and loss of genetic material to cellular lethality due to repair impairment and lesion or repair-intermediate persistency (Figure 4). In fact, the deleterious effect of repair intermediates may be amplified at replication and cause cell killing. Tandem lesions have been isolated and have been shown to be poorly repaired or by-passed by DNA polymerases, and may be lethal lesions. Bistranded AP sites are more likely to result in DSBs, the outcome of which will depend on the presence of damage bases around the DSB, and subsequent deletions have been observed in human cells. The repair of bistranded oxidized bases is mostly impaired, causing a point mutation at the unrepaired damaged base. DSBs are unlikely to be formed by BER at these clusters, but may occur at replication forks if the unrepaired damage is a block to replication as is the case for thymidine glycol or an AP site. Homologous recombination or single strand annealing will overcome the collapse of the replication fork and may produce deletions. Importantly, the cell will survive with mutations or loss of genetic material. The more complex the non-DSB cluster (i.e. several oxidized base damages or SSB/gap), the more complex the subsequent mutations. The presence of two or more base substitutions or +/− 1 insertions/deletions can be considered the signature of complex non-DSB clusters. Indeed, this is clear from yeast data and multiple mutagenic events have also been observed in human cells (Sage, Kozmin, Sedletska unpublished data). The repair of complex DSBs, either formed by the radiation track or originating from “repair” of non-DSB clusters, is likely compromised and very slow, potentially forming large deletions. Lethal events may be expected from such lesions, even though this has not yet been demonstrated.
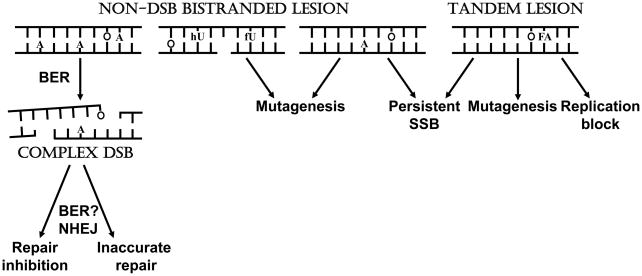
Figure 4. Biological Consequences of Clustered DNA Damage in Eukaryotes.
Clustered damage in the form of non-DSB bistranded lesions, tandem lesions or complex DSBs can consist of AP sites (A), SSBs or base damage (O is 8-oxoG, fU is 5-formyluracil, hU is 5-hydroxyuracil, and FA is formylamine). Two opposing AP sites can be converted to a DSB and hence complex DSBs can be formed either directly by the DNA damaging agent or by abortive BER. Complex DSBs can decrease the efficiency of NHEJ and be inaccurately repaired. Lesions of high complexity can result in mutagenesis: DSBs are not formed due to inhibition of repair enzymes by near-by damage. Tandem lesions can block DNA replication, but can also be mutagenic as found for bistranded lesions consisting of base damage. Partial processing of these latter two types of lesions could also result in persistent SSBs. Clustered lesions can therefore be mutagenic, result in DSBs and inaccurate repair, or block replication, and could be cytotoxic to the eukaryote cell.
What did we learn from studies on the effects of low vs high LET radiation on human cells or animal tissues and how much does it corroborate with the above observations? Early observations showed that densely ionizing radiation was more cytotoxic than sparsely ionizing radiation and it was anticipated that a greater proportion of non-repairable strand breaks originating from clustered lesions was responsible for the increased biological effectiveness of densely ionizing radiation. The distinct effects of the two types of radiation have been extensively reviewed in Blakely and Kronenberg [97]. Larger proportions of DSBs remain unrejoined after exposure to high LET radiation than after exposure to sparsely ionizing radiation, and chromosomal damage is more severe and complex. The frequency of chromosome breaks and of complex rearrangements increases up to a LET of 100–150 keV/μm and seems to plateau at higher LETs. In most cellular systems examined but not all, high LET radiation is observed to generate larger deletions (over Mbp size). A high frequency of complex deletion events and complex rearrangements at deletion junctions have been observed with high LET radiation and have not been reported for low LET radiation [98–100]. An elevated mutation burden was maintained in kidney epithelium of mice exposed to high energy iron ion [101]. Notably, an unusually high proportion of radon-induced mutants exhibited two or more base subtitutions and insertions/deletions within 3–14 bp at the HPRT locus in T lymphocytes and this has been suggested as a signature of exposure to densely ionizing radiation [102]. The carcinogenic potential of high LET radiation has also been proven. In addition, exposures to densely ionizing radiation have been shown to lead to a persistent, transmissible genomic instability in a variety of biological systems. With regard to DNA repair, it has recently been shown that DNA-PKcs participates in the repair of some frank DSBs and some non-DSB clustered damages that are converted into DSB by replication in tumor cells exposed to high LET radiation [103]. In addition, intact homologous recombination is also required to ensure DNA repair and cell survival after exposure to high-energy iron ions [104]. It appears that the biological features specific to high LET radiation do relate well to the known processing of the clustered lesions examined in this review. We still need to reconstitute the path leading to complex large deletions.
Radiation exposure has been associated with risk of diseases and in particular cancer. On the other hand, radiotherapy for cancer treatment is used advantageously for about 70% of cancer patients. The clinical applications of high LET radiation have a long history. High LET radiotherapy is increasing worldwide, with respect to proton therapy and hadron therapy, even though a complete understanding of the mechanisms underlying the biological action has not yet been determined. An important feature of high LET radiation which can be anticipated from studies on clustered DNA lesions is the increase of point mutations and clusters of mutations at non-DSB clusters. If a tumor suppressor gene is thus inactivated in normal tissue located near the irradiated tumor and hit by the beam, it may be a step towards the development of a secondary, radio-induced cancer. This situation would particularly apply to proliferating tissues. Even though the energy deposition of high LET radiation in normal tissue situated near the tumor is not elevated, the effect is not yet known. In tumor cells, such mutations may contribute to increase genomic instability and eventually lead to cell death. Low LET radiation also induces point mutations from dispersed base damages, but the probability of formation and the complexity of clustered lesions are lower. The frequency of mutation in normal tissue is expected to be higher than with high LET radiation. Delayed repair of clustered DNA damage possibly induced by high LET radiation could cause mutations but may also generate DSBs from stalled replication forks [105], as in rapidly replicating tumor cells. Such complex DSBs may undergo mutagenic repair via homologous recombination and may reflect the large (over Mbp) and complex deletions observed in culture cells exposed to high LET radiation. Such genome loss will contribute to genome instability of the tumor cells, and a probable final outcome is cell death. High LET-induced non-DSB clusters containing opposing AP sites would be particularly toxic via the formation of DSBs which will be poorly repaired, or due to the presence of unrepaired AP sites. A dead cell is a good cell not only for tumor eradication, but also for normal tissue since it prevents replication of damaged cells with radiation–induced mutations. With respect to the biological effectiveness of clustered DNA damage, high LET radiotherapy seems to present a relatively better benefit over risk to the patient than low LET radiotherapy. Indeed, in combination with a low dose deposited in the entrance channel, fewer as well as more easily repairable damages are produced in normal tissue, whereas a large dose is deposited in the tumor, accompanied by complex and poorly repairable lesion production. In addition, our understanding of repair processes at clustered lesions lets us predict that inhibiting the late steps of BER should increase toxic DSBs and repair intermediates and consequently would be beneficial for tumor cell killing and a combination of inhibitors for the late steps of BER, HR and/or NHEJ would lead to additional cell killing. Such inhibitors may be used in conjunction with hadron therapy to maximize the differences in biological efficiency between normal and tumor tissues . Since bistranded clustered lesions are generated by low LET radiation, manipultaion of the cells repair capacity may also be beneficial to most standard radiotherapy regimes. This was demonstrated by the cell studies over-expressing BER enzymes. In summary, the research on clustered lesions has progressed substantially since they were first proposed to exist in the 1990's. This increase in knowledge has uncovered more about how tumor cells die by radiotherapy and chemotherapy, and should lead in the future to mechanisms to improve cancer treatment.
Acknowledgments
Work in Figure 1 was performed by the Sage laboratory, while work in Figure 3 was performed by Svitlana Malyarchuk and Reneau Castore in the Harrison lab. Work in the Harrison lab is supported by NCI grant numbers CA085693 and CA085693-09S1. Work in the Sage lab is supported by Électricité de France, CNRS, Institut Curie and Institut National du Cancer. The mouse WT and Ku80−/− cells were obtained from Dr. D. Chen, Ph.D. (UT Southwestern, TX, USA), the mouse WT and polβ−/− cells were obtained from Dr. R. Sobol (University of Pittsburgh, PA, USA), the HeLa dox-inducible Ung2 over-expression cell line was obtained from Dr. H.E. Krokan (Norwegian University of Science and Technology, Norway), and CH12F3 was obtained from Dr. K. Yu (Michiogan State University) with permission from Dr. T. Honjo (Kyoto University, Japan).
Abbreviations
- DSB
double strand break
- MDS
multiply damaged sites
- SSB
single strand break
- LET
linear energy transfer
- BER
base excision repair
- NER
nucleotide excision repair
- HR
homologous recombination
- NHEJ
non-homologous end joining
- PRR
post-replication repair
- TLS
translesion DNA synthesis
- 8-oxoG
7,8-dihydro-8-oxoguanine
- oA
7,8-dihydro-8-oxoadenine
- hU
5-hydroxyuracil
- fU
5-formyluracil
- AP sites
abasic sites
- oligonucleotides
deoxyoligonucleotides
- Fpg
formamidopyrimidine DNA N-glycosylase
- Ung
uracil DNA glycosylase
- furan
tetrahydrofuran
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Teoule R. Radiation induced DNA damage and its repair. Int J Radiat Biol. 1987;51:573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 2.Ward JF, Evans JW, Limoli CL, Calabro-Jones PM. Radiation and hydrogen peroxide induced free radical damage to DNA. Cancer. 1987;55:105–112. [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation and repairability. Progress in Nucleic Acids & Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 4.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;61:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 5.Brenner DJ, Ward JF. Constraints on energy deposition and target size of multiply damaged sites associated with DNA double-strand breaks. Int J Radiat Biol. 1992;61:737–748. doi: 10.1080/09553009214551591. [DOI] [PubMed] [Google Scholar]
- 6.Muller WEG, Zahn RK. Bleomycin, an antibiotic that removes thymine from double-stranded DNA. In: Cohn WE, editor. Prog Nucleic Acid Res And Mol Biol. Academic Press; NY: 1977. pp. 21–57. [DOI] [PubMed] [Google Scholar]
- 7.Sausville EA, Horwitz SB. A mechanism for the degradation of DNA by bleomycin. In: Gordon M, Cooke ST, editors. Bristol-Meyers Cancer Symposia. NY: Academic Press; 1979. pp. 181–205. [Google Scholar]
- 8.von Sonntag CV. The chemical basis of radiation biology. Taylor & Francis Ltd; UK: 1987. [Google Scholar]
- 9.Povirk LF, Houlgrave CW. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988;27:3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- 10.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 11.Olive PL, Johnston PJ. DNA damage from oxidants: Influence of lesion complexity and chromatin organization. Oncol Res. 1997;9:287–294. [PubMed] [Google Scholar]
- 12.Ward JF. Radiation mutagenesis: the initial DNA lesions responsible. Radiat Res. 1995;142:362–368. [PubMed] [Google Scholar]
- 13.Hong IS, Carter KN, Sato K, Greenburg MM. Characterization and mechanism of formation of tandem lesions in DNA by a nucleobase peroxyl radical. J Am Chem Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- 14.Box HC, Patrzyc HB, Dawidzik JB, Wallace JC, Freund HG, Iijima H, Budzinski EE. Double base lesions in DNA X-irradiated in the presence or absence of oxygen. Radiat Res. 2000;153:442–446. doi: 10.1667/0033-7587(2000)153[0442:dblidx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Bourdat A-G, Douki T, Frelon S, Gasparutto D, Cadet J. Tandem base lesions are generated by hydroxyl radical within isolated DNA in aerated aqueous solution. J Am Chem Soc. 2000;122:4549–4556. [Google Scholar]
- 16.Douki T, Rivière J, Cadet J. DNA tandem lesions containing 8-oxo-7,8-dihydroguanine and formamido residues arise from intramolecular addition of thymine peroxyl radical to guanine. Chem Res Toxicol. 2002;15:445–454. doi: 10.1021/tx0155909. [DOI] [PubMed] [Google Scholar]
- 17.Bergeron F, Auvré F, Radicella JP, Ravanat JL. HO• radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc Natl Acad Sci USA. 2010;107:5528–5533. doi: 10.1073/pnas.1000193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawidzik JB, Patrzyc HB, Iijima H, Budzinski EE, Higbee AJ, Cheng HC, Box HC. DNA damage measured by liquid chromatography-mass spectrometry in mouse fibroblast cells exposed to oxidative stress. Biochim Biophys Acta. 2003;1621:211–217. doi: 10.1016/s0304-4165(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 19.Box HC, Budzinski EE, Dawidzik J, Patrzyc HB, Freund HG. A novel double lesion in X-irradiated DNA consists of a strand break and a base modification. Radiat Res. 2001;156:215–219. doi: 10.1667/0033-7587(2001)156[0215:andlix]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic Biol Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 21.Regulus P, Duroux B, Bayle P, Favier A, Cadet J, Ravanat JL. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc Natl Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikjoo H, O’Neill PO, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Rydberg B. Clusters of DNA damage induced by ionizing radiation: Formation of short DNA fragments. II. Experimental Detection. Radiat Res. 1996;145:200–209. [PubMed] [Google Scholar]
- 24.Ahnstrom G, Bryant PE. DNA double-strand breaks generated by the repair of X-ray damage in Chinese hamster cells. Int J Radiat Biol. 1982;41:671–676. doi: 10.1080/09553008214550761. [DOI] [PubMed] [Google Scholar]
- 25.Bonura T, Smith KC, Kaplan HS. Enzymatic induction of DNA double-strand breaks in gamma-irradiated E.coli K-12. Proc Natl Acad Sci USA. 1975;72:4265–4269. doi: 10.1073/pnas.72.11.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland BM, Georgakilas AG, Bennett PV, Laval J, Sutherland JC. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutation Research. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Barnes DE, Lindahl T. Repair and genetic consequences of endogeneous base damage in mammalian cells. Ann Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 29.Georgakilas AG, Bennett PV, Sutherland BM. High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acid Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated cells and human cells by low doses of ionizing radiation. Proc Natl Acad Sci USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by X-rays in human cells. Radiat Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Hada M, Sutherland BM. Spectrum of complex DNA damages depends on the incident radiation. Radiat Res. 2006;165:223–230. doi: 10.1667/rr3498.1. [DOI] [PubMed] [Google Scholar]
- 33.Campa A, Ballarini F, Belli M, Cherubini R, Dini V, Esposito G, Friedland W, Gerardi S, Molinelli S, Ottolenghi A, Paretzke H, Simone G, Tabocchini MA. DNA DSB in human cells by charged particles and gamma rays: Experimental results and theoretical approaches. Int J Radiat Biol. 2005;81:841–854. doi: 10.1080/09553000500530888. [DOI] [PubMed] [Google Scholar]
- 34.Semenenko VA, Stewart RD. A fast Monte Carlo algorithm to simulate the spectrum of DNA damages formed by ionizing radiation. Radiat Res. 2004;161:451–457. doi: 10.1667/rr3140. [DOI] [PubMed] [Google Scholar]
- 35.Semenenko VA, Stewart RD. Fast Monte Carlo simulation of DNA damage formed by electrons and light ions. Phys Med Biol. 2006;51:1693–1706. doi: 10.1088/0031-9155/51/7/004. [DOI] [PubMed] [Google Scholar]
- 36.Bennett PV, Cuomo NL, Paul S, Tafrov ST, Sutherland BM. Endogenous DNA damage clusters in human skin, 3-D model, and cultured cells. Free Rad Biol Med. 2005;39:832–839. doi: 10.1016/j.freeradbiomed.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett P, Ishchenko AA, Laval J, Paap B, Sutherland BM. Endogenous DNA damage clusters in human hematopoietic stem and progenitor cells. Free Rad Biol Med. 2008;45:1352–1359. doi: 10.1016/j.freeradbiomed.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Gollapalle E, Wang R, Adetolu R, Tsao D, Francisco D, Sigounas G, Georgakilas AG. Detection of oxidative clustered DNA lesions in X-irradiated mouse skin tissues and human MCF-7 breast cancer cells. Radiat Res. 2007;167:207–216. doi: 10.1667/rr0659.1. [DOI] [PubMed] [Google Scholar]
- 39.Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutation Res./Genetic Tox. Environ Mut. 2009;674:131–136. doi: 10.1016/j.mrgentox.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redon CE, Dickey JS, Nakamura AJ, Kareva IG, Naf D, Nowsheen S, Kryston TB, Bonner WM, Gerorgakilas AG, Sedelnikova OA. Tumors induce complex DNA damage in distant proliferative tissues in vivo. Proc Natl Acad Sci USA. 2010;107:17992–17997. doi: 10.1073/pnas.1008260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA repair. 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Yang N, Chaudhry MA, Wallace SS. Base excision repair by hNTH1 and hOGG1: A two edged sword in the processing of DNA damage in γ-irradiated human cells. DNA repair. 2006;5:43–51. doi: 10.1016/j.dnarep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Holt SM, Georgakilas AG. Detection of complex DNA damage in gamma-irradiated acute lymphoblastic leukemia pre-B NALM-6 cells. Radiat Res. 2007;168:527–534. doi: 10.1667/RR0974.1. [DOI] [PubMed] [Google Scholar]
- 45.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 46.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson CG, Shikazono N, Thacker J, O’Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shikazono N, Pearson C, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budworth H, Dianov GL. Mode of inhibition of short-patch base excision repair by thymine glycol within clustered DNA lesions. J Biol Chem. 2003;278:9378–9381. doi: 10.1074/jbc.M212068200. [DOI] [PubMed] [Google Scholar]
- 50.Lomax ME, Cunniffe S, O’Neill P. 8-oxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Budworth H, Matthewman G, O'Neill P, Dianov GL. Repair of tandem base lesions in DNA by human cell extracts generates persisting single-strand breaks. J Mol Biol. 2005;351:1020–1029. doi: 10.1016/j.jmb.2005.06.069. [DOI] [PubMed] [Google Scholar]
- 52.Eot-Houillier G, Eon-Marchais S, Gasparutto D, Sage E. Processing of a complex multiply damaged site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eot-Houillier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between DNA N-glycosylases/AP lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paap G, Wilson DM, III, Sutherland BM. Human abasic endonuclease action on multilesion abasic clusters: implications for radiation-induced biological damage. Nucleic Acids Res. 2008;36:2717–2727. doi: 10.1093/nar/gkn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons JL, Zharkov DO, Dianov GL. NEIL1 excises 3' end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons JL, Dianova II, Boswell E, Weinfeld M, Dianov GL. End-damage-specific proteins facilitate recruitment or stability of X-ray cross-complementing protein 1 at the sites of DNA single-strand break repair. FEBS J. 2005;272:5753–5763. doi: 10.1111/j.1742-4658.2005.04962.x. [DOI] [PubMed] [Google Scholar]
- 57.Dobbs TA, Palmer P, Maniou Z, Lomax ME, O'Neill P. Interplay of two major repair pathways in the processing of complex double-strand DNA breaks. DNA Repair. 2008;7:1372–1383. doi: 10.1016/j.dnarep.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Kozmin S, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moscariello M, Sutherland B. Saccharomyces cerevisiae-based system for studying clustered DNA damages. Radiat Environ Biophys. 2010;49:447–456. doi: 10.1007/s00411-010-0303-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bao G, Kow YW. Effect of sequence context and direction of replication on AP site bypass in Saccharomyces cerevisiae. Mutation Research. 2009;669:147–154. doi: 10.1016/j.mrfmmm.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cappelli E, Degan P, Frosina G. Comparative repair of the endogenous lesions 8-oxo-7, 8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis. 2000;21:1135–1141. [PubMed] [Google Scholar]
- 62.Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis. 2001;22:387–393. doi: 10.1093/carcin/22.3.387. [DOI] [PubMed] [Google Scholar]
- 63.D'Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shikazono N, O'Neill P. Biological consequences of potential repair intermediates of clustered base damage site in Escherichia coli. Mutat Res. 2009;669:162–168. doi: 10.1016/j.mrfmmm.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol Gen Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 67.Bellon S, Shikazono N, Cunniffe S, Lomax M, O'Neill P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucleic Acids Res. 2009;37:4430–4440. doi: 10.1093/nar/gkp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cuniffe SMT, Lomax ME, O’Neill P. An AP site can protect against the mutagenic potential of 8-oxoG when present within a tandem clustered site in E.coli. DNA Repair. 2007;6:1839–1849. doi: 10.1016/j.dnarep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Yuan B, Jiang Y, Wang Y, Wang Y. Efficient formation of the tandem thymine glycol/ 8-oxo-7,8-dihydroguanine lesion in isolated DNA and the mutagenic and cytotoxic properties of the tandem lesions in Escherichia coli cells. Chem Res Toxicol. 2010;23:11–19. doi: 10.1021/tx9004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bourdat A-G, Gasparutto D, Cadet J. Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res. 1999;27:1015–1024. doi: 10.1093/nar/27.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gentil A, Le Page F, Cadet J, Sarasin A. Mutation spectra induced by replication of two vicinal oxidative DNA lesions in mammalian cells. Mutation Res. 2000;452:51–56. doi: 10.1016/s0027-5107(00)00034-8. [DOI] [PubMed] [Google Scholar]
- 73.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Georgakilas AG, Bennett PV, Wilson DM, III, Sutherland BM. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrison L, Skorvaga M, Cunningham RP, Hendry JH, Margison GP. Transfection of the E.coli nth gene into radiosensitive Chinese Hamster cells: Effects on sensitivity to radiation, hydrogen peroxide and bleomycin sulfate. Radiation Research. 1992;132:30–39. [PubMed] [Google Scholar]
- 76.Paul S, Gros L, Laval J, Sutherland BM. Expression of the E.coli Fpg protein in CHO cells lowers the endogenous oxypurine clustered damage levels and decreases the accumulation of endogenous Hprt mutations. Environmental and Molecular Mutagenesis. 2006;47:311–319. doi: 10.1002/em.20208. [DOI] [PubMed] [Google Scholar]
- 77.Malyarchuk S, Castore R, Harrison L. Apex1 can cleave complex clustered lesions in cells. DNA Repair. 2009;8:1343–1354. doi: 10.1016/j.dnarep.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malyarchuk S, Harrison L. DNA repair of clustered uracils in HeLa cells. J Mol Biol. 2005;345:731–743. doi: 10.1016/j.jmb.2004.10.079. [DOI] [PubMed] [Google Scholar]
- 80.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.David-Cordonnier M-H, Boiteux S, O’Neill P. Efficiency of excision of 8-oxoguanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 82.Hashimoto M, Donald CD, Yannone SM, Chen DJ, Roy R, Kow YW. A possible role of Ku in mediating sequential repair of closely opposed lesions. J Biol Chem. 2001;276:12827–12831. doi: 10.1074/jbc.M010906200. [DOI] [PubMed] [Google Scholar]
- 83.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Ann Rev Biochem. 2007;76:16.1–16.22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 84.Schrader CE, Guikema JEJ, Wu X, Stavnezer J. The roles of APE1, APE2, DNA polymerase β and mismatch repair in creating S region DNA breaks during antibody class switch. Phil Trans R Soc B. 2009;364:645–652. doi: 10.1098/rstb.2008.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakamura M, Kondo S, Sugai M, Nazarea M, Imamura S, Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. International Immunology. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 87.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 88.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 89.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of theDNAglycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bennett RA, Wilson DM, III, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc Natl Acad Sci USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 93.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III as a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 95.Peddi P, Loftin CW, Dickey JS, Hair JM, Burns KJ, Aziz K, Francisco DC, Panayiotidis MI, Sedelnikova OA, Bonner WM, Winters TA, Georgakilas AG. DNA-PKcs deficiency leads to persistence of oxidatively induced clustered DNA lesions in human tumor cells. Free Radical Biol Med. 2010;48:1435–1443. doi: 10.1016/j.freeradbiomed.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Covo S, de Villartay J-P, Jeggo PA, Livneh Z. Translesion synthesis-assisted non-homologous end-joining of complex double-strand breaks prevents loss of DNA sequences in mammalian cells. Nucleic Acids Res. 2009;37:6737–6745. doi: 10.1093/nar/gkp703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blakely EA, Kronenberg A. Heavy-ion radiobiology: new approaches to delineate mechanisms underlying enhanced biological effectiveness. Radiat Res. 1998;150(suppl):S126–S145. [PubMed] [Google Scholar]
- 98.Kagawa Y, Shimazu T, Gordon AJ, Fukunishi N, Inabe N, Suzuki M, Hirano M, Kato T, Watanabe M, Hanaoka F, Yatagai F. Complex hprt deletion events are recovered after exposure of human lymphoblastoid cells to high-LET carbon and neon ion beams. Mutagenesis. 1999;14:199–205. doi: 10.1093/mutage/14.2.199. [DOI] [PubMed] [Google Scholar]
- 99.Masumura K, Kuniya K, Kurobe T, Fukuoka M, Yatagai F, Nohmi T. Heavy-ion-induced mutations in the gpt delta transgenic mouse: comparison of mutation spectra induced by heavy-ion, X-ray, and gamma-ray radiation. Environ Mol Mutagen. 2002;40:207–215. doi: 10.1002/em.10108. [DOI] [PubMed] [Google Scholar]
- 100.Singleton BK, Griffin CS, Thacker J. Clustered DNA damage leads to complex genetic changes in irradiated human cells. Cancer Res. 2002;62:6263–6269. [PubMed] [Google Scholar]
- 101.Kronenberg A, Gauny S, Kwoh E, Connolly L, Dan C, Lasarev M, Turker MS. Comparative analysis of cell killing and autosomal mutation in mouse kidney epithelium exposed to 1 GeV/nucleon iron ions in vitro or in situ. Radiat Res. 2009;172:550–557. doi: 10.1667/RR1804.1. [DOI] [PubMed] [Google Scholar]
- 102.Albertini RJ, Clark LS, Nicklas JA, O'Neill P, Hui TE, Jostes R. Radiation quality affects the efficiency of induction and the molecular spectrum of HPRT mutations in human T cells. Radiat Res. 1997;148(Suppl):S76–S86. [PubMed] [Google Scholar]
- 103.Anderson JA, Harper JV, Cucinotta FA, O'Neill P. Participation of DNA-PKcs in DSB repair after exposure to high- and low-LET radiation. Radiat Res. 2010;174:195–205. doi: 10.1667/RR2071.1. [DOI] [PubMed] [Google Scholar]
- 104.Zafar F, Seidler SB, Kronenberg A, Schild D, Wiese C. Homologous recombination contributes to the repair of DNA double-strand breaks induced by high-energy iron ions. Radiat Res. 2010;173:27–39. doi: 10.1667/RR1910.1. [DOI] [PubMed] [Google Scholar]
- 105.Harper JV, Anderson JA, O'Neill P. Radiation induced DNA DSB: contribution from stalled replication forks? DNA repair. 2010;9:907–913. doi: 10.1016/j.dnarep.2010.06.002. [DOI] [PubMed] [Google Scholar]